Abstract
Bacterial cryptic prophage (defective prophage) genes are known to drastically influence host physiology, such as causing cell growth arrest or lysis, upon expression. Many phages encode lytic proteins to destroy the cell envelope. As natural antibiotics, only a few lysis target proteins were identified. ydfD is a lytic gene from the Qin cryptic prophage that encodes a 63-amino-acid protein, the ectopic expression of which in Escherichia coli can cause nearly complete cell lysis rapidly. The bacterial 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway is responsible for synthesizing the isoprenoids uniquely required for sustaining bacterial growth. In this study, we provide evidence that YdfD can interact with IspG, a key enzyme involved in the MEP pathway, both in vivo and in vitro. We show that intact YdfD is required for the interaction with IspG to perform its lysis function and that the mRNA levels of ydfD increase significantly under certain stress conditions. Crucially, the cell lysis induced by YdfD can be abolished by the overexpression of ispG or the complementation of the IspG enzyme catalysis product methylerythritol 2,4-cyclodiphosphate. We propose that YdfD from the Qin cryptic prophage inhibits IspG to block the MEP pathway, leading to a compromised cell membrane and cell wall biosynthesis and eventual cell lysis.
1. Introduction
Cryptic prophages, also known as defective prophages, are segments of phage DNA integrated and maintained in bacterial chromosomes. It has been shown that bacterial genomes can harbor multiple cryptic prophages [1,2,3,4]. In fact, as much as 10–20% of bacterial chromosome segments are estimated to be prophage genes, and a growing body of evidence suggests that the proportion may be even greater [1,3]. For example, it was in the 1950s that the E. coli K-12 genomes were first found to carry phage genes [5] and now there are 10 known cryptic prophages residing in the chromosome of E. coli K-12 [4]. The expression of most of the prophage genes is usually suppressed under normal conditions, as certain proteins they encode are typically lytic and, therefore, toxic to the host [6]. However, there is evidence that some prophage genes can not only endow their hosts with traits and behaviors beneficial to surviving harsh environmental conditions but also expand the hosts’ genetic diversity, which plays a key role in microbial evolution [7,8,9,10]. Despite recent progress made in understanding cryptic prophages, the relationship between cryptic prophages and hosts is still largely elusive.
Investigation of cryptic prophages may provide hints to understand the process of phage infection, especially phage-induced cell lysis. As far as presently known, phages establish canonical lytic pathways to escape from hosts by two proteins, holin and endolysin [11,12,13]. Holins form holes in the cell membrane, and endolysins are a class of hydrolases that degrade bacterial cell wall. Therefore, holins are considered to control the access of endolysins to the cell periplasm. Apart from degradation of the cell wall, some phage genes encode proteins to inhibit the key enzyme to prevent cell wall biosynthesis. These proteins were dubbed “single-gene lysis” (Sgl) [11]. Many phages encode lytic protein. However, there are only three phage targets that have been proposed: (1) the Leviviridae Qβ uses the protein A2 to bind and inhibit MurA, which is the first step in bacterial cell wall biosynthesis [14]; (2) the protein E from phage ΦX174 inhibits peptidoglycan synthesis by interaction with MraY, which involves the peptidoglycan (PG) biosynthesis pathway [15]; and (3) LysM inhibits the translocation of the final lipid-linked PG precursor across the cytoplasmic membrane by interfering with MurJ [16]. Therefore, phages being natural antibiotics, exploring the lysis targets of phages is timely and of considerable importance.
Q-independent (Qin) prophage was the third prophage identified, found in E. coli K-12 [17]. As a part of Qin prophage, ydfD encodes a 63-amino-acid protein and ydfD is located downstream of dicB. Both ydfD and dicB are regulated by dicA [18]. Overexpression of ydfD induces cell lysis in E. coli, whereas lysis is prevented by coexpression with the cell division inhibitor dicB or sulA. It is now known that DicB as well as SulA prevent cell division by inhibiting FtsZ polymerization [19,20]. This indicates that the cell lysis caused by YdfD depends on functional cell division. Based on what is currently known, YdfD is speculated to promote cell lysis by targeting an unknown unique host protein, although the identity of the target protein of YdfD has not been established [21].
Our preliminary binding assays indicate that YdfD interacts directly with IspG, also known as GcpE [22]. IspG is a part of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway, also called the 1-deoxy-D-xylulose 5-phosphate (DOXP) pathway, that occurs in eubacteria [23]. Most bacteria synthesize ubiquitous isoprenoid compounds via the MEP pathway. The MEP pathway products are isopentenyl pyrophosphate (IPP) and its isomer dimethylallyl pyrophosphate (DMAPP), both fundamental and unique five-carbon blocks for the synthesis of terpenoid compounds, such as dolichols and cholesterol, which are required for peptidoglycan synthesis and maintenance of cell membrane stability, respectively [24,25,26]. The MEP pathway proceeds through a linear sequence of seven enzymes. Understandably, the MEP pathway is completely absent in humans, and each of the seven enzymes has potential as a drug target [27,28,29,30,31,32]. Blocking the MEP pathway can be bactericidal, and multiple inhibitors have been developed [33,34,35]. Fosmidomycin, an inhibitor of 1-deoxy-D-xylulose 5-phosphate reductoisomerase (Dxr), is the most anticipated inhibitor because it is the sole MEP pathway inhibitor that is being investigated clinically [36,37,38]. The sixth, IspG, converts 4-diphosphocytidyl-methylerythritol 2-phosphate (ME-cPP) into methylerythritol 2,4-cyclodiphosphate (HMBPP) assisted by an Fe/S cluster [39] (Scheme 1). The conversion to HMBPP is an irreversible committed step in the MEP pathway. HMBPP is an essential precursor of cell wall and membrane synthesis, and, predictably, E. coli cells are unable to survive when the ispG gene is deleted [40,41,42].
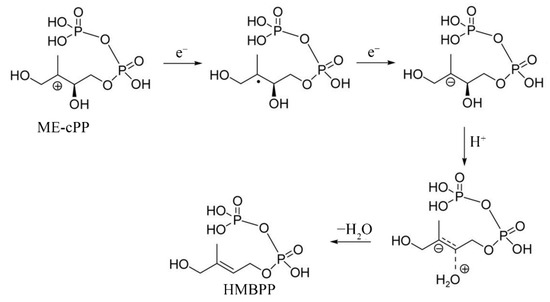
Scheme 1.
One of the proposed reaction mechanisms of IspG catalysis. (Adapted from Ref. [39]).
Herein, we present the results of several in vitro experiments demonstrating the interaction between YdfD and IspG, including pull-down assays, size exclusion chromatography (SEC) analysis, and isothermal titration calorimetry (ITC). We confirm this interaction further in vivo using fluorescence resonance energy transfer (FRET) and by showing that the overexpression of ispG or the compensation of the IspG enzyme catalysis product HMBPP rescues cells from the rapid lysis induced by YdfD. Taking these together, we suggest that YdfD is a specific inhibitor of IspG and inhibits IspG-mediated synthesis of HMBPP. Thus, by targeting IspG, YdfD compromises cell wall biosynthesis and causes cell lysis.
2. Results
2.1. Protein-Protein Binding between YdfD and IspG
Pull-down experiments were conducted to identify specific cellular targets of YdfD. His-YdfD was synthesized in E. coli BL21(DE3) cells as the bait for the whole cell lysate on Ni-NTA. After the Ni-NTA resin was washed, the eluted protein was subjected to SDS-PAGE and identified via mass spectrometry (MS). Dozens of proteins were identified, as shown in Table S1. Given YdfD’s function in inducing cell lysis, potential target proteins related to the cell membrane or cell wall synthesis were screened out. Next, we recombinantly expressed and purified these proteins and tested their interaction with YdfD with another pull-down assay (Table 1, Figure 1 and Figure S1).

Table 1.
Pull-down confirmation of interaction with YdfD of target proteins identified via MS.
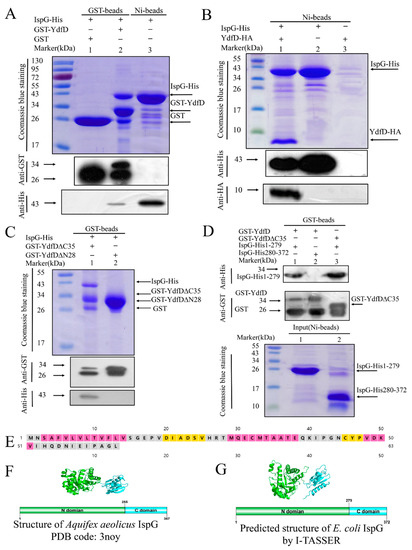
Figure 1.
Pull-down assays between YdfD, IspG, and their derivatives. (A) Lane 1: GST control; lane 2: GST-YdfD pulled down IspG; lane 3: IspG was purified by Ni beads as a control; the arrows indicate the positions of the corresponding proteins. (B) Lane 1: IspG pulled down YdfD-HA; lane 2: IspG-His was purified by Ni beads as a control; lane 3: YdfD-HA was purified by Ni beads as a control; the arrows indicate the positions of the corresponding proteins. (C) Lane 1: GST-YdfD∆C35 pulled down IspG; lane 2: GST-YdfD∆N28 pulled down IspG. (D) Lane 1: GST-YdfD pulled down IspG 1-279; lane 2: GST-YdfD pulled down IspG 280-372; lane 3: GST-YdfD∆C35 pulled down IspG 1-279. (E) Predicted secondary structure of YdfD by the PSIPRED server. (F) The structure of Aquifex aeolicus IspG, PDB code: 3noy. (G) The predicted structure of E. coli IspG by I-TASSER.
Based on the MS and pulled-down results (Figure 1 and Figure S1), IspG stood out as the most likely to interact with YdfD. IspG, an oxidoreductase, is an essential enzyme in the MEP pathway that synthesizes vital precursors of the cell wall and membrane [41]. Given YdfD’s strong activity in inducing cell lysis [21], we selected IspG for further characterization. Further pull-down experiments using N-GST-tagged ydfD (Table S2) coexpressed with His-tagged ispG showed that both proteins were present in the elute following glutathione and Ni-NTA affinity chromatography, as confirmed by SDS-PAGE and Western blotting (Figure 1A,B). YdfD, therefore, appeared to exhibit strong binding to IspG, warranting further investigation of their interaction.
To identify the exact segments of YdfD and IspG involved in their interaction, multiple truncation derivatives of YdfD and IspG were generated (Table S2). YdfD is predicted by the PSIPRED server to consist of distinct two domains, C-terminal (residues 29–63) and N-terminal (residues 1–28) domains [43] (Figure 1E), so a C-terminal deletion mutant (YdfD∆C35) and an N-terminal deletion mutant (YdfD∆N28) were synthesized and purified. Pull-down results showed it was GST-YdfD∆C35 rather than GST-YdfD∆N28 that exhibited interaction with IspG (Figure 1C).
Similarly, IspG constructs were designed using the structure of Aquifex aeolicus IspG (PDB code: 3noy) as a reference as it shares a 51.9% sequence identity with E. coli IspG. Aquifex aeolicus IspG has two separate domains, and as such, E. coli IspG is predicted using I-TASSER [44] to also consist of two distinct N-terminal (residues 1–279) and C-terminal (residues 280–372) domains (Figure 1F,G). As shown in Figure 1D, YdfD∆C35 showed interaction with IspG1-279. Although interactions of varying strength were detected for multiple derivatives, the interaction between the full-length YdfD and the full-length IspG was much stronger than that between the mutant combinations (Figure 1A,D), suggesting that competent IspG-YdfD interaction requires two full-length proteins.
2.2. YdfD Forms Stable Complex with IspG in Solution
SEC was used to purify and analyze the YdfD/IspG complex. N-MBP-tagged YdfD and IspG-His were co-synthesized in E. coli B21(DE3) cells to purify a large amount of the YdfD and IspG complex (Figure 2A). MBP-YdfD, IspG-His, and their complex were individually loaded on a Superdex 200 SEC column. Individual IspG-His was observed at a retention volume of approximately 81 mL (Figure 2B). SDS-PAGE of the individual MBP-YdfD elution profile showed that the first peak was MBP-YdfD and the subsequent peak was MBP (Figure 2C). Compared with IspG-His and MBP-YdfD, the retention volume of the MBP-YdfD/IspG-His complex was 69 mL, earlier than either protein alone (Figure 2D,E), though the relative band intensities in SDS-PAGE gel for the two proteins co-eluted from the column were consistent. These results indicate a stable MBP-YdfD/IspG-His complex in solution.
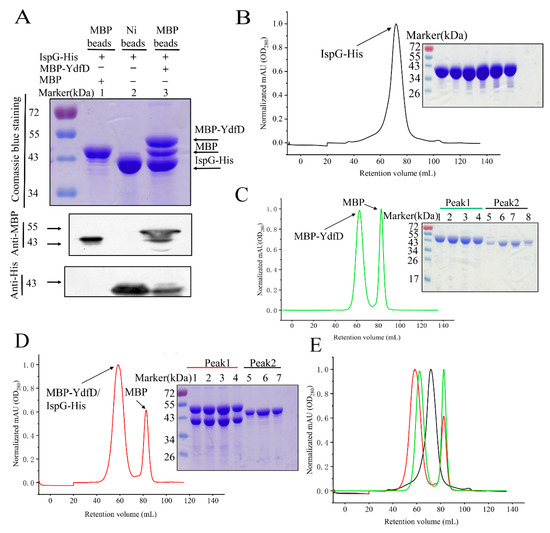
Figure 2.
SEC (Superdex 200 10/300 GL; flow rate 1.0 mL/min; 280 nm) elution profiles of IspG, MBP-YdfD, and the IspG/MBP-YdfD complex. (A) Pull-down SDS and Western blotting; lane 1: MBP pulled down IspG and was purified by maltose protein binding resin as a control; lane 2: IspG protein was purified by Ni-NTA resin; lane 3: A complex of fusion protein MBP-YdfD and IspG was purified by the maltose protein binding resin; the arrows indicate the positions of the corresponding proteins. (B) Elution profile (left) and SDS-PAGE (right) of IspG. (C) Elution profile of MBP-YdfD (left) and SDS-PAGE (right) of the two peaks; lanes 1–4 correspond to the first peak (right), and lanes 5–8 correspond to the second peak (left). (D) Elution profile (left) of the MBP-YdfD/IspG complex and SDS-PAGE (right) of the two peaks; lanes 1–4 correspond to the first peak (right), and lanes 5–7 correspond to the second peak (left). (E) merged (B–D) elution profiles.
2.3. YdfD and IspG Interactions In Vitro and Vivo
ITC is a powerful tool to measure enthalpy changes accompanying the binding of a compound or a protein to a protein [45]. ITC assays were used to quantify the affinity and thermodynamic parameters of the YdfD–IspG interaction. A considerable endothermic heat change was observed when YdfD was titrated with IspG via a single injection (Figure 3A). The best fit for the processed ITC data was obtained using a one-site binding equation [46]. The thermodynamic parameters between YdfD and IspG (Kd, n, ∆H, and ∆S) were 18.38 μM, 0.63, 1573 cal/mol, and 26.9 cal/mol/deg, respectively. The association constant (Kd, 18.38 μM) between YdfD and IspG is in micromolar range, indicating a relatively strong binding between YdfD and IspG.
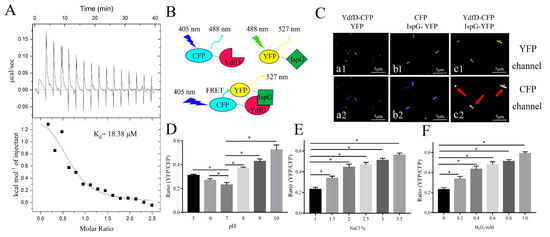
Figure 3.
YdfD–IspG interactions in vitro and in vivo. (A) Binding curve of IspG titrated into YdfD via a single injection in identical buffer (20 mM Tris, 150 mM NaCl, pH = 8.0) detected by ITC; Kd of 18.38 μM was obtained by ITC. (B) A schematic representation of FRET. (C) Confocal imaging of FRET cells. (a1), YdfD-CFP and YFP were coexpressed and observed in the YFP channel; (a2), Ydfd-CFP and YFP were coexpressed and observed in the CFP channel; (b1), CFP and IspG-YFP were coexpressed and observed in the YFP channel; (b2), CFP and IspG-YFP were coexpressed and observed in the CFP channel; (c1), YdfD-CFP and IspG-YFP were coexpressed and observed in the YFP channel; (c2), YdfD-CFP and IspG-YFP were coexpressed and observed in the CFP channel; (a1,a2,b1,b2) were control. (D) Different pH values affected the emission intensity ratio (527/488 nm ratio) in living cells. (E) Different osmotic pressures affected the emission intensity ratio (527/488 nm ratio) in living cells. (F) Different oxidative levels affected the emission intensity ratio (527/488 nm ratio) in living cells. * refers to p < 0.05.
Pull-down assays and ITC experiments demonstrated that YdfD associates with IspG in vitro. Further FRET experiments also demonstrated this interaction in vivo. E. coli BL21(DE3) cells were cotransformed with plasmids carrying ydfD-cfp and ispG-yfp and induced by IPTG for 8 h. Compared to the control (Figure 3C(a1,a2,b1,b2), the cells that were transformed with pET22b- ydfD-cfp and pET28b- ispG-yfp had distinct multiplex fluorescence signals under the CFP channel (Figure 3C(c2), indicated by the red arrow). The mixture fluorescence signals consisted of two components: the direct emission of CFP (Figure 3C(a2,b2)) and the emission of YFP excited by energy transferred from CFP, meaning that an energy transfer from CFP to YFP occurred upon YdfD interaction with IspG. In addition, FRET efficiency (ratio of YFP/CFP) increased when the cells were exposed to adverse conditions (Figure 3D–F).
2.4. The Intact YdfD Protein Is Required for Its Cell Lysis Function
To investigate which segment of YdfD is responsible for its lysis activity, the N-terminal and C-terminal residue(s) was omitted stepwise and a series of YdfD mutants (YdfD∆C1, YdfD∆C2, YdfD∆C5, YdfD∆N1, YdfD∆N2, YdfD∆N5, YdfD N1A, and YdfD L63A) were produced (Figure 4A). Owing to the strong lysis activity of the IPTG-induced intact YdfD, cells overexpressing ydfD could not survive on LB plates (Figure 4C(a1)). However, all mutants survived on LB plates and longer deletions produced more CFUs (Figure 4B,C). In addition, compared with the intact YdfD, the strains with more residues omitted had lag phases shorter than those with fewer residues omitted and all strains reached similar OD600 maxima (Figure 4D–F). Although the N-terminal domain of YdfD interacted with IspG somewhat in vitro (Figure 1), it could not function individually in cells. Since the C-terminal domain of YdfD was also indispensable, it is plausible that the N-terminal domain and C-terminal domain of YdfD cooperate closely in its lysis function.
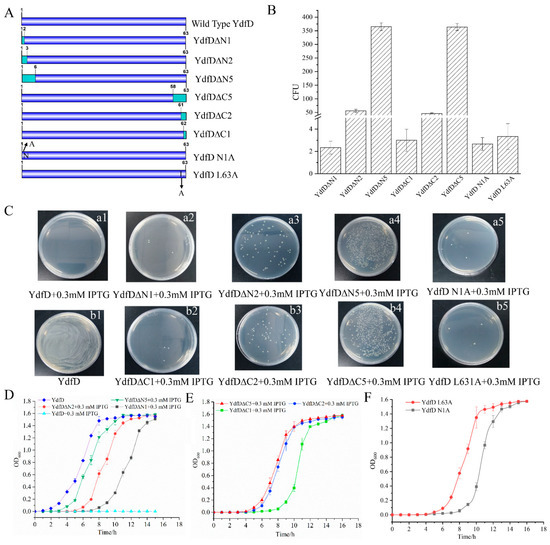
Figure 4.
The YdfD N-terminus and C-terminus are critical to the lysis function. (A) Schematic of YdfD mutants. (B) CFU of YdfD mutants that survived on the plates. (C), (a1): Wild-type YdfD; cells could not survive on the plate; (a2): YdfD∆N1; (a3): YdfD∆N2; (a4): YdfD∆N5; (a5): YdfD N1A; (b1): YdfD (no IPTG induction); (b2): YdfD∆C1; (b3): YdfD∆C2; (b4): YdfD∆C5; (b5) YdfD L63A. (D) Growth curves of YdfD and mutants with induction by IPTG, wild-type in the absence of IPTG (blue), YdfD∆N5 (green), YdfD∆N2 (red), YdfD∆N1 (black), and wild-type YdfD (cyan). (E) Growth curves of YdfD mutants with induction by IPTG, YdfD∆C5 (red), YdfD∆C2 (blue), YdfD∆C1 (green), and wild-type YdfD (cyan). (F) Growth curves of YdfD mutants with induction by IPTG, YdfD N1A (red), and YdfD L63A (gray). All protein expression was induced by IPTG unless otherwise stated.
2.5. mRNA Levels of ydfD Increased under Stress
Lysogenic bacteriophages enter the lytic cycle when the host is exposed to UV irradiation, X-ray irradiation, and mutagens [47,48,49], because the phage genes are activated, transcribed, and translated. To evaluate the effects of adverse conditions on the transcription of ydfD as part of prophage, we tested mRNA levels of ydfD in response to various stressors. RT-PCR showed that when E. coli cells were exposed to non-acidic stress conditions, including alkalinity (Figure 5A), oxidative stress (H2O2; Figure 5B), and high salinity (NaCl; Figure 5C), mRNA levels of ydfD increased compared to standard growth conditions (pH = 7, NaCl 1%, and absence of oxidative stress).
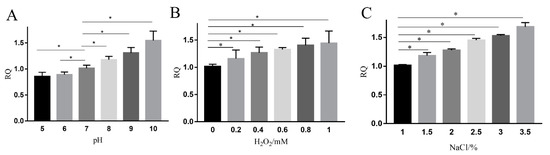
Figure 5.
RT-PCR of ydfD in stress conditions. (A) pH gradient effect on the transcription levels of ydfD. (B) H2O2 gradient effect on the transcription levels of ydfD. (C) NaCl gradient effect on the transcription levels of ydfD; * p < 0.05.
2.6. Compensation Experiments
After confirming the interaction between YdfD and IspG using multiple approaches, we attempted to determine if it was possible to offset YdfD-induced lysis either by coexpressing ispG with ydfD or by supplementing cells with the IspG enzyme catalysis product HMBPP (Figure 6A). Overexpressing ispG neutralized the lysis induced by overexpressed ydfD (Figure 6B), and supplementing with HMBPP completely restored normal cell growth (Figure 6C). The cell morphology was observed using SEM. The cell morphology was seriously compromised upon YdfD induction (Figure 6D–F) but remained unchanged in the presence of HMBPP (Figure 6G,H).
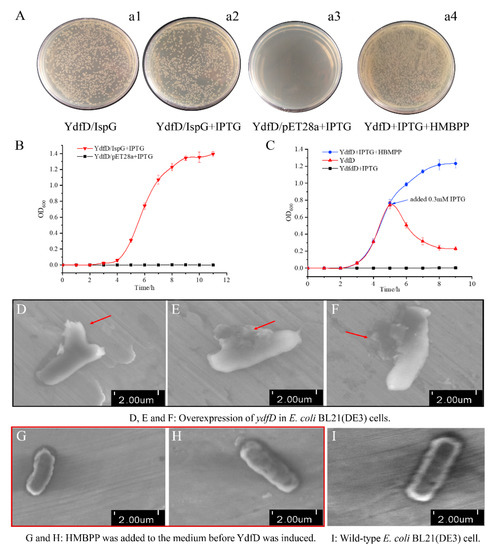
Figure 6.
IspG or its catalysis product HMBPP were able to offset the lysis induced by YdfD. (A). (a1): ydfD and ispG were not induced; (a2): ydfD and ispG were induced; (a3): ydfD and an empty pET28a vector were co-induced; (a4): YdfD was synthesized in the presence of HMBPP. (B) Growth curves of the ydfD/ispG and ydfD/pET28a vector induced by IPTG. (C) Growth curves of ydfD-expressed cells with variations. (D–F) Overexpression of ydfD in E. coli BL21(DE3) cells, and as the red arrow shows, the cellular morphology became incomplete and abnormal. ((G,H): as the red box indicate) HMBPP was added to the medium before ydfD was induced, and HMBPP was able to rescue cell lysis. (I) Wild-type E. coli BL21(DE3) cell. Scale bar 2 μm.
3. Discussion
The number of prophages identified in bacterial genomes or plasmids has increased substantially in recent years [4,45,46], although the underlying functional mechanisms of their host interactions have remained unclear [50]. Nevertheless, prophages appear to play key roles in host metabolism. Cryptic prophages have been shown to help bacteria resist antibiotic stress [50] and dicB, an upstream gene of ydfD, suppresses cell division in E. coli once induced [51]. Prophages can also prevent different categories of phages from infecting the host. DicB specifically inhibited infection by λ and other phages that use ManYZ membrane proteins to prevent phage DNA entrance [52], provide population-level benefits by promoting biofilm formation, and influence the evolution of their hosts [7,8,9].
A previous study showed that overexpression of ydfD caused cell lysis of E. coli [21], although the molecular mechanism was not illustrated. In this study, we attempted to characterize YdfD and its underlying mechanism of cell lysis activity. Herein, we report that YdfD interacts with the essential MEP enzyme IspG. The MEP pathway synthesizes precursors of the cell wall and membrane, and a variety of inhibitors have already been designed and screened against it [53,54]. It is thus tempting to suggest that YdfD-mediated cell lysis may be caused by YdfD inhibition of MEP production. This phenomenon may be regarded as a strategy for the prophage precursor to escape from the host under adverse conditions. This is analogous to the lytic function of protein E from phage ΦX174, which inhibits peptidoglycan synthesis by the MraY enzyme [15].
YdfD contains two well-conserved cysteine residues among bacterial species [21], and IspG has three conserved cysteine residues involved in the binding of the iron–sulfur cluster, responsible for the electron transfer of the catalytic process, as shown in Figure 7. The catalytic activity was significantly reduced when any cysteine that involved the cluster was replaced by serine [55]. However, the YdfD mutants lacking cysteine residue(s) had no effect on its binding to IspG (Figure S2), suggesting that the YdfD–IspG interaction is not through a disulfide bond. As the [4Fe-4S]2+ cluster is indispensable for IspG catalytic activity, all experimental procedures need to be performed under strict anerobic conditions [56]. In addition, ME-cPP, the substrate of IspG, is not commercially available. It is thus difficult to assay the effects of YdfD on IspG enzyme kinetics.
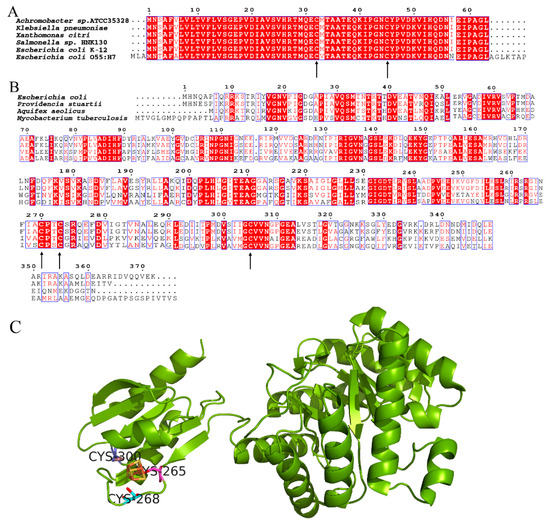
Figure 7.
Conserved cysteine residues of YdfD and IspG. (A) Alignment of YdfD sequences from Achromobacter sp. ATCC35328, Klebsiella pneumoniae, Xanthomonas citri, Salmonella sp. HNK130, E. coli K-12, and E. coli O55:H7; the arrows indicate conserved cysteine residues. (B) Alignment of IspG sequences from E. coli, Providencia stuartii, Aquifex aeolicus, and Mycobacterium tuberculosis; the arrows indicate conserved cysteine residues. (C) Aquifex aeolicus IspG; three conserved cysteine residues binding the iron–sulfur cluster are responsible for the electron transfer of the catalytic process (PDB code: 3noy).
While it appears that the N-terminal domain of YdfD was responsible for binding to the N-terminal domain of IspG, intact YdfD was necessary for its full activity in the cell (Figure 4). It is possible that the N-terminal domain of YdfD recruits the C-terminal domain to IspG, which inhibits IspG’s activity (Figure 4), which is why both N-terminal and C-terminal domains of YdfD are essential for its cellular activity. This agrees with previous studies showing that the C-terminal domain of YdfD is essential for lysis but that the killing activity of the C-terminal domain alone is significantly reduced compared to that of the intact protein [21]. The Aquifex aeolicus IspG is known to fold into two domains: an N-terminal and a C-terminal domain [56]. In the Aquifex aeolicus IspG catalytic process, the substrate binds to the positively charged surface region at the N-terminal domain [57]. Our ProtParam model (https://web.expasy.org/protparam/, accessed on 10 November 2021) [58] shows that the C-terminal domain of YdfD is negatively charged. Hence, it is possible that the C-terminal domain of YdfD competitively inhibits IspG by obstructing the binding of its substrate, ME-cPP, to the positively charged surface region of the IspG N-terminal domain. As a result, cooperation of both YdfD domains would be essential to inhibit IspG activity.
Under normal conditions, lysogenic phage levels remain steady. However, once the environment worsens, the transcription of lysis genes increases as the lysogenic phages look for ways to escape from the host [59,60,61]. Prophages also respond in this manner: although they cannot successfully assemble into viable phage particles, the lysis genes (ydfD) of Qin would increase transcription under adverse conditions. As expected, the RT-PCR of E. coli grown under stress conditions showed that mRNA levels of ydfD dramatically increases in non-acidic stress conditions and only slightly decreases under acidic conditions (Figure 5). The slight transcription decrease in the acidic medium is likely due to the bacteria’s general preference for more alkaline conditions and the corresponding reduction in the growth rate in a medium with a pH of 5.0 [62,63]. Consequently, the transcription of all unessential genes would be expected to decrease when bacteria are grown in an acidic environment.
The products of the MEP pathway are IPP and DMAPP, both of which are sole precursors for vital cellular components, including hopanoids [64]; menaquinone [65]; and polyprenyl phosphates, particularly undecaprenyl diphosphate (Und-p), a lipid carrier to synthesize peptidoglycan cell wall precursors that are essential to maintain cell integrity [66,67,68]. No matter how complex the terpenoid compounds are, all of them share a common five-carbon isoprene building block. A likely mechanism for YdfD-induced lysis is that YdfD blocks the MEP pathway in the cell membrane and cell wall biosynthesis. This would result in a shortage of key precursors for the cell wall and the cell membrane, leading to the abortion of cell wall formation, especially during cell division. A previous study by Masudo et al. found that YdfD cannot induce cell lysis when cell division is restrained [21]. We suppose that YdfD cuts off the MEP pathway by interaction with IspG, resulting in deficient cell components and cell wall membrane. When E. coli cells divide normally, as cytomembrane and cell wall element synthesis is most intense at sites of division, YdfD blocks terpenoid synthesis, thus leading to cell lysis. However, when cell division is slowed or stalled, the need for terpenoids (e.g., Und-P) is reduced. Therefore, YdfD cannot lyse the nondividing cells, i.e., YdfD-mediated effects would depend on cell division [21]. Further studies are needed to explore the direct effects of YdfD on IspG. HMBPP synthesis is a committed step in the MEP pathway, and IspG is strongly conserved across bacterial species [39], so it is unsurprising that phages would adapt to target IspG specifically to disrupt the host physiology and exert their function. IspG will be the fourth identified target of the phage lysis proteins; the details of how YdfD binds IspG and interferes with HMBPP will likely require structure studies of the YdfD/IspG complex.
It is clear that DicB binds to MinC and prevents FtsZ ring formation. Moreover, DicB specifically inhibits infection by λ and other phages that use ManYZ membrane proteins to prevent phage DNA entrance [52]. dicB and ydfD are located in the same operon, with partial overlap. As mentioned before, DicB was able to offset YdfD-induced lysis [21]. We propose the following model: in normal conditions, DicB inhibits cell division and prevents phage infection; when the environment gets worse, the transcription and expression of ydfD are activated, eventually leading to cell lysis (Figure 8).
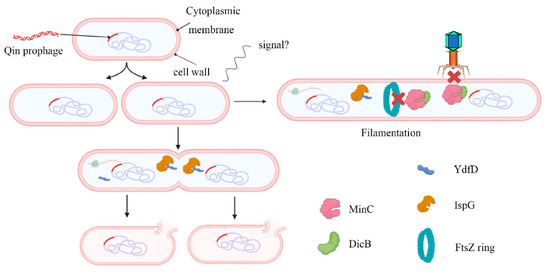
Figure 8.
Working model for the regulation of host morphology by YdfD and DicB. The Qin cryptic prophage encodes the protein DicB and YdfD. When cell division is inhibited by DicB, demand for isoprenoid compounds is decreased and lysis is prevented (right). When YdfD is ectopically expressed, YdfD interacts with IspG, reducing isoprenoid production and causing the cells to lyse (left).
The growing severity of antibiotic resistance means there is a growing need to target novel bacterial pathways. The MEP pathway is absent in humans, making it an attractive target for antibiotic drug development [50]. The mechanisms used by prophages such as the Qin YdfD protein have proven to have a robust and widespread capacity to co-opt bacterial physiology. In addition to offering new insight into the particularities of host-prophage interactions, YdfD represents a promising new avenue for generating lead compounds for designing antibacterial agents.
4. Materials and Methods
4.1. Cell Strains and Plasmids Construction
E. coli DH5α and E. coli BL21(DE3) strains were purchased from Invitrogen. Plasmids were constructed and maintained in E. coli DH5α. All plasmids and primers used in this study are listed in Tables S2 and S3. ydfD and ispG were amplified by PCR from the E. coli BL21(DE3) cell genome. The DNA sequences of ydfD and ispG are shown in Table S3.
4.2. Protein Expression and Purification
All proteins were produced in E. coli BL21(DE3) cells. Competent cells were transformed by plasmids. The transformants were scraped from the plate and cultured in the Luria Bertani (LB) liquid medium (1% peptone, 0.5% yeast extract, 1% NaCl, pH = 7.0). Isopropyl-beta-D-thiogalactopyranoside (IPTG) was added to the final concentration of 0.3 mM when the optical density 600 nm (OD600) was about 0.8, and the strain was incubated for 20 h at 16 °C. Next, the cells were harvested and resuspended in lysis buffer (20 mM Tris, 150 mM NaCl, pH = 8.0) for high-pressure cell disruption. The debris was centrifugated at 15,000× g at 4 °C for 40 min, while the supernatants were incubated with a corresponding purification affinity beads resin. Maltose binding protein (MBP)-tagged, His-tagged, and glutathione S-transferase (GST)-tagged proteins were purified by amylose resin (NEB, Beverly, MA, USA), Ni-NTA (Qiagen, Duesseldorf, Germany) resin, and GST-tag (Beaver, Guangzhou, China) purification beads, respectively.
4.3. Mass Spectrometry
Mass spectrometry (MS) was used to identify the proteins bound to YdfD. E. coli BL21(DE3) cells synthesizing His-YdfD were subjected to induction with 0.3 mM IPTG at 16 °C for 4 h. The cells were harvested and resuspended in lysis buffer for cell disruption. The debris was centrifugated and the supernatants were subjected to incubation with Ni-NTA resin. Subsequently, the beads were washed 3 times with wash buffer (20 mM Tris, 250 mM NaCl, pH = 8.0) to remove non-specific binding proteins. The beads were analyzed by SDS-PAGE and digested with trypsin (Thermo Fisher Scientific, Waltham, MA, USA). The MS was performed as described [69]. After digestion, the tryptic peptides were desalted using C18 stage tips and separated by a C18 column (75 μm inner diameter, 150 mm length, 5 μm, 300 Å) with a Thermo-Dionex Ultimate 3000 HPLC system that was directly connected with a Thermo Scientific Q-Exactive HF-X Hybrid Quadrupole-Orbitrap mass spectrometer. The generated MS/MS spectra were searched against the Uniprot E. coli k-12(Taxon ID: 83333) database using the SEQUEST search engine with Proteome Discoverer 2.2 software. The search criteria were as follows: full tryptic specificity was required; one missed cleavage was allowed. Peptide spectral matches were validated using the percolator provided by Proteome Discoverer software based on the q values at a 1% false discovery rate.
4.4. Pull-Down Assays
Pull-down assays were performed to assess the interaction between YdfD and IspG. YdfD and its various mutants (Table S1) were attached to GST for the identification of YdfD–IspG interactions. For GST-YdfD pulled-down IspG-His, E. coli BL21(DE3) cells were cotransformed by pGEX-6P-1-ydfD and pET28a-ispG-his and induced by IPTG. After bacterial cells were harvested, they were lysed and centrifuged and the supernatants were incubated with the GST beads. Finally, the protein sample beads were directly subjected to SDS-PAGE and Western blotting (diluted 1:10) after 3 washes using wash buffer and heat denaturation (98 °C for 10 min). Other samples were prepared by the same method. To prepare large amounts of protein, MBP tag was fused to the N-terminus of YdfD. The complex of MBP-YdfD and IspG was purified via amylose resin; after the resin treatment with wash buffer, the protein was eluted by wash buffer containing 20 mM maltose. IspG protein was purified using His-tag by Ni-NTA resin and eluted by wash buffer containing 300 mM imidazole. Finally, the elution was concentrated by a centrifugal filter (Millipore, Billerica, MA, USA, 10 kD) to 1 mL and loaded onto a Superdex 200 column for SEC analysis. SEC runs were conducted at 4 °C at a flow rate of 1.0 mL/min using a buffer containing 20 mM Tris, 150 mM NaCl, and 0.5 mM DTT at pH = 8.0.
4.5. Cell Survival Assays
Cell survival assays were conducted to assess which structural segment(s) of YdfD is responsible for its lysis activity. E. coli BL21(DE3) cells were transformed by plasmids carrying ydfD or its mutants (Table S1). After being cultured overnight, the clones were transferred to the LB liquid medium and the culture was continued for 12 h. Next, the media were diluted to the same OD600. Colony forming unit (CFU) assays were performed by plating cells onto LB plates and counting the CFUs after culturing for 12 h. Similarly, for the growth assay, the strains were cultured in a fresh medium with the same inoculum. OD600 was measured every half an hour. The cell growth was evaluated by OD600 and CFUs. All cells were cultured at 37 °C unless otherwise specified. All assays were performed in triplicate.
4.6. Stress Experiments
Reverse transcriptase quantitative PCR assays were performed to measure the mRNA level of ydfD under different stress conditions. E. coli BL21(DE3) cells were cultured in an LB medium for 12 h, and subsequently the cultures were transferred to the LB medium (1:100) under different stress conditions and cultured for 12 h at 37 °C. The stress conditions tested included pH values of the medium changing from 5.0 to 10.0, with a gradient of 1.0; increased H2O2 concentration of the medium from 0 to 1 mM, with a gradient of 0.2 mM; and increased NaCl concentration of the medium from 1 to 3.5%, with a gradient of 0.5% [47,60,67]. mRNA isolation was performed according to kit protocols (Tiangen Biotech, Beijing, China). Next, the DNAase was added to the mRNA to digest the DNA. Finally, the mRNA was reverse-transcripted at 42 °C for 30 min and real-time PCR assays were performed as described [70]. The primer sequences are listed in Table S3, and the 16S rRNA gene was used as a control. All assays were performed in triplicate.
4.7. Fluorescence Resonance Energy Transfer
FRET assays were performed to measure the interaction between YdfD and IspG in cells. The donor cyan fluorescent protein (CFP) was fused to the C-terminus of YdfD, and the receptor yellow fluorescent protein (YFP) was attached to the C-terminus of IspG (Table S2). E. coli BL21(DE3) cells were cotransformed by the plasmids of pET22b-ydfD-cfp and pET28a-ispG-yfp, and the fusion proteins were induced by IPTG for 8 h at 37 °C. The cells were dropped onto microslides for observation. For CFP, the excitation and emission wavelengths were 405 and 488 nm, respectively; for YFP, the excitation and emission wavelengths were 488 and 527 nm, respectively; and for FRET, the excitation and emission wavelengths were 488 and 527 nm, respectively (Zeiss LSM980 Airyscan 2, Oberkochen, Germany). Additionally, we quantified the YFP/CFP ratio by detecting the fluorescence intensity at 488 and 527 nm for 30 s.
4.8. Isothermal Titration Calorimetry
ITC (GE, Microcal iTC-200, Boston, MA, USA) assays were used to further characterize and quantify the interaction between YdfD and IspG. MBP-YdfD and IspG protein were prepared as mentioned above. For YdfD, the MBP tag was cleaved by PreScission proteases. YdfD and IspG were purified and concentrated in buffer (20 mM Tris, 150 mM NaCl, pH = 8.0) to 90 μM and 1.1 mM, respectively. Single-injection measurements were collected by injecting 2 μL of 1.1 mM IspG into 0.9 μM YdfD at a spacing of 120 s. All assays were run at 25 °C at a stirring speed of 300 rpm. The raw data were processed by Origin Pro 2018 to obtain the binding parameters between YdfD and IspG, including the association constant (Kd), the enthalpy value (∆H), and the entropy value (∆S). All assays were performed in triplicate.
4.9. Compensation Experiment
Rescue experiments contained two independent experiments. The first experiment was performed to test whether the overexpression of ispG in vivo could prevent the cell lysis induced by YdfD. To achieve this goal, ydfD and ispG were overexpressed into E. coli BL21(DE3) cells and their expression were induced simultaneously by IPTG as above. In the second experiment, to explore whether the IspG catalysis product was able to prevent cell lysis, the IspG enzyme catalysis product HMBPP (Sigma, St. Louis, MO, USA) was added to the medium. Cell growth was evaluated by monitoring OD600 and CFUs in plates. For the test, 0.2 mg of HMBPP was added to 10 mL of the LB culture medium. Since HMBPP is largely insoluble in the LB medium, we did not note its accurate concentration.
4.10. Scanning Electron Microscopy
SEM analysis was used to observe the bacterial morphology [71]. The plasmids were transformed into E. coli BL21(DE3) cells and induced by 0.3 mM IPTG for 8 h. The cells were collected by centrifugation at 3000× g for 10 min. After the cells were washed with PBS (pH = 7.4), they were resuspended using 2.5% glutaraldehyde in 0.1 M cacodylate buffer and gently shaken at 37 °C for 30 min. The cells were dropped onto a foil after dehydration by gradient acetone solutions (10%, 30%, 50%, and 70%, 20 min each). Eventually, the foil was fastened onto a sample holder by conductive tape and the pictures of bacteria were captured by SEM (Hitachi SU8010, Toyko, Japan) at 5 kV and 3 μA.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23031560/s1.
Author Contributions
Z.L. and B.W. performed the experiments; Z.Q. and R.Z. conceived the study; J.Z. and Z.J. wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (NO. 21773014) and the Natural Sciences and Engineering Research Council of Canada (RGPIN-2018-04427).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The datasets of this study are available from the corresponding author on reasonable request.
Acknowledgments
We thank Greg Hicks for helping edit this manuscript.
Conflicts of Interest
The authors declare that they have no conflict of interest with the contents of this article. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
References
- Arndt, D.; Marcu, A.; Liang, Y.J.; Wishart, D.S. PHAST, PHASTER and PHASTEST: Tools for finding prophage in bacterial genomes. Brief. Bioinform. 2019, 20, 1560–1567. [Google Scholar] [CrossRef] [PubMed]
- Reis-Cunha, J.L.; Bartholomeu, D.C.; Manson, A.L.; Earl, A.M.; Cerqueira, G.C. ProphET, prophage estimation tool: A stand-alone prophage sequence prediction tool with self-updating reference database. PLoS ONE 2019, 14, e0223364. [Google Scholar] [CrossRef] [PubMed]
- Song, W.C.; Sun, H.X.; Zhang, C.; Cheng, L.; Peng, Y.; Deng, Z.Q.; Wang, D.; Wang, Y.; Hu, M.; Liu, W.E.; et al. Prophage Hunter: An integrative hunting tool for active prophages. Nucleic Acids Res. 2019, 47, W74–W80. [Google Scholar] [CrossRef] [PubMed]
- Blattner, F.R.; Plunkett, G.; Bloch, C.A.; Perna, N.T.; Burland, V.; Riley, M.; Collado-Vides, J.; Glasner, J.D.; Rode, C.K.; Mayhew, G.F.; et al. The complete genome sequence of Escherichia coli K-12. Science 1997, 277, 1453–1642. [Google Scholar] [CrossRef]
- Lederberg, E.M.; Lederberg, J. Genetic Studies of Lysogenicity in Escherichia-Coli. Genetics 1953, 38, 51–64. [Google Scholar] [CrossRef]
- Rueggeberg, K.G.; Toba, F.A.; Bird, J.G.; Franck, N.; Thompson, M.G.; Hay, A.G. The lysis cassette of DLP12 defective prophage is regulated by RpoE. Microbiology 2015, 161, 1683–1693. [Google Scholar] [CrossRef]
- Stern, A.; Sorek, R. The phage-host arms race: Shaping the evolution of microbes. Bioessays 2011, 33, 43–51. [Google Scholar] [CrossRef]
- Obeng, N.; Pratama, A.A.; van Elsas, J.D. The Significance of Mutualistic Phages for bacterial Ecology and Evolution. Trends Microbiol. 2016, 24, 440–449. [Google Scholar] [CrossRef]
- Labrie, S.J.; Samson, J.E.; Moineau, S. Bacteriophage resistance mechanisms. Nat. Rev. Microbiol. 2010, 8, 317–327. [Google Scholar] [CrossRef]
- Casjens, S. Prophages and bacterial genomics: What have we learned so far? Mol. Microbiol. 2003, 49, 277–300. [Google Scholar] [CrossRef]
- Chamakura, K.; Young, R. Phage single-gene lysis: Finding the weak spot in the bacterial cell wall. J. Biol. Chem. 2019, 294, 3350–3358. [Google Scholar] [CrossRef] [PubMed]
- Wang, I.N.; Smith, D.L.; Young, R. Holins: The protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 2000, 54, 799–825. [Google Scholar] [CrossRef] [PubMed]
- Zampara, A.; Sørensen, M.C.H.; Grimon, D.; Antenucci, F.; Vitt, A.R.; Bortolaia, V.; Briers, Y.; Brøndsted, L. Exploiting phage receptor binding proteins to enable endolysins to kill Gram-negative bacteria. Sci. Rep. 2020, 10, 12087. [Google Scholar] [CrossRef] [PubMed]
- Reed, C.A.; Langlais, C.; Kuznetsov, V.; Young, R. Inhibitory mechanism of the Qβ lysis protein A2. Mol. Microbiol. 2012, 86, 836–844. [Google Scholar] [CrossRef] [PubMed]
- Bernhardt, T.G.; Struck, D.K.; Young, R. The lysis protein E of phi X174 is a specific inhibitor of the MraY-catalyzed step in peptidoglycan synthesis. J. Biol. Chem. 2001, 276, 6093–6097. [Google Scholar] [CrossRef]
- Chamakura, K.R.; Sham, L.T.; Davis, R.M.; Min, L.; Cho, H.; Ruiz, N.; Bernhardt, T.G.; Young, R. A viral protein antibiotic inhibits lipid II flippase activity. Nat. Microbiol. 2017, 2, 1480–1484. [Google Scholar] [CrossRef]
- Espion, D.; Kaiser, K.; Dambly-Chaudiere, C. A third defective lambdoid prophage of Escherichia coli K12 defined by the lambda derivative, lambdaqin111. J. Mol. Biol. 1983, 170, 611–633. [Google Scholar] [CrossRef]
- Béjar, S.; Cam, K.; Bouché, J.P. Control of cell division in Escherichia coli. DNA sequence of dicA and of a second gene complementing mutation dicA1, dicC. Nucleic Acids Res. 1986, 14, 6821–6833. [Google Scholar] [CrossRef]
- Chen, Y.; Milam, S.L.; Erickson, H.P. SulA inhibits assembly of FtsZ by a simple sequestration mechanism. Biochemistry 2012, 51, 3100–3109. [Google Scholar] [CrossRef]
- Johnson, J.E.; Lackner, L.L.; de Boer, P.A. Targeting of (D)MinC/MinD and (D)MinC/DicB complexes to septal rings in Escherichia coli suggests a multistep mechanism for MinC-mediated destruction of nascent FtsZ rings. J. Bacteriol. 2002, 184, 2951–2962. [Google Scholar] [CrossRef]
- Masuda, H.; Awano, N.; Inouye, M. ydfD encodes a novel lytic protein in Escherichia coli. FEMS Microbiol. Lett. 2016, 363, fnw039. [Google Scholar] [CrossRef][Green Version]
- Hecht, S.; Eisenreich, W.; Adam, P.; Amslinger, S.; Kis, K.; Bacher, A.; Arigoni, D.; Rohdich, F. Studies on the nonmevalonate pathway to terpenes: The role of the GcpE (IspG) protein. Proc. Natl. Acad. Sci. USA 2001, 98, 14837–14842. [Google Scholar] [CrossRef] [PubMed]
- Rohmer, M. From Molecular Fossils of Bacterial Hopanoids to the Formation of Isoprene Units: Discovery and Elucidation of the Methylerythritol Phosphate Pathway. Lipids 2008, 43, 1095–1107. [Google Scholar] [CrossRef] [PubMed]
- Odom, A.R. Five questions about non-mevalonate isoprenoid biosynthesis. PLoS Pathog. 2011, 7, e1002323. [Google Scholar] [CrossRef] [PubMed]
- Anderson, R.G.; Hussey, H.; Baddiley, J. The mechanism of wall synthesis in bacteria. The organization of enzymes and isoprenoid phosphates in the membrane. Biochem. J. 1972, 127, 11–25. [Google Scholar] [CrossRef]
- Bouhss, A.; Trunkfield, A.E.; Bugg, T.D.; Mengin-Lecreulx, D. The biosynthesis of peptidoglycan lipid-linked intermediates. FEMS Microbiol. Rev. 2008, 32, 208–233. [Google Scholar] [CrossRef]
- Wang, X.; Dowd, C.S. The Methylerythritol Phosphate Pathway: Promising Drug Targets in the Fight against Tuberculosis. ACS Infect. Dis. 2018, 4, 278–290. [Google Scholar] [CrossRef] [PubMed]
- He, L.; He, P.; Luo, X.; Li, M.; Yu, L.; Guo, J.; Zhan, X.; Zhu, G.; Zhao, J. The MEP pathway in Babesia orientalis apicoplast, a potential target for anti-babesiosis drug development. Parasites Vectors 2018, 11, 452. [Google Scholar] [CrossRef]
- Rohmer, M.; Grosdemange-Billiard, C.; Seemann, M.; Tritsch, D. Isoprenoid biosynthesis as a novel target for antibacterial and antiparasitic drugs. Curr. Opin. Investig. Drugs 2004, 5, 154–162. [Google Scholar]
- Hunter, W.N. The non-mevalonate pathway of isoprenoid precursor biosynthesis. J. Biol. Chem. 2007, 282, 21573–21577. [Google Scholar] [CrossRef]
- Masini, T.; Hirsch, A.K. Development of inhibitors of the 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway enzymes as potential anti-infective agents. J. Med. Chem. 2014, 57, 9740–9763. [Google Scholar] [CrossRef] [PubMed]
- Frank, A.; Groll, M. The Methylerythritol Phosphate Pathway to Isoprenoids. Chem. Rev. 2017, 117, 5675–5703. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.; Nemeria, N.S.; Brammer, L.A.; Freel Meyers, C.L.; Jordan, F. Observation of Thiamin-Bound Intermediates and Microscopic Rate Constants for Their Interconversion on 1-Deoxy-d-xylulose 5-Phosphate Synthase: 600-Fold Rate Acceleration of Pyruvate Decarboxylation by d-Glyceraldehyde-3-phosphate. J. Am. Chem. Soc. 2012, 134, 18374–18379. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yang, L.; DeColli, A.; Freel Meyers, C.; Nemeria, N.S.; Jordan, F. Conformational dynamics of 1-deoxy-d-xylulose 5-phosphate synthase on ligand binding revealed by H/D exchange MS. Proc. Natl. Acad. Sci. USA 2017, 114, 9355–9360. [Google Scholar] [CrossRef] [PubMed]
- Masini, T.; Kroezen, B.S.; Hirsch, A.K. Druggability of the enzymes of the non-mevalonate-pathway. Drug Discov. Today 2013, 18, 1256–1262. [Google Scholar] [CrossRef]
- Borrmann, S.; Lundgren, I.; Oyakhirome, S.; Impouma, B.; Matsiegui, P.B.; Adegnika, A.A.; Issifou, S.; Kun, J.F.; Hutchinson, D.; Wiesner, J.; et al. Fosmidomycin plus clindamycin for treatment of pediatric patients aged 1 to 14 years with Plasmodium falciparum malaria. Antimicrob. Agents Chemother. 2006, 50, 2713–2718. [Google Scholar] [CrossRef]
- Missinou, M.A.; Borrmann, S.; Schindler, A.; Issifou, S.; Adegnika, A.A.; Matsiegui, P.-B.; Binder, R.; Lell, B.; Wiesner, J.; Baranek, T.; et al. Fosmidomycin for malaria. Lancet 2002, 360, 1941–1942. [Google Scholar] [CrossRef]
- Borrmann, S.; Adegnika, A.A.; Moussavou, F.; Oyakhirome, S.; Esser, G.; Matsiegui, P.B.; Ramharter, M.; Lundgren, I.; Kombila, M.; Issifou, S.; et al. Short-course regimens of artesunate-fosmidomycin in treatment of uncomplicated Plasmodium falciparum malaria. Antimicrob. Agents Chemother. 2005, 49, 3749–3754. [Google Scholar] [CrossRef]
- Wang, W.; Oldfield, E. Bioorganometallic chemistry with IspG and IspH: Structure, function, and inhibition of the [Fe4S4] proteins involved in isoprenoid biosynthesis. Angew. Chem. Int. Ed. 2014, 53, 4294–4310. [Google Scholar] [CrossRef]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006–2008. [Google Scholar] [CrossRef]
- Campos, N.; Rodriguez-Concepcion, M.; Seemann, M.; Rohmer, M.; Boronat, A. Identification of gcpE as a novel gene of the 2-C-methyl-D-erythritol 4-phosphate pathway for isoprenoid biosynthesis in Escherichia coli. FEBS Lett. 2001, 488, 170–173. [Google Scholar] [CrossRef]
- Sauret-Gueto, S.; Ramos-Valdivia, A.; Ibanez, E.; Boronat, A.; Rodriguez-Concepcion, M. Identification of lethal mutations in Escherichia coli genes encoding enzymes of the methylerythritol phosphate pathway. Biochem. Biophys. Res. Commun. 2003, 307, 408–415. [Google Scholar] [CrossRef]
- Buchan, D.W.A.; Jones, D.T. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019, 47, W402–W407. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.Y.; Zhang, Y. I-TASSER server: New development for protein structure and function predictions. Nucleic Acids Res. 2015, 43, W174–W181. [Google Scholar] [CrossRef]
- Blaskovich, M.A.T.; Hansford, K.A.; Gong, Y.; Butler, M.S.; Muldoon, C.; Huang, J.X.; Ramu, S.; Silva, A.B.; Cheng, M.; Kavanagh, A.M.; et al. Protein-inspired antibiotics active against vancomycin- and daptomycin-resistant bacteria. Nat. Commun. 2018, 9, 22. [Google Scholar] [CrossRef]
- Miura, K. An Overview of Current Methods to Confirm Protein-Protein Interactions. Protein Pept. Lett. 2018, 25, 728–733. [Google Scholar] [CrossRef]
- Kondo, S.; Ichikawa, H.; Iwo, K.; Kato, T. Base-change mutagenesis and prophage induction in strains of Escherichia coli with different DNA repair capacities. Genetics 1970, 66, 187–217. [Google Scholar] [CrossRef]
- Nanda Arun, M.; Thormann, K.; Frunzke, J.; Margolin, W. Impact of Spontaneous Prophage Induction on the Fitness of Bacterial Populations and Host-Microbe Interactions. J. Bacteriol. 2015, 197, 410–419. [Google Scholar] [CrossRef]
- Barnhart, B.J.; Cox, S.H.; Jett, J.H. Prophage induction and inactivation by UV light. J. Virol. 1976, 18, 950–955. [Google Scholar] [CrossRef]
- Wang, X.X.; Kim, Y.; Ma, Q.; Hong, S.H.; Pokusaeva, K.; Sturino, J.M.; Wood, T.K. Cryptic prophages help bacteria cope with adverse environments. Nat. Commun. 2010, 1, 147. [Google Scholar] [CrossRef]
- Cam, K.; Béjar, S.; Gil, D.; Bouché, J.P. Identification and sequence of gene dicB: Translation of the division inhibitor from an in-phase internal start. Nucleic Acids Res. 1988, 16, 6327–6338. [Google Scholar] [CrossRef] [PubMed]
- Ragunathan, P.T.; Vanderpool, C.K. Cryptic-Prophage-Encoded Small Protein DicB Protects Escherichia coli from Phage Infection by Inhibiting Inner Membrane Receptor Proteins. J. Bacteriol. 2019, 201, e00475-19. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.; Wang, K.; Li, J.K.; Wang, W.X.; Liu, Y.L.; Amin, S.; Oldfield, E. Inhibition of the 4Fe-4S proteins IspG and IspH: An EPR, ENDOR and HYSCORE investigation. Chem. Sci. 2014, 5, 1642–1649. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, J.; Wang, K.; Huang, C.; Zhang, Y.; Oldfield, E. Organometallic mechanism of action and inhibition of the 4Fe-4S isoprenoid biosynthesis protein GcpE (IspG). Proc. Natl. Acad. Sci. USA 2010, 107, 11189–11193. [Google Scholar] [CrossRef] [PubMed]
- Zepeck, F.; Gräwert, T.; Kaiser, J.; Schramek, N.; Eisenreich, W.; Bacher, A.; Rohdich, F. Biosynthesis of Isoprenoids. Purification and Properties of IspG Protein from Escherichia coli. J. Org. Chem. 2005, 70, 9168–9174. [Google Scholar] [CrossRef]
- Quitterer, F.; Frank, A.; Wang, K.; Rao, G.; O’Dowd, B.; Li, J.; Guerra, F.; Abdel-Azeim, S.; Bacher, A.; Eppinger, J.; et al. Atomic-Resolution Structures of Discrete Stages on the Reaction Coordinate of the [Fe4S4] Enzyme IspG (GcpE). J. Mol. Biol. 2015, 427, 2220–2228. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.; Gräwert, T.; Quitterer, F.; Rohdich, F.; Eppinger, J.; Eisenreich, W.; Bacher, A.; Groll, M. Biosynthesis of isoprenoids: Crystal structure of the [4Fe-4S] cluster protein IspG. J. Mol. Biol. 2010, 404, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.H.C.; Gattiker, A.; Duvaud, S.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein Identification and Analysis Tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Oppenheim, A.B.; Kobiler, O.; Stavans, J.; Court, D.L.; Adhya, S. Switches in bacteriophage lambda development. Annu. Rev. Genet. 2005, 39, 409–429. [Google Scholar] [CrossRef]
- Erez, Z.; Steinberger-Levy, I.; Shamir, M.; Doron, S.; Stokar-Avihail, A.; Peleg, Y.; Melamed, S.; Leavitt, A.; Savidor, A.; Albeck, S.; et al. Communication between viruses guides lysis-lysogeny decisions. Nature 2017, 541, 488–493. [Google Scholar] [CrossRef]
- Zong, C.H.; So, L.H.; Sepulveda, L.A.; Skinner, S.O.; Golding, I. Lysogen stability is determined by the frequency of activity bursts from the fate-determining gene. Mol. Syst. Biol. 2010, 6, 440. [Google Scholar] [CrossRef]
- Hickey, E.W.; Hirshfield, I.N. Low-pH-induced effects on patterns of protein synthesis and on internal pH in Escherichia coli and Salmonella typhimurium. Appl. Environ. Microbiol. 1990, 56, 1038–1045. [Google Scholar] [CrossRef] [PubMed]
- Booth, I.R. Regulation of cytoplasmic pH in bacteria. Microbiol. Rev. 1985, 49, 359–378. [Google Scholar] [CrossRef] [PubMed]
- Sandoval-Calderón, M.; Guan, Z.; Sohlenkamp, C. Knowns and unknowns of membrane lipid synthesis in streptomycetes. Biochimie 2017, 141, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Upadhyay, A.; Fontes, F.L.; Gonzalez-Juarrero, M.; McNeil, M.R.; Crans, D.C.; Jackson, M.; Crick, D.C. Partial Saturation of Menaquinone in Mycobacterium tuberculosis: Function and Essentiality of a Novel Reductase, MenJ. ACS Cent. Sci. 2015, 1, 292–302. [Google Scholar] [CrossRef]
- Crick, D.C.; Schulbach, M.C.; Zink, E.E.; Macchia, M.; Barontini, S.; Besra, G.S.; Brennan, P.J. Polyprenyl phosphate biosynthesis in Mycobacterium tuberculosis and Mycobacterium smegmatis. J. Bacteriol. 2000, 182, 5771–5778. [Google Scholar] [CrossRef]
- Bongers, M.; Chrysanthopoulos, P.K.; Behrendorff, J.B.Y.H.; Hodson, M.P.; Vickers, C.E.; Nielsen, L.K. Systems analysis of methylerythritol-phosphate pathway flux in E. coli: Insights into the role of oxidative stress and the validity of lycopene as an isoprenoid reporter metabolite. Microb. Cell Factories 2015, 14, 193. [Google Scholar] [CrossRef]
- Chang, S.-Y.; Ko, T.-P.; Liang, P.-H.; Wang, A.H.J. Catalytic Mechanism Revealed by the Crystal Structure of Undecaprenyl Pyrophosphate Synthase in Complex with Sulfate, Magnesium, and Triton. J. Biol. Chem. 2003, 278, 29298–29307. [Google Scholar] [CrossRef]
- Jiang, D.; Jiang, Z.; Lu, D.; Wang, X.; Liang, H.; Zhang, J.; Meng, Y.; Li, Y.; Wu, D.; Huang, Y.; et al. Migrasomes provide regional cues for organ morphogenesis during zebrafish gastrulation. Nat. Cell Biol. 2019, 21, 966–977. [Google Scholar] [CrossRef]
- Chakravorty, S.; Helb, D.; Burday, M.; Connell, N.; Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J. Microbiol. Methods 2007, 69, 330–339. [Google Scholar] [CrossRef]
- Yang, S.; Pei, H.; Zhang, X.; Wei, Q.; Zhu, J.; Zheng, J.; Jia, Z. Characterization of DicB by partially masking its potent inhibitory activity of cell division. Open Biol. 2016, 6, 160082. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).