Isolation and Sequencing of Chromosome Arm 7RS of Rye, Secale cereale
Abstract
1. Introduction
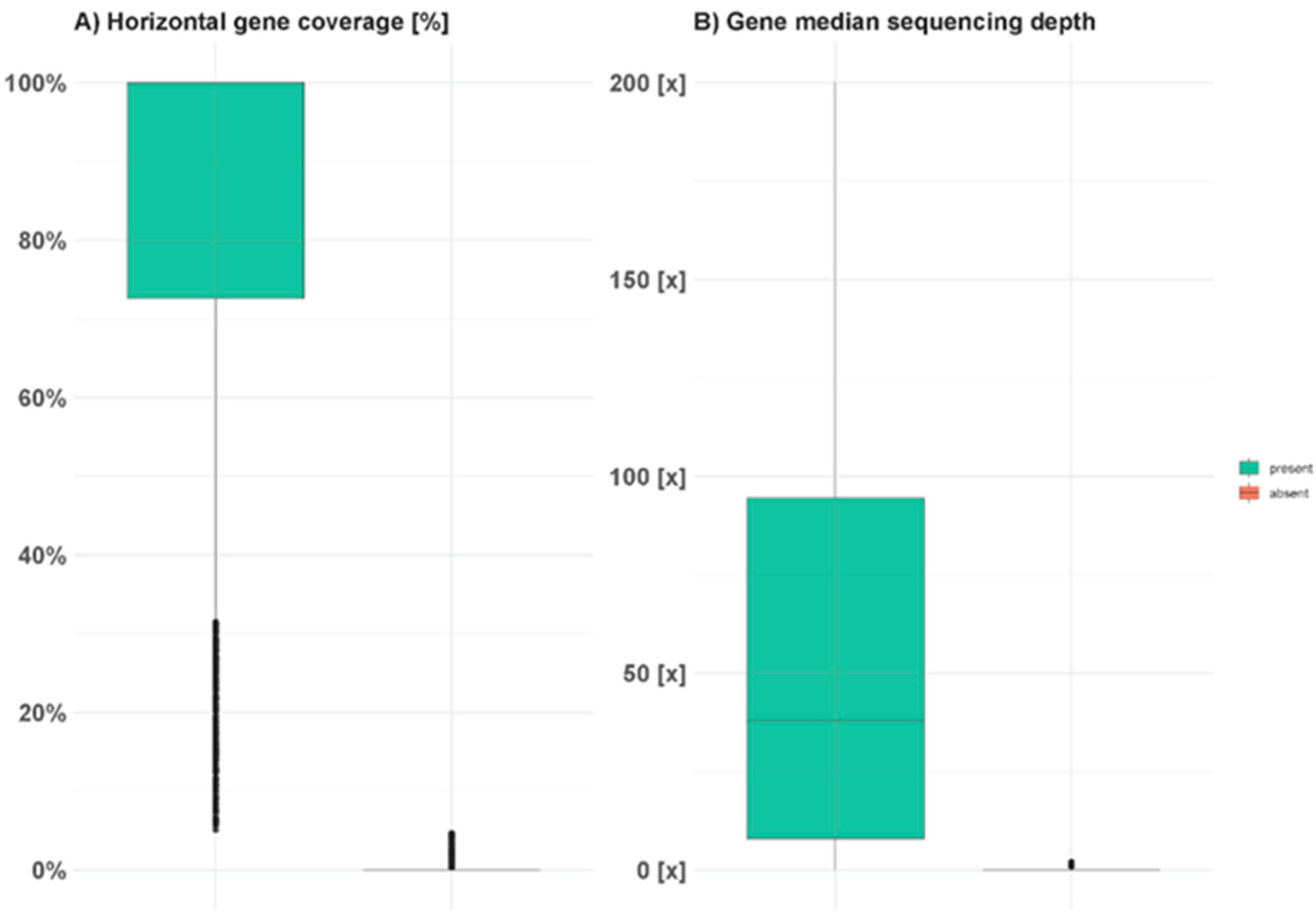
2. Results and Discussion
3. Materials and Methods
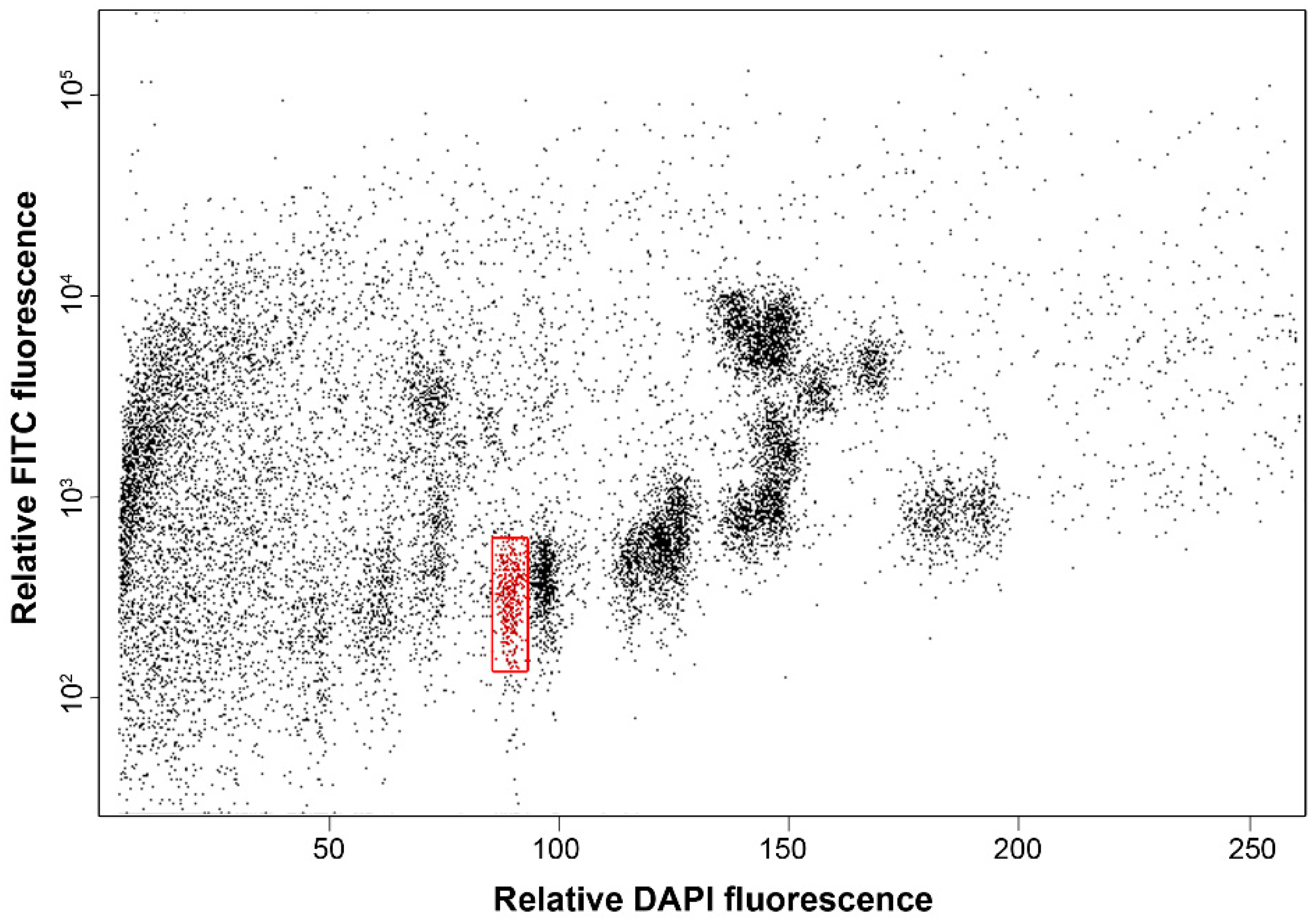
3.1. Purification of Chromosome Arm 7RS
3.2. Sequencing Flow-Sorted 7RS
3.3. Presence-Absence Variation Analysis
3.4. Data Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weipert, D. Rye and triticale. In Cereal Grain Quality; Springer: Manhattan, NY, USA, 1996; pp. 205–224. [Google Scholar]
- Fowler, D.; Carles, R. Growth, development, and cold tolerence of fall-acclimated cereal grains 1. Crop Sci. 1979, 19, 915–922. [Google Scholar] [CrossRef]
- Schittenhelm, S.; Kraft, M.; Wittich, K.-P. Performance of winter cereals grown on field-stored soil moisture only. Eur. J. Agron. 2014, 52, 247–258. [Google Scholar] [CrossRef]
- Gawroński, P.; Pawełkowicz, M.; Tofil, K.; Uszyński, G.; Sharifova, S.; Ahluwalia, S.; Tyrka, M.; Wędzony, M.; Kilian, A.; Bolibok-Brągoszewska, H. DArT markers effectively target gene space in the rye genome. Front. Plant Sci. 2016, 7, 1600. [Google Scholar] [CrossRef]
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774. [Google Scholar] [CrossRef]
- Huang, X.; Börner, A.; Röder, M.; Ganal, M. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002, 105, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Riley, R.; Chapman, V.; Johnson, R. The incorporation of alien disease resistance in wheat by genetic interference with the regulation of meiotic chromosome synapsis. Genet. Res. 1968, 12, 199–219. [Google Scholar] [CrossRef]
- Sears, E. Transfer of alien genetic material to wheat. Wheat Sci. Today Tomorrow 1981, 75–89. [Google Scholar]
- Rabinovich, S. Importance of wheat-rye translocations for breeding modern cultivar of Triticum aestivum L. Euphytica 1998, 100, 323–340. [Google Scholar] [CrossRef]
- Mater, Y.; Baenziger, S.; Gill, K.; Graybosch, R.; Whitcher, L.; Baker, C.; Specht, J.; Dweikat, I. Linkage mapping of powdery mildew and greenbug resistance genes on recombinant 1RS from ‘Amigo’ and ‘Kavkaz’ wheat–rye translocations of chromosome 1RS.1AL. Genome 2004, 47, 292–298. [Google Scholar] [CrossRef]
- Jung, W.J.; Seo, Y.W. Employment of wheat-rye translocation in wheat improvement and broadening its genetic basis. J. Crop Sci. Biotechnol. 2014, 17, 305–313. [Google Scholar] [CrossRef]
- Bennett, M.D.; Smith, J.B.; Barclay, I. Early seed development in the Triticeae. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1975, 272, 199–227. [Google Scholar]
- Bartoš, J.; Paux, E.; Kofler, R.; Havránková, M.; Kopecký, D.; Suchánková, P.; Šafář, J.; Šimková, H.; Town, C.; Lelley, T.; et al. A first survey of the rye (Secale cereale) genome composition through BAC end sequencing of the short arm of chromosome 1R. BMC Plant Biol. 2008, 8, 95. [Google Scholar] [CrossRef] [PubMed]
- Appels, R.; Francki, M.; Chibbar, R. Advances in cereal functional genomics. Funct. Integr. Genom. 2003, 3, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Bennetzen, J. Transposable element contributions to plant gene and genome evolution. Plant Mol. Biol. 2000, 42, 251–269. [Google Scholar] [CrossRef] [PubMed]
- Rabanus-Wallace, M.T.; Hackauf, B.; Mascher, M.; Lux, T.; Wicker, T.; Gundlach, H.; Baez, M.; Houben, A.; Mayer, K.F.X.; Guo, L.; et al. Chromosome-scale genome assembly provides insights into rye biology, evolution and agronomic potential. Nat. Genet. 2021, 53, 564–573. [Google Scholar] [CrossRef]
- Li, G.; Wang, L.; Yang, J.; He, H.; Jin, H.; Li, X.; Ren, T.; Ren, Z.; Li, F.; Han, X.; et al. A high-quality genome assembly highlights rye genomic characteristics and agronomically important genes. Nat. Genet. 2021, 53, 574–584. [Google Scholar] [CrossRef]
- Parat, F.; Schwertfirm, G.; Rudolph, U.; Miedaner, T.; Korzun, V.; Bauer, E.; Schön, C.C.; Tellier, A. Geography and end use drive the diversification of worldwide winter rye populations. Mol. Ecol. 2016, 25, 500–514. [Google Scholar] [CrossRef]
- Yang, M.; Ren, T.; Yan, B.; Li, Z.; Ren, Z. Diversity resistance to Puccinia striiformis f. sp tritici in rye chromosome arm 1RS expressed in wheat. Genet. Mol. Res. 2014, 13, 8783–8793. [Google Scholar] [CrossRef]
- Ren, T.; Tang, Z.; Fu, S.; Yan, B.; Tan, F.; Ren, Z.; Li, Z. Molecular cytogenetic characterization of novel wheat-rye T1RS.1BL translocation lines with high resistance to diseases and great agronomic traits. Front. Plant Sci. 2017, 8, 799. [Google Scholar] [CrossRef]
- Ruperao, P.; Chan, C.K.K.; Azam, S.; Karafiátová, M.; Hayashi, S.; Čížková, J.; Saxena, R.K.; Šimková, H.; Song, C.; Vrána, J.; et al. A chromosomal genomics approach to assess and validate the desi and kabuli draft chickpea genome assemblies. Plant Biotechnol. J. 2014, 12, 778–786. [Google Scholar] [CrossRef]
- Berkman, P.J.; Skarshewski, A.; Lorenc, M.T.; Lai, K.; Duran, C.; Ling, E.Y.; Stiller, J.; Smits, L.; Imelfort, M.; Manoli, S.; et al. Sequencing and assembly of low copy and genic regions of isolated Triticum aestivum chromosome arm 7DS. Plant Biotechnol. J. 2011, 9, 768–775. [Google Scholar] [PubMed]
- Berkman, P.J.; Skarshewski, A.; Manoli, S.; Lorenc, M.T.; Stiller, J.; Smits, L.; Lai, K.; Campbell, E.; Kubaláková, M.; Šimková, H.; et al. Sequencing wheat chromosome arm 7BS delimits the 7BS/4AL translocation and reveals homoeologous gene conservation. Theor. Appl. Genet. 2012, 124, 423–432. [Google Scholar] [CrossRef] [PubMed]
- Berkman, P.J.; Visendi, P.; Lee, H.C.; Stiller, J.; Manoli, S.; Lorenc, M.T.; Lai, K.; Batley, J.; Fleury, D.; Šimková, H.; et al. Dispersion and domestication shaped the genome of bread wheat. Plant Biotechnol. J. 2013, 11, 564–571. [Google Scholar]
- Kreplak, J.; Madoui, M.-A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- Doležel, J.; Vrána, J.; Šafář, J.; Bartoš, J.; Kubaláková, M.; Šimková, H. Chromosomes in the flow to simplify genome analysis. Funct. Integr. Genom. 2012, 12, 397–416. [Google Scholar] [CrossRef]
- Masojć, P.; Banek-Tabor, A.; Milczarski, P.; Twardowska, M. TLs for resistance to preharvest sprouting in rye (Secale cereale L.). J. Appl. Genet. 2007, 48, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Masojæ, P.; Milczarski, P. Mapping QTLs for α-amylase activity in rye grain. J. Appl. Genet. 2005, 46, 115–123. [Google Scholar]
- Hackauf, B.; Haffke, S.; Fromme, F.J.; Roux, S.R.; Kusterer, B.; Musmann, D.; Kilian, A.; Miedaner, T. QTL mapping and comparative genome analysis of agronomic traits including grain yield in winter rye. Theor. Appl. Genet. 2017, 130, 1801–1817. [Google Scholar] [CrossRef]
- Milczarski, P.; Masojć, P. The mapping of QTLS for chlorophyll content and responsiveness to gibberellic (GA3) and abscisic (ABA) acids in rye. Cell. Mol. Biol. Lett. 2002, 7, 449–456. [Google Scholar]
- Miedaner, T.; Laidig, F. Hybrid breeding in rye (Secale cereale L.). In Advances in Plant Breeding Strategies: Cereals; Springer: Manhattan, NY, USA, 2019; pp. 343–372. [Google Scholar]
- Arseniuk, E. Triticale Abiotic Stresses—An Overview. In Triticale; Eudes, F., Ed.; Springer International Publishing: Manhattan, NY, USA, 2015; pp. 69–81. [Google Scholar]
- Alkhimova, A.G.; Heslop-Harrison, J.S.; Shchapova, A.I.; Vershinin, A.V. Rye chromosome variability in wheat–rye addition and substitution lines. Chromosom. Res. 1999, 7, 205–212. [Google Scholar]
- Fu, S.; Lv, Z.; Guo, X.; Zhang, X.; Han, F. Alteration of Terminal Heterochromatin and Chromosome Rearrangements in Derivatives of Wheat-Rye Hybrids. J. Genet. Genom. 2013, 40, 413–420. [Google Scholar]
- Niwa, K.; Sakamoto, S. Origin of B chromosomes in cultivated rye. Genome 1995, 38, 307–312. [Google Scholar] [PubMed]
- Marques, A.; Banaei-Moghaddam, A.M.; Klemme, S.; Blattner, F.R.; Niwa, K.; Guerra, M.; Houben, A. B chromosomes of rye are highly conserved and accompanied the development of early agriculture. Ann. Bot. 2013, 112, 527–534. [Google Scholar] [CrossRef] [PubMed]
- Figueiras, A.; González-Jaén, M.; Candela, M.; Benito, C. Genic heterozygosity, chromosomal interchanges and fitness in rye: Any relationship? Genetica 2006, 128, 273–286. [Google Scholar] [PubMed]
- Li, J.; Endo, T.R.; Saito, M.; Ishikawa, G.; Nakamura, T.; Nasuda, S. Homoeologous relationship of rye chromosome arms as detected with wheat PLUG markers. Chromosoma 2013, 122, 555–564. [Google Scholar] [CrossRef] [PubMed]
- Golicz, A.A.; Martinez, P.A.; Zander, M.; Patel, D.A.; Van De Wouw, A.P.; Visendi, P.; Fitzgerald, T.L.; Edwards, D.; Batley, J. Gene loss in the fungal canola pathogen Leptosphaeria maculans. Funct. Integr. Genom. 2015, 15, 189–196. [Google Scholar]
- Vrána, J.; Kubaláková, M.; Šimková, H.; Číhalíková, J.; Lysák, M.; Doležel, J. Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.). Genetics 2000, 156, 2033–2041. [Google Scholar]
- Kubaláková, M.; Vrána, J.; Číhalíková, J.; Šimková, H.; Doležel, J. Flow karyotyping and chromosome sorting in bread wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 104, 1362–1372. [Google Scholar]
- Doležel, J.; Binarová, P.; Lucretti, S. Analysis of Nuclear DNA content in plant cells by flow cytometry. Biol. Plant. 1989, 31, 113–120. [Google Scholar]
- Giorgi, D.; Farina, A.; Grosso, V.; Gennaro, A.; Ceoloni, C.; Lucretti, S. FISHIS: Fluorescence in situ hybridization in suspension and chromosome flow sorting made easy. PLoS ONE 2013, 8, e57994. [Google Scholar]
- Kubaláková, M.; Macas, J.; Doležel, J. Mapping of repeated DNA sequences in plant chromosomes by PRINS and C-PRINS. Theor. Appl. Genet. 1997, 94, 758–763. [Google Scholar] [CrossRef]
- Šimková, H.; Svensson, J.T.; Condamine, P.; Hřibová, E.; Suchánková, P.; Bhat, P.R.; Bartoš, J.; Šafář, J.; Close, T.J.; Doležel, J. Coupling amplified DNA from flow-sorted chromosomes to high-density SNP mapping in barley. BMC Genom. 2008, 9, 294. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar]
- R Core Team. A language and environment for statistical computing. Open J. Stat. 2013, 6, 3. [Google Scholar]
- Allaire, J. Integrated Development Environment for R; RStudio: Boston, MA, USA, 2012; Volume 770, pp. 165–171. [Google Scholar]
- Wickham, H.; Averick, M.; Bryan, J.; Chang, W.; McGowan, L.D.A.; François, R.; Grolemund, G.; Hayes, A.; Henry, L.; Hester, J. Welcome to the Tidyverse. J. Open Source Softw. 2019, 4, 1686. [Google Scholar] [CrossRef]
- Dowle, M.; Srinivasan, A.; Gorecki, J.; Chirico, M.; Stetsenko, P.; Short, T.; Lianoglou, S.; Antonyan, E.; Bonsch, M.; Parsonage, H. Package Data. Table; Extension of Data. Frame. 2019. Available online: https://CRAN.R-project.org/package=data.table (accessed on 7 August 2022).

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Petereit, J.; Tay Fernandez, C.; Marsh, J.I.; Bayer, P.E.; Thomas, W.J.W.; Aliyeva, A.J.; Karafiátová, M.; Doležel, J.; Batley, J.; Edwards, D. Isolation and Sequencing of Chromosome Arm 7RS of Rye, Secale cereale. Int. J. Mol. Sci. 2022, 23, 11106. https://doi.org/10.3390/ijms231911106
Petereit J, Tay Fernandez C, Marsh JI, Bayer PE, Thomas WJW, Aliyeva AJ, Karafiátová M, Doležel J, Batley J, Edwards D. Isolation and Sequencing of Chromosome Arm 7RS of Rye, Secale cereale. International Journal of Molecular Sciences. 2022; 23(19):11106. https://doi.org/10.3390/ijms231911106
Chicago/Turabian StylePetereit, Jakob, Cassandria Tay Fernandez, Jacob I. Marsh, Philipp E. Bayer, William J. W. Thomas, Aybeniz Javad Aliyeva, Miroslava Karafiátová, Jaroslav Doležel, Jacqueline Batley, and David Edwards. 2022. "Isolation and Sequencing of Chromosome Arm 7RS of Rye, Secale cereale" International Journal of Molecular Sciences 23, no. 19: 11106. https://doi.org/10.3390/ijms231911106
APA StylePetereit, J., Tay Fernandez, C., Marsh, J. I., Bayer, P. E., Thomas, W. J. W., Aliyeva, A. J., Karafiátová, M., Doležel, J., Batley, J., & Edwards, D. (2022). Isolation and Sequencing of Chromosome Arm 7RS of Rye, Secale cereale. International Journal of Molecular Sciences, 23(19), 11106. https://doi.org/10.3390/ijms231911106









