Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade
Abstract
1. Introduction
2. Results
2.1. Chronic, Static Effects of ARB Treatment
2.1.1. OLETF Rats Present with Hepatic Hyperglycemia and Hypertriglyceridemia
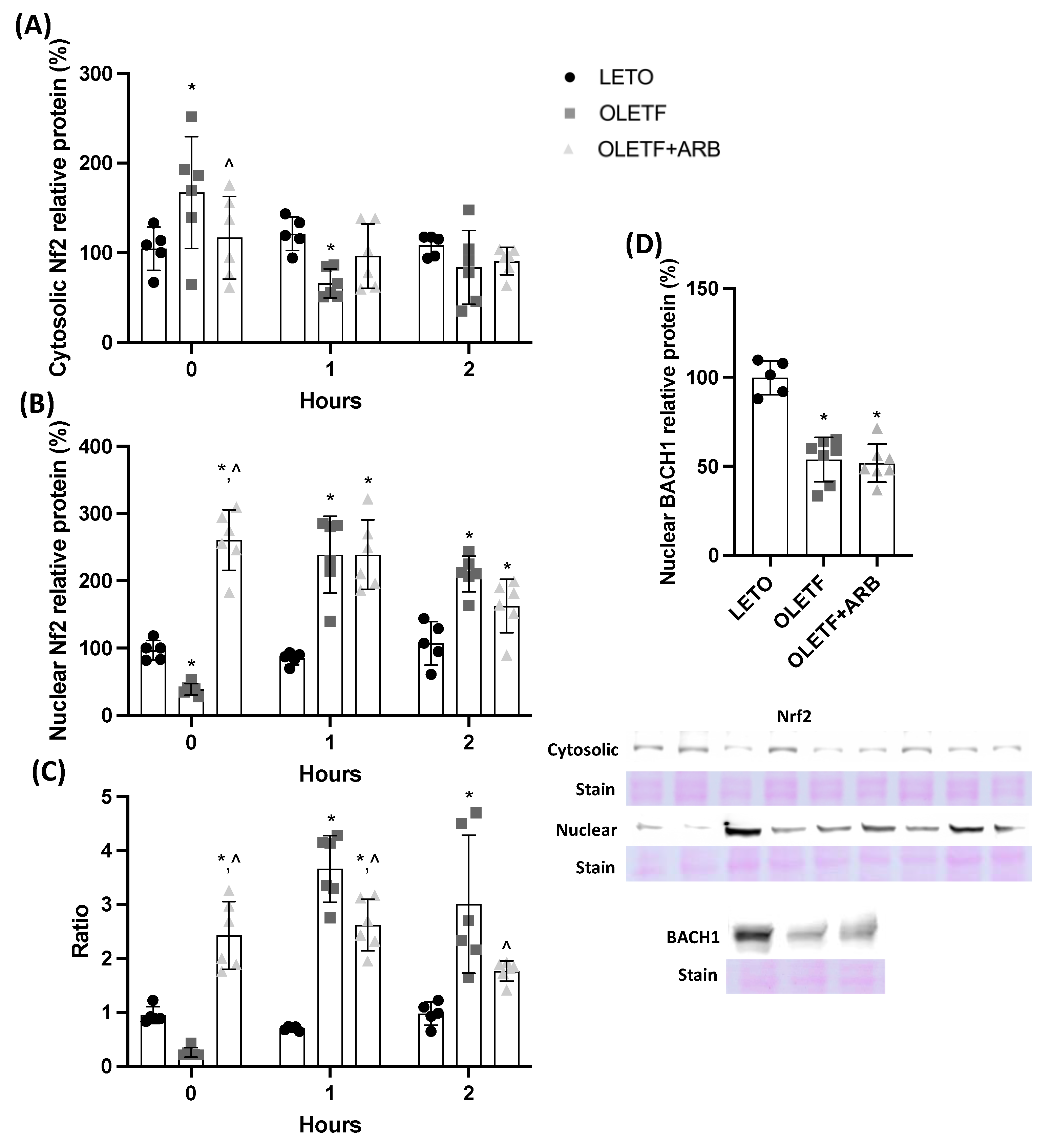
2.1.2. AT1 Blockade Increases Nuclear Hepatic Nrf2 Levels
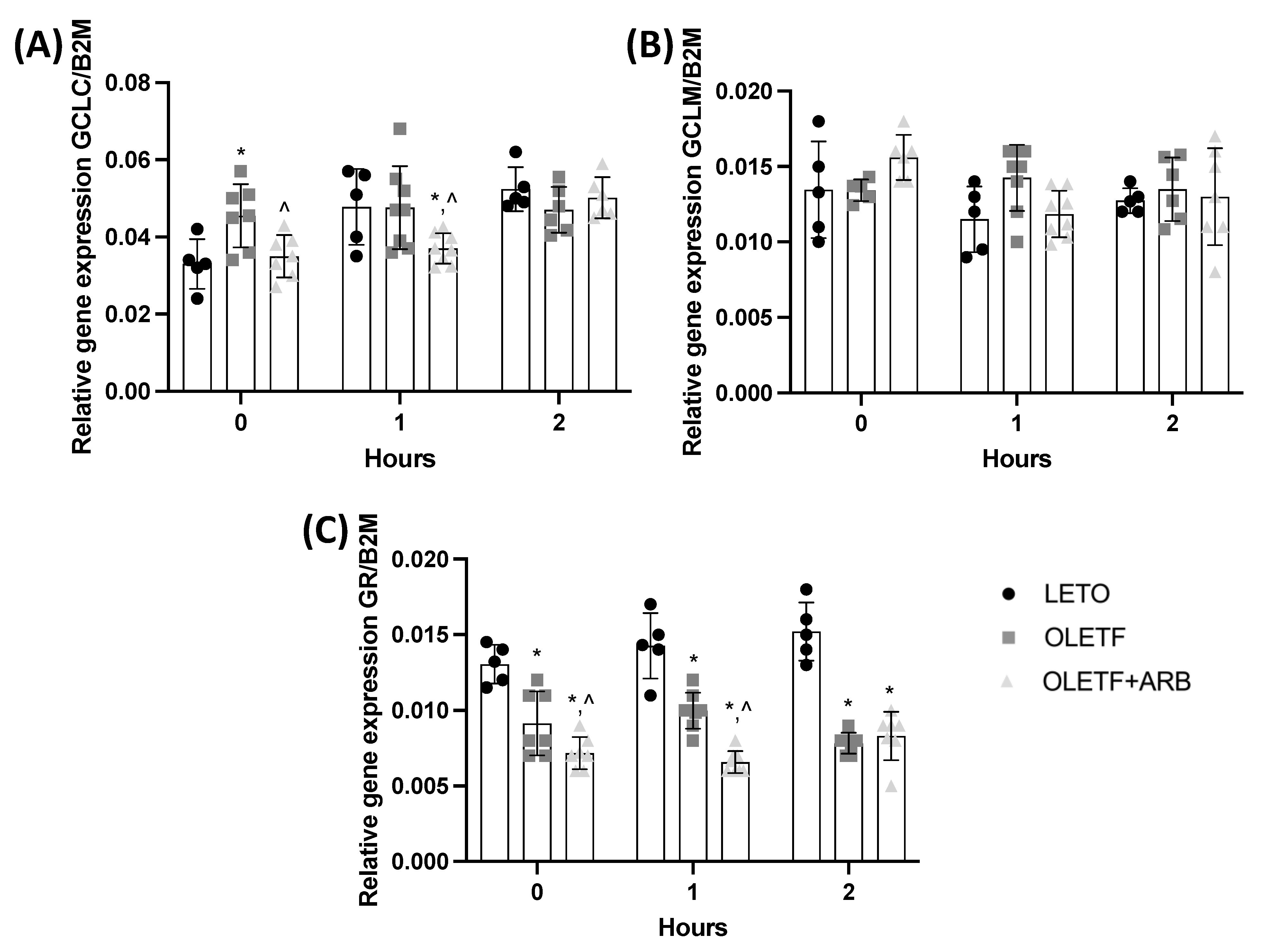
2.1.3. Hepatic Expressions of GCLC and GR Were Decreased with AT1 Blockade
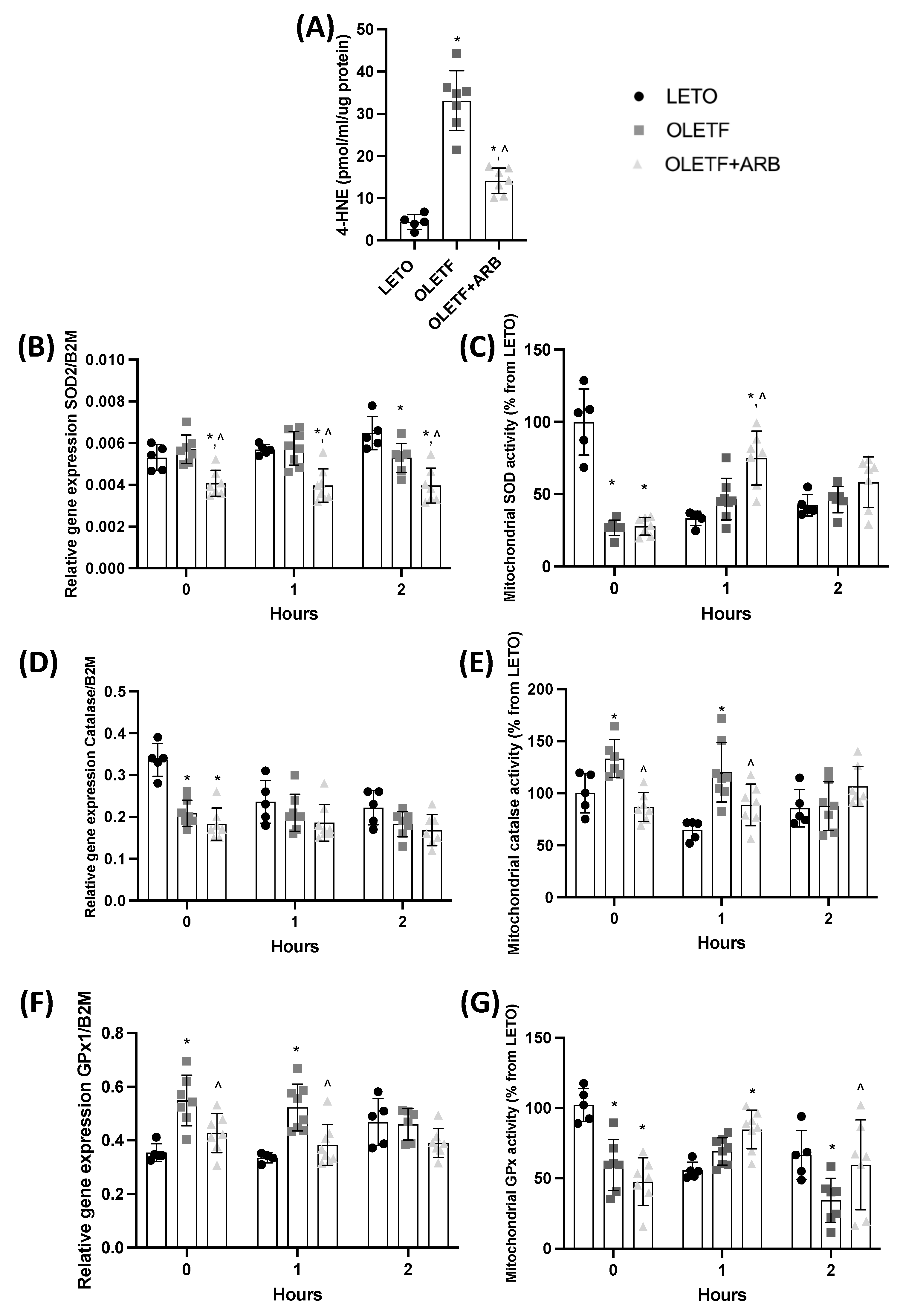
2.1.4. Total Hepatic 4-Hydroxynonenal Levels and Mitochondrial Catalase Activity Were Decreased with AT1 Blockade
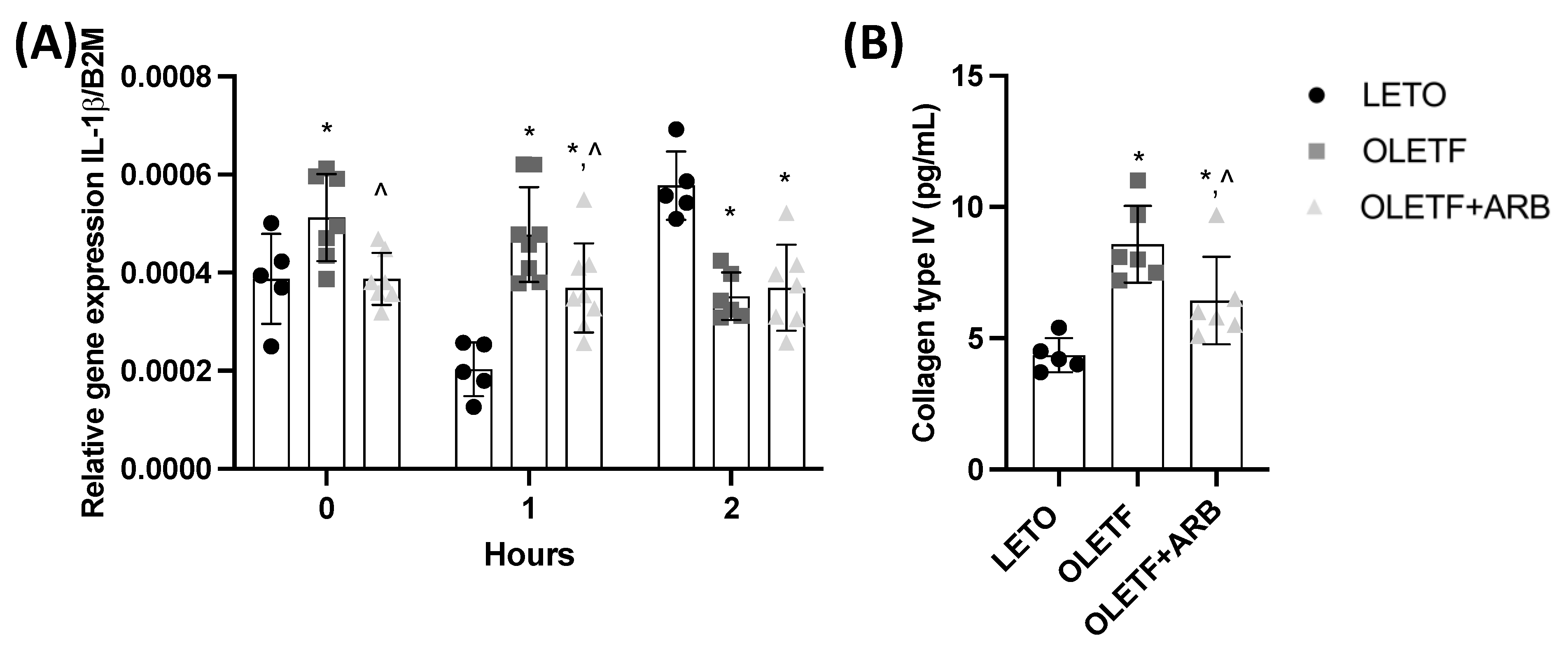
2.1.5. Hepatic Expression of IL-1β and Plasma Levels of Col4 Decreased with AT1 Blockade
2.2. Acute, Dynamic Responses to Glucose Challenge
2.2.1. Glucose Stimulated Nrf2 Translocation in OLETF
2.2.2. Hepatic GCLC, GCLM, and GR Expressions Remained Constant during the Acute Glucose Challenge
2.2.3. SOD and GPx Activities Increased in Response to the Glucose Challenge
2.2.4. Chronic ARB Treatment Stabilized the Expression of IL-1β in Response to a Glucose Challenge
3. Discussion
4. Methods
4.1. Animals
4.2. Glucose Challenge
4.3. Plasma Marker of Hepatic Fibrosis
4.4. Western Blots
4.5. Biochemical Analyses
4.6. qPCR mRNA Quantification
4.7. Statistics
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sanyal, A.J. Past, Present and Future Perspectives in Nonalcoholic Fatty Liver Disease. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Kader, S.M.; El-Den Ashmawy, E.M.S. Non-Alcoholic Fatty Liver Disease: The Diagnosis and Management. World J. Hepatol. 2015, 7, 846–858. [Google Scholar] [CrossRef] [PubMed]
- Eslam, M.; Sanyal, A.J.; George, J.; Sanyal, A.; Neuschwander-Tetri, B.; Tiribelli, C.; Kleiner, D.E.; Brunt, E.; Bugianesi, E.; Yki-Järvinen, H.; et al. MAFLD: A Consensus-Driven Proposed Nomenclature for Metabolic Associated Fatty Liver Disease. Gastroenterology 2020, 158, 1999–2014.e1. [Google Scholar] [CrossRef]
- Kalupahana, N.S.; Moustaid-Moussa, N. The Renin-Angiotensin System: A Link between Obesity, Inflammation and Insulin Resistance. Obes. Rev. Off. J. Int. Assoc. Study Obes. 2012, 13, 136–149. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-H.; Li, F.; Takahashi, N. The Renin Angiotensin System and the Metabolic Syndrome. Open Hypertens. J. 2010, 3, 1–13. [Google Scholar] [CrossRef]
- Friedman, S.L.; Neuschwander-Tetri, B.A.; Rinella, M.; Sanyal, A.J. Mechanisms of NAFLD Development and Therapeutic Strategies. Nat. Med. 2018, 24, 908–922. [Google Scholar] [CrossRef]
- Latif, M.U.; Schmidt, G.E.; Mercan, S.; Rahman, R.; Gibhardt, C.S.; Stejerean-Todoran, I.; Reutlinger, K.; Hessmann, E.; Singh, S.K.; Moeed, A.; et al. NFATc1 Signaling Drives Chronic ER Stress Responses to Promote NAFLD Progression. Gut 2022. [Google Scholar] [CrossRef]
- Prasun, P.; Ginevic, I.; Oishi, K. Mitochondrial Dysfunction in Nonalcoholic Fatty Liver Disease and Alcohol Related Liver Disease. Transl. Gastroenterol. Hepatol. 2021, 6, 4. [Google Scholar] [CrossRef]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-Mediated Dysbiosis Regulates Progression of NAFLD and Obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef]
- Anderson, M.E. Glutathione: An Overview of Biosynthesis and Modulation. Chem. Biol. Interact. 1998, 111–112, 1–14. [Google Scholar] [CrossRef]
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. J. Amino Acids 2012, 2012, 736837. [Google Scholar] [CrossRef] [PubMed]
- Ballatori, N.; Krance, S.M.; Notenboom, S.; Shi, S.; Tieu, K.; Hammond, C.L. Glutathione Dysregulation and the Etiology and Progression of Human Diseases. Biol. Chem. 2009, 390, 191–214. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Sharma, A.; Duseja, A.; Das, A.; Dhiman, R.K.; Chawla, Y.K.; Kohli, K.K.; Bhansali, A. Patients with Nonalcoholic Fatty Liver Disease (NAFLD) Have Higher Oxidative Stress in Comparison to Chronic Viral Hepatitis. J. Clin. Exp. Hepatol. 2013, 3, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Irie, M.; Sohda, T.; Anan, A.; Fukunaga, A.; Takata, K.; Tanaka, T.; Yokoyama, K.; Morihara, D.; Takeyama, Y.; Shakado, S.; et al. Reduced Glutathione Suppresses Oxidative Stress in Nonalcoholic Fatty Liver Disease. Euroasian J. Hepatogastroenterol. 2016, 6, 13–18. [Google Scholar] [CrossRef] [PubMed]
- Forman, H.J.; Ursini, F.; Maiorino, M. An Overview of Mechanisms of Redox Signaling. J. Mol. Cell Cardiol. 2014, 73, 2–9. [Google Scholar] [CrossRef]
- Goutzourelas, N.; Orfanou, M.; Charizanis, I.; Leon, G.; Spandidos, D.A.; Kouretas, D. GSH Levels Affect Weight Loss in Individuals with Metabolic Syndrome and Obesity Following Dietary Therapy. Exp. Ther. Med. 2018, 16, 635–642. [Google Scholar] [CrossRef]
- Lu, S.C. Glutathione Synthesis. Biochim. Biophys. Acta 2013, 1830, 3143–3153. [Google Scholar] [CrossRef]
- Seelig, G.F.; Simondsen, R.P.; Meister, A. Reversible Dissociation of Gamma-Glutamylcysteine Synthetase into Two Subunits. J. Biol. Chem. 1984, 259, 9345–9347. [Google Scholar] [CrossRef]
- Huang, C.S.; Chang, L.S.; Anderson, M.E.; Meister, A. Catalytic and Regulatory Properties of the Heavy Subunit of Rat Kidney Gamma-Glutamylcysteine Synthetase. J. Biol. Chem. 1993, 268, 19675–19680. [Google Scholar] [CrossRef]
- Zhang, H.; Forman, H.J.; Choi, J. Gamma-Glutamyl Transpeptidase in Glutathione Biosynthesis. Methods Enzym. 2005, 401, 468–483. [Google Scholar] [CrossRef]
- Lauterburg, B.H.; Adams, J.D.; Mitchell, J.R. Hepatic Glutathione Homeostasis in the Rat: Efflux Accounts for Glutathione Turnover. Hepatology 1984, 4, 586–590. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Abramov, A.Y. The Emerging Role of Nrf2 in Mitochondrial Function. Free Radic. Biol. Med. 2015, 88, 179–188. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The Emerging Role of the Nrf2–Keap1 Signaling Pathway in Cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Levonen, A.-L.; Hill, B.G.; Kansanen, E.; Zhang, J.; Darley-Usmar, V.M. Redox Regulation of Antioxidants, Autophagy, and the Response to Stress: Implications for Electrophile Therapeutics. Free Radic. Biol. Med. 2014, 71, 196–207. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.; Itoh, K.; Yamamoto, M.; Zweier, J.L.; Li, Y. Role of Nrf2 Signaling in Regulation of Antioxidants and Phase 2 Enzymes in Cardiac Fibroblasts: Protection against Reactive Oxygen and Nitrogen Species-Induced Cell Injury. FEBS Lett. 2005, 579, 3029–3036. [Google Scholar] [CrossRef]
- Ramadori, P.; Drescher, H.; Erschfeld, S.; Schumacher, F.; Berger, C.; Fragoulis, A.; Schenkel, J.; Kensler, T.W.; Wruck, C.J.; Trautwein, C.; et al. Hepatocyte-Specific Keap1 Deletion Reduces Liver Steatosis but Not Inflammation during Non-Alcoholic Steatohepatitis Development. Free Radic. Biol. Med. 2016, 91, 114–126. [Google Scholar] [CrossRef]
- Du, J.; Zhang, M.; Lu, J.; Zhang, X.; Xiong, Q.; Xu, Y.; Bao, Y.; Jia, W. Osteocalcin Improves Nonalcoholic Fatty Liver Disease in Mice through Activation of Nrf2 and Inhibition of JNK. Endocrine 2016, 53, 701–709. [Google Scholar] [CrossRef]
- Thorwald, M.A.; Godoy-Lugo, J.A.; Rodriguez, R.; Stanhope, K.L.; Graham, J.L.; Havel, P.J.; Forman, H.J.; Ortiz, R.M. Cardiac NF-ΚB Acetylation Increases While Nrf2-Related Gene Expression and Mitochondrial Activity Are Impaired during the Progression of Diabetes in UCD-T2DM Rats. Antioxidants 2022, 11, 927. [Google Scholar] [CrossRef]
- Thorwald, M.; Rodriguez, R.; Lee, A.; Martinez, B.; Peti-Peterdi, J.; Nakano, D.; Nishiyama, A.; Ortiz, R.M. Angiotensin Receptor Blockade Improves Cardiac Mitochondrial Activity in Response to an Acute Glucose Load in Obese Insulin Resistant Rats. Redox Biol. 2018, 14, 371–378. [Google Scholar] [CrossRef]
- Vazquez-Medina, J.P.; Popovich, I.; Thorwald, M.A.; Viscarra, J.A.; Rodriguez, R.; Sonanez-Organis, J.G.; Lam, L.; Peti-Peterdi, J.; Nakano, D.; Nishiyama, A.; et al. Angiotensin Receptor-Mediated Oxidative Stress Is Associated with Impaired Cardiac Redox Signaling and Mitochondrial Function in Insulin-Resistant Rats. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H599–H607. [Google Scholar] [CrossRef]
- Triguero, A.; Barber, T.; Garcia, C.; Puertes, I.R.; Sastre, J.; Vina, J.R. Liver Intracellular L-Cysteine Concentration Is Maintained after Inhibition of the Trans-Sulfuration Pathway by Propargylglycine in Rats. Br. J. Nutr. 1997, 78, 823–831. [Google Scholar] [CrossRef] [PubMed]
- Perlemuter, G.; Davit-Spraul, A.; Cosson, C.; Conti, M.; Bigorgne, A.; Paradis, V.; Corre, M.-P.; Prat, L.; Kuoch, V.; Basdevant, A.; et al. Increase in Liver Antioxidant Enzyme Activities in Non-Alcoholic Fatty Liver Disease. Liver Int. Off. J. Int. Assoc. Study Liver 2005, 25, 946–953. [Google Scholar] [CrossRef] [PubMed]
- Bekyarova, G.; Tzaneva, M.; Bratoeva, K.; Ivanova, I.; Kotzev, A.; Hristova, M.; Krastev, D.; Kindekov, I.; Mileva, M. 4-Hydroxynonenal (HNE) and Hepatic Injury Related to Chronic Oxidative Stress. Biotechnol. Biotechnol. Equip. 2019, 33, 1544–1552. [Google Scholar] [CrossRef]
- Meier, R.P.H.; Meyer, J.; Montanari, E.; Lacotte, S.; Balaphas, A.; Muller, Y.D.; Clément, S.; Negro, F.; Toso, C.; Morel, P.; et al. Interleukin-1 Receptor Antagonist Modulates Liver Inflammation and Fibrosis in Mice in a Model-Dependent Manner. Int. J. Mol. Sci. 2019, 20, 1295. [Google Scholar] [CrossRef]
- Li, Y.; Xu, H.; Wu, W.; Ye, J.; Fang, D.; Shi, D.; Li, L. Clinical Application of Angiotensin Receptor Blockers in Patients with Non-Alcoholic Fatty Liver Disease: A Systematic Review and Meta-Analysis. Oncotarget 2018, 9, 24155–24167. [Google Scholar] [CrossRef]
- Buzzetti, E.; Pinzani, M.; Tsochatzis, E.A. The Multiple-Hit Pathogenesis of Non-Alcoholic Fatty Liver Disease (NAFLD). Metabolism 2016, 65, 1038–1048. [Google Scholar] [CrossRef]
- Fattahi, M.R.; Niknam, R.; Safarpour, A.; Sepehrimanesh, M.; Lotfi, M. The Prevalence of Metabolic Syndrome In Non-Alcoholic Fatty Liver Disease; A Population-Based Study. Middle East J. Dig. Dis. 2016, 8, 131–137. [Google Scholar] [CrossRef]
- García-Ruiz, C.; Fernández-Checa, J.C. Mitochondrial Oxidative Stress and Antioxidants Balance in Fatty Liver Disease. Hepatol. Commun. 2018, 2, 1425–1439. [Google Scholar] [CrossRef]
- Cui, R.; Gao, M.; Qu, S.; Liu, D. Overexpression of Superoxide Dismutase 3 Gene Blocks High-Fat Diet-Induced Obesity, Fatty Liver and Insulin Resistance. Gene Ther. 2014, 21, 840–848. [Google Scholar] [CrossRef]
- Chambel, S.S.; Santos-Gonçalves, A.; Duarte, T.L. The Dual Role of Nrf2 in Nonalcoholic Fatty Liver Disease: Regulation of Antioxidant Defenses and Hepatic Lipid Metabolism. BioMed Res. Int. 2015, 2015, 597134. [Google Scholar] [CrossRef]
- Thorwald, M.A.; Godoy-Lugo, J.A.; Rodriguez, G.J.; Rodriguez, M.A.; Jamal, M.; Kinoshita, H.; Nakano, D.; Nishiyama, A.; Forman, H.J.; Ortiz, R.M. Nrf2-Related Gene Expression Is Impaired during a Glucose Challenge in Type II Diabetic Rat Hearts. Free Radic Biol. Med. 2019, 130, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Dhakshinamoorthy, S.; Jain, A.K.; Bloom, D.A.; Jaiswal, A.K. Bach1 Competes with Nrf2 Leading to Negative Regulation of the Antioxidant Response Element (ARE)-Mediated NAD(P)H:Quinone Oxidoreductase 1 Gene Expression and Induction in Response to Antioxidants. J. Biol. Chem. 2005, 280, 16891–16900. [Google Scholar] [CrossRef] [PubMed]
- Fukai, T.; Ushio-Fukai, M. Superoxide Dismutases: Role in Redox Signaling, Vascular Function, and Diseases. Antioxid. Redox Signal. 2011, 15, 1583–1606. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Oxidative Stress: Oxidants and Antioxidants. Exp. Physiol. 1997, 82, 291–295. [Google Scholar] [CrossRef]
- Salvi, M.; Battaglia, V.; Brunati, A.M.; La Rocca, N.; Tibaldi, E.; Pietrangeli, P.; Marcocci, L.; Mondovi, B.; Rossi, C.A.; Toninello, A. Catalase Takes Part in Rat Liver Mitochondria Oxidative Stress Defense. J. Biol. Chem. 2007, 282, 24407–24415. [Google Scholar] [CrossRef]
- Lubos, E.; Loscalzo, J.; Handy, D.E. Glutathione Peroxidase-1 in Health and Disease: From Molecular Mechanisms to Therapeutic Opportunities. Antioxid. Redox Signal. 2011, 15, 1957–1997. [Google Scholar] [CrossRef]
- Cianchetti, S.; Del Fiorentino, A.; Colognato, R.; Di Stefano, R.; Franzoni, F.; Pedrinelli, R. Anti-Inflammatory and Anti-Oxidant Properties of Telmisartan in Cultured Human Umbilical Vein Endothelial Cells. Atherosclerosis 2008, 198, 22–28. [Google Scholar] [CrossRef]
- Song, Y.S.; Fang, C.H.; So, B.I.; Park, J.Y.; Lee, Y.; Shin, J.H.; Jun, D.W.; Kim, H.; Kim, K.S. Time Course of the Development of Nonalcoholic Fatty Liver Disease in the Otsuka Long-Evans Tokushima Fatty Rat. Gastroenterol. Res. Pract. 2013, 2013, 342648. [Google Scholar] [CrossRef][Green Version]
- Godoy-Lugo, J.A.; Mendez, D.A.; Rodriguez, R.; Nishiyama, A.; Nakano, D.; Soñanez-Organis, J.G.; Ortiz, R.M. Improved Lipogenesis Gene Expression in Liver Is Associated with Elevated Plasma Angiotensin 1-7 after AT1 Receptor Blockade in Insulin-Resistant OLETF Rats. Mol. Cell. Endocrinol. 2022, 555, 111729. [Google Scholar] [CrossRef]
- Tan, Y.; Ichikawa, T.; Li, J.; Si, Q.; Yang, H.; Chen, X.; Goldblatt, C.S.; Meyer, C.J.; Li, X.; Cai, L.; et al. Diabetic Downregulation of Nrf2 Activity via ERK Contributes to Oxidative Stress-Induced Insulin Resistance in Cardiac Cells in Vitro and in Vivo. Diabetes 2011, 60, 625–633. [Google Scholar] [CrossRef]
- Swiderska, M.; Maciejczyk, M.; Zalewska, A.; Pogorzelska, J.; Flisiak, R.; Chabowski, A. Oxidative Stress Biomarkers in the Serum and Plasma of Patients with Non-Alcoholic Fatty Liver Disease (NAFLD). Can Plasma AGE Be a Marker of NAFLD? Oxidative Stress Biomarkers in NAFLD Patients. Free Radic Res. 2019, 53, 841–850. [Google Scholar] [CrossRef]
- Chen, Z.; Tian, R.; She, Z.; Cai, J.; Li, H. Role of Oxidative Stress in the Pathogenesis of Nonalcoholic Fatty Liver Disease. Free Radic. Biol. Med. 2020, 152, 116–141. [Google Scholar] [CrossRef] [PubMed]
- Mota, M.; Banini, B.A.; Cazanave, S.C.; Sanyal, A.J. Molecular Mechanisms of Lipotoxicity and Glucotoxicity in Nonalcoholic Fatty Liver Disease. Metabolism 2016, 65, 1049–1061. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Valle, V.; Chavez-Tapia, N.C.; Uribe, M.; Mendez-Sanchez, N. Role of Oxidative Stress and Molecular Changes in Liver Fibrosis: A Review. Curr. Med. Chem. 2012, 19, 4850–4860. [Google Scholar] [CrossRef] [PubMed]
- Duan, Y.; Pan, X.; Luo, J.; Xiao, X.; Li, J.; Bestman, P.L.; Luo, M. Association of Inflammatory Cytokines With Non-Alcoholic Fatty Liver Disease. Front. Immunol. 2022, 13, 880298. [Google Scholar] [CrossRef]
- Stefano, J.T.; Guedes, L.V.; de Souza, A.A.A.; Vanni, D.S.; Alves, V.A.F.; Carrilho, F.J.; Largura, A.; Arrese, M.; Oliveira, C.P. Usefulness of Collagen Type IV in the Detection of Significant Liver Fibrosis in Nonalcoholic Fatty Liver Disease. Ann. Hepatol. 2021, 20, 100253. [Google Scholar] [CrossRef]
- Mizuno, M.; Shima, T.; Oya, H.; Mitsumoto, Y.; Mizuno, C.; Isoda, S.; Kuramoto, M.; Taniguchi, M.; Noda, M.; Sakai, K.; et al. Classification of Patients with Non-Alcoholic Fatty Liver Disease Using Rapid Immunoassay of Serum Type IV Collagen Compared with Liver Histology and Other Fibrosis Markers. Hepatol. Res. 2017, 47, 216–225. [Google Scholar] [CrossRef]
- Chiarelli, F.; Di Marzio, D.; Santilli, F.; Mohn, A.; Blasetti, A.; Cipollone, F.; Mezzetti, A.; Verrotti, A. Effects of Irbesartan on Intracellular Antioxidant Enzyme Expression and Activity in Adolescents and Young Adults with Early Diabetic Angiopathy. Diabetes Care 2005, 28, 1690–1697. [Google Scholar] [CrossRef]
- Onozato, M.L.; Tojo, A.; Goto, A.; Fujita, T.; Wilcox, C.S. Oxidative Stress and Nitric Oxide Synthase in Rat Diabetic Nephropathy: Effects of ACEI and ARB. Kidney Int. 2002, 61, 186–194. [Google Scholar] [CrossRef]
- Godoy-Lugo, J.A.; Thorwald, M.A.; Hui, D.Y.; Nishiyama, A.; Nakano, D.; Sonanez-Organis, J.G.; Ortiz, R.M. Chronic Angiotensin Receptor Activation Promotes Hepatic Triacylglycerol Accumulation during an Acute Glucose Challenge in Obese-Insulin-Resistant OLETF Rats. Endocrine 2022, 75, 92–107. [Google Scholar] [CrossRef]
- Ahmed, I.A.; Mikail, M.A.; Mustafa, M.R.; Ibrahim, M.; Othman, R. Lifestyle Interventions for Non-Alcoholic Fatty Liver Disease. Saudi J. Biol. Sci. 2019, 26, 1519–1524. [Google Scholar] [CrossRef] [PubMed]
- Takaki, A.; Kawai, D.; Yamamoto, K. Multiple Hits, Including Oxidative Stress, as Pathogenesis and Treatment Target in Non-Alcoholic Steatohepatitis (NASH). Int. J. Mol. Sci. 2013, 14, 20704–20728. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.Y.; Kobori, H.; Nakano, D.; Hitomi, H.; Mori, H.; Masaki, T.; Sun, Y.X.; Zhi, N.; Zhang, L.; Huang, W.; et al. Aberrant Activation of the Intrarenal Renin-Angiotensin System in the Developing Kidneys of Type 2 Diabetic Rats. Horm. Metab. Res. 2013, 45, 338–343. [Google Scholar] [CrossRef] [PubMed]
- Dimauro, I.; Pearson, T.; Caporossi, D.; Jackson, M.J. A Simple Protocol for the Subcellular Fractionation of Skeletal Muscle Cells and Tissue. BMC Res. Notes 2012, 5, 513. [Google Scholar] [CrossRef]
- Nakamura, K.; Tanaka, T.; Kuwahara, A.; Takeo, K. Microassay for Proteins on Nitrocellulose Filter Using Protein Dye-Staining Procedure. Anal. Biochem. 1985, 148, 311–319. [Google Scholar] [CrossRef]
- Holden, P.; Horton, W.A. Crude Subcellular Fractionation of Cultured Mammalian Cell Lines. BMC Res. Notes 2009, 2, 243. [Google Scholar] [CrossRef]
- Ghasemi, A.; Zahediasl, S. Normality Tests for Statistical Analysis: A Guide for Non-Statisticians. Int. J. Endocrinol. Metab. 2012, 10, 486–489. [Google Scholar] [CrossRef]
- Mukaka, M.M. Statistics Corner: A Guide to Appropriate Use of Correlation Coefficient in Medical Research. Malawi Med. J. 2012, 24, 69–71. [Google Scholar]
- Motulsky, H.J.; Brown, R.E. Detecting Outliers When Fitting Data with Nonlinear Regression—A New Method Based on Robust Nonlinear Regression and the False Discovery Rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef]
- Kwak, S.K.; Kim, J.H. Statistical Data Preparation: Management of Missing Values and Outliers. Korean J. Anesth. 2017, 70, 407–411. [Google Scholar] [CrossRef]
| Primer Name | Sequence | NCBI Reference Sequence |
|---|---|---|
| GCLC F GCLC R | AGTGGAGGTAAAAGCGACCC TCACTTGTGGGCAACTGGAA | NM_012815.2 |
| GCLM F GCLM R | GAAAAAGTGTCCGTCCACGC CCACTGCATGGGACATGGTA | NM_017305.2 |
| GR F GR R | CGAGGAAGACGAAATGCGTG CGAAGCCCTGAAGCATCTCA | NM_053906.2 |
| SOD2 F SOD2 R | TTGCTGGAGGCTATCAAGCG CGGCAATCTGTAAGCGACCT | NM_017051.2 |
| Catalase F Catalase R | CACTCAGGTGCGGACATTCT CAGGGTGGACGTCAGTGAAA | NM_012520.2 |
| GPx1 F GPx1 R | GGTGTTCCAGTGCGCAGATA CTTAGGGGTTGCTAGGCTGC | NM_030826.4 |
| IL-1B F IL-1B R | CCTATGTCTTGCCCGTGGAG TCCTGGGGAAGGCATTAGGA | NM_031512.2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Godoy-Lugo, J.A.; Thorwald, M.A.; Mendez, D.A.; Rodriguez, R.; Nakano, D.; Nishiyama, A.; Ortiz, R.M. Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade. Int. J. Mol. Sci. 2022, 23, 10897. https://doi.org/10.3390/ijms231810897
Godoy-Lugo JA, Thorwald MA, Mendez DA, Rodriguez R, Nakano D, Nishiyama A, Ortiz RM. Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade. International Journal of Molecular Sciences. 2022; 23(18):10897. https://doi.org/10.3390/ijms231810897
Chicago/Turabian StyleGodoy-Lugo, Jose A., Max A. Thorwald, Dora A. Mendez, Ruben Rodriguez, Daisuke Nakano, Akira Nishiyama, and Rudy M. Ortiz. 2022. "Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade" International Journal of Molecular Sciences 23, no. 18: 10897. https://doi.org/10.3390/ijms231810897
APA StyleGodoy-Lugo, J. A., Thorwald, M. A., Mendez, D. A., Rodriguez, R., Nakano, D., Nishiyama, A., & Ortiz, R. M. (2022). Glucose Increases Hepatic Mitochondrial Antioxidant Enzyme Activities in Insulin Resistant Rats Following Chronic Angiotensin Receptor Blockade. International Journal of Molecular Sciences, 23(18), 10897. https://doi.org/10.3390/ijms231810897






