A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae
Abstract
1. Introduction
2. Results
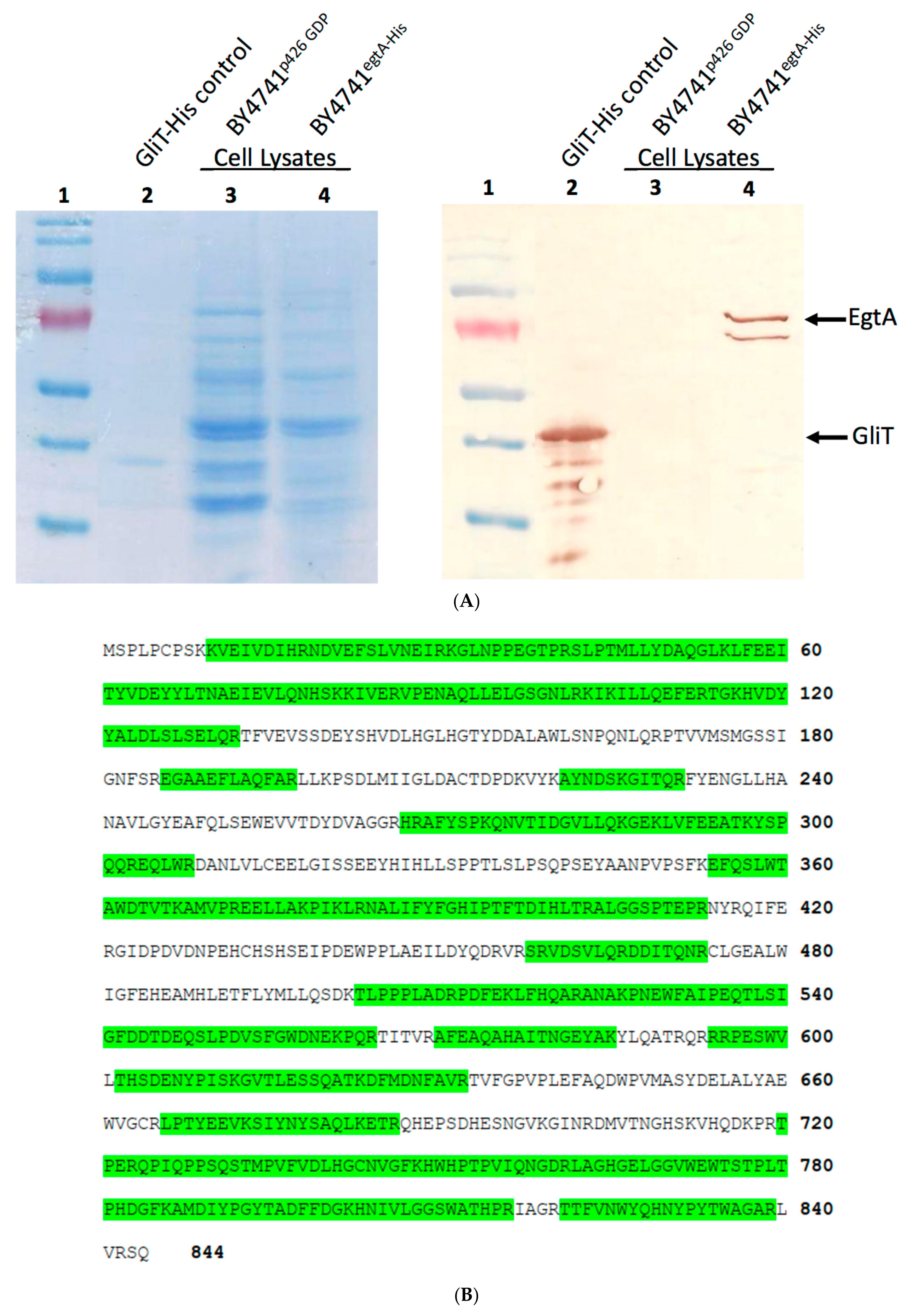
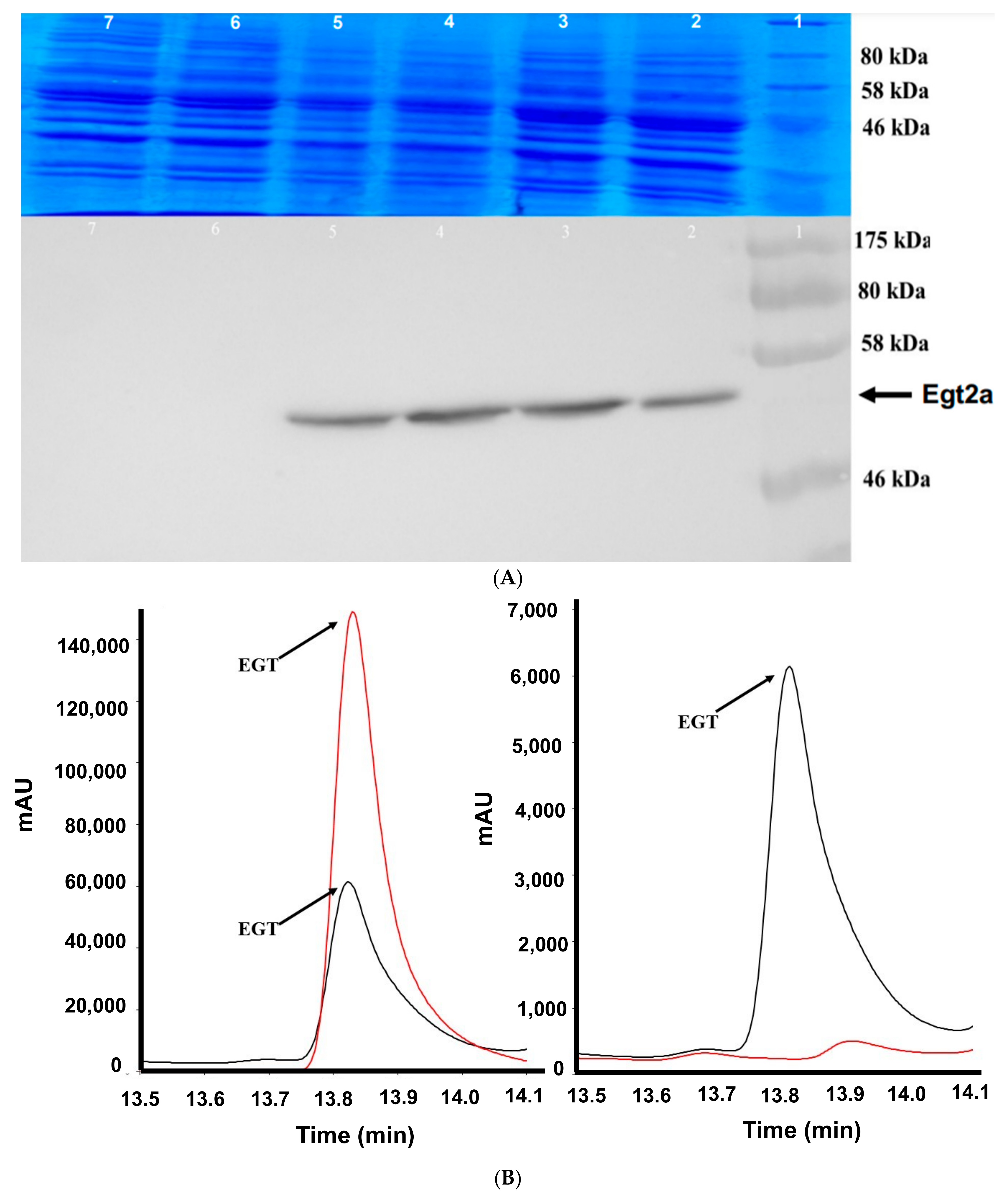
2.1. EgtA Expression in Saccharomyces Cerevisiae
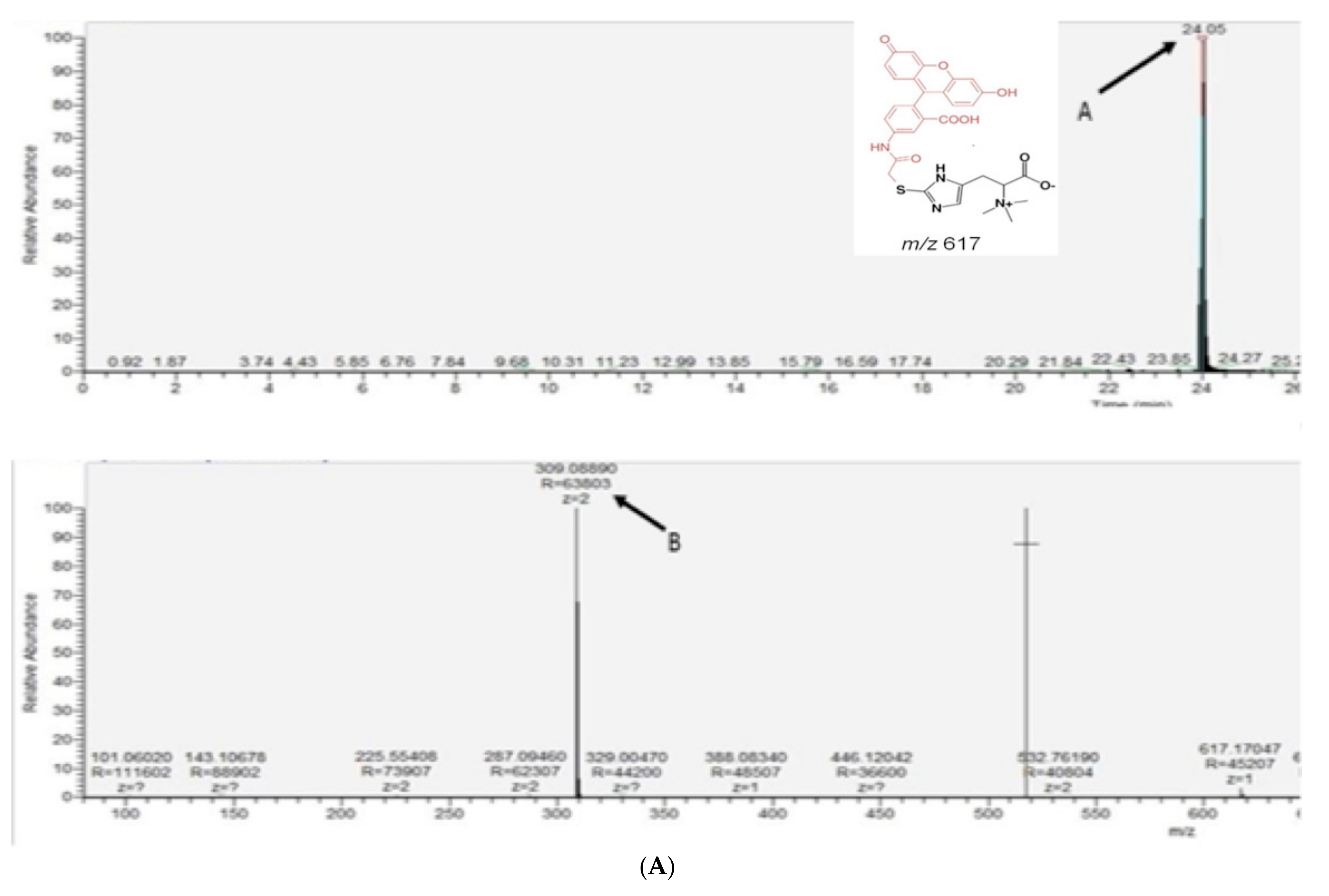
2.2. Intracellular Ergothioneine Production in Recombinant S. cerevisiae–EgtA Using LC–MS Analysis
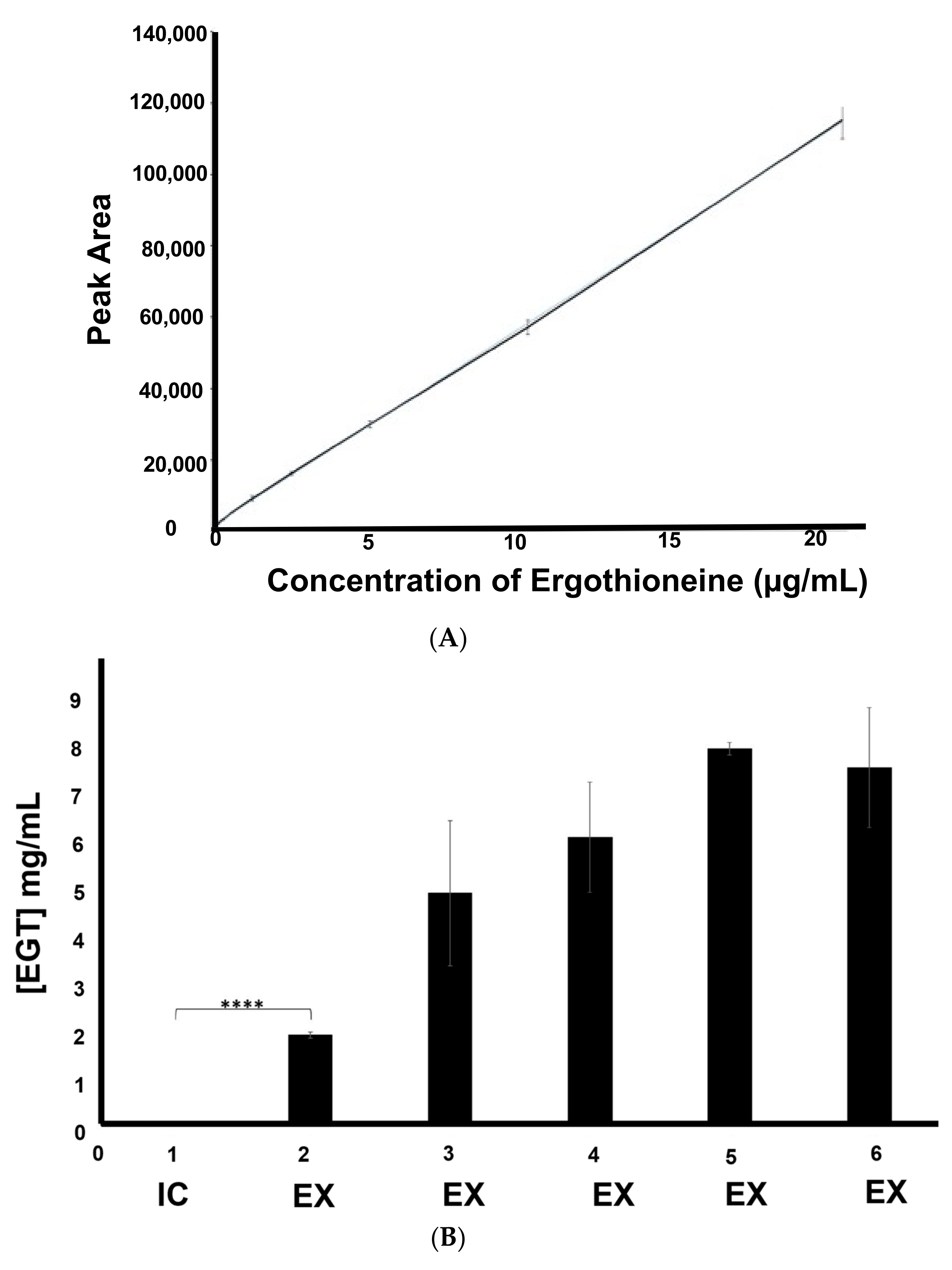
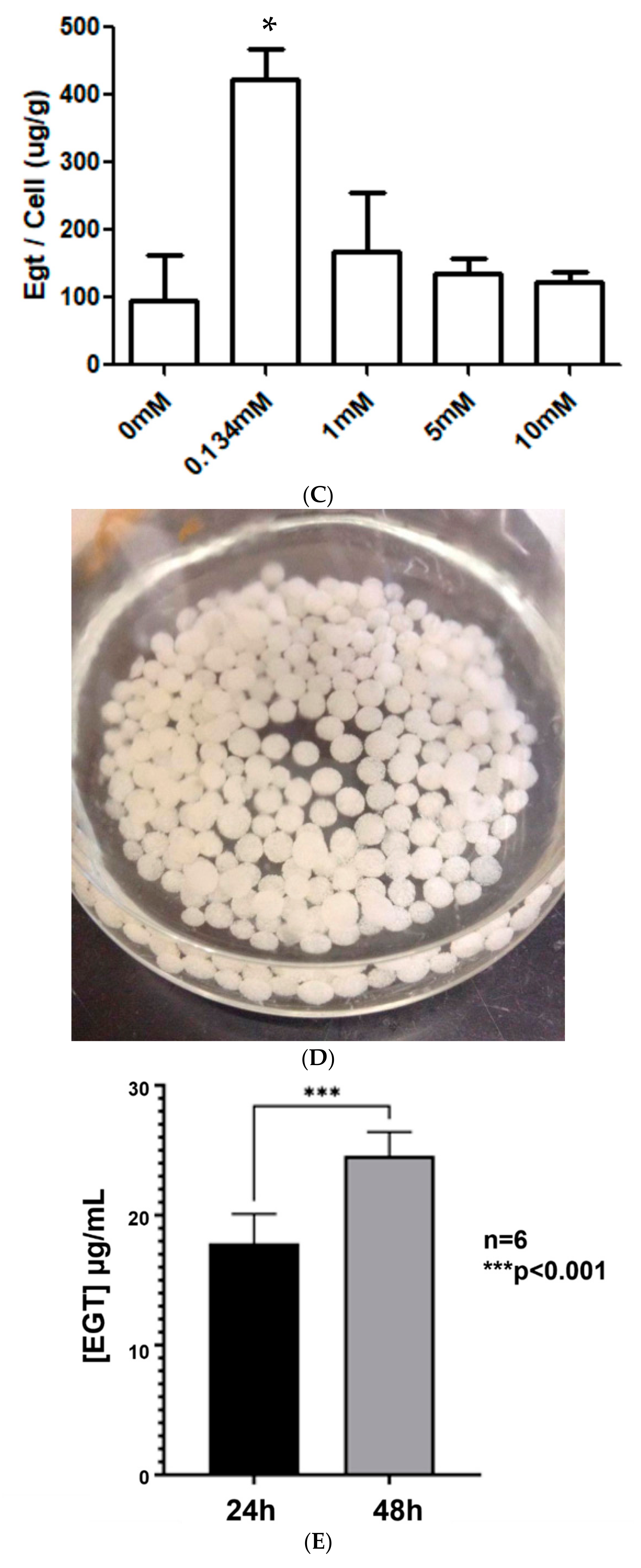
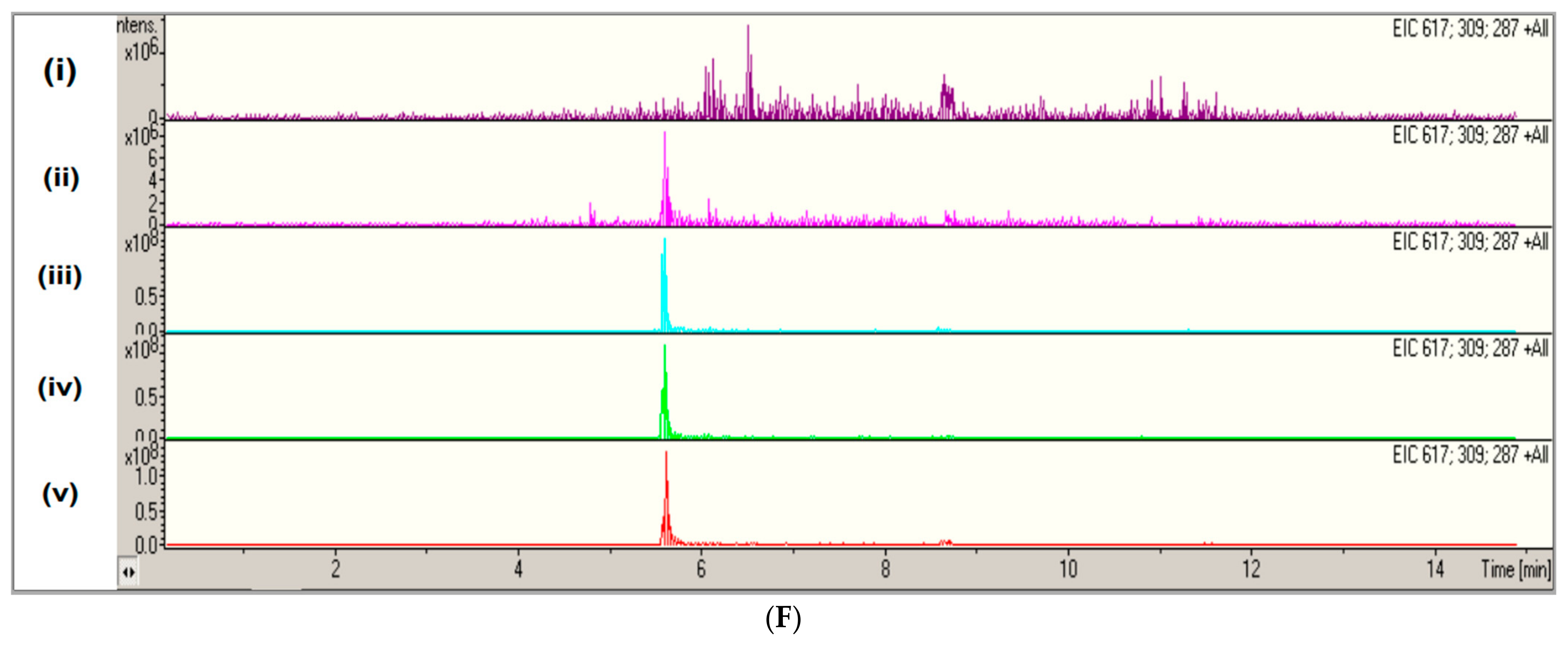
2.3. Extracellular Ergothioneine Production in Recombinant S. cerevisiae–EgtA Using LC–MS Analysis
2.4. Detection of Mainly Extracellular Ergothioneine from Recombinant S. cerevisiae–EgtA Using RP–HPLC Analysis
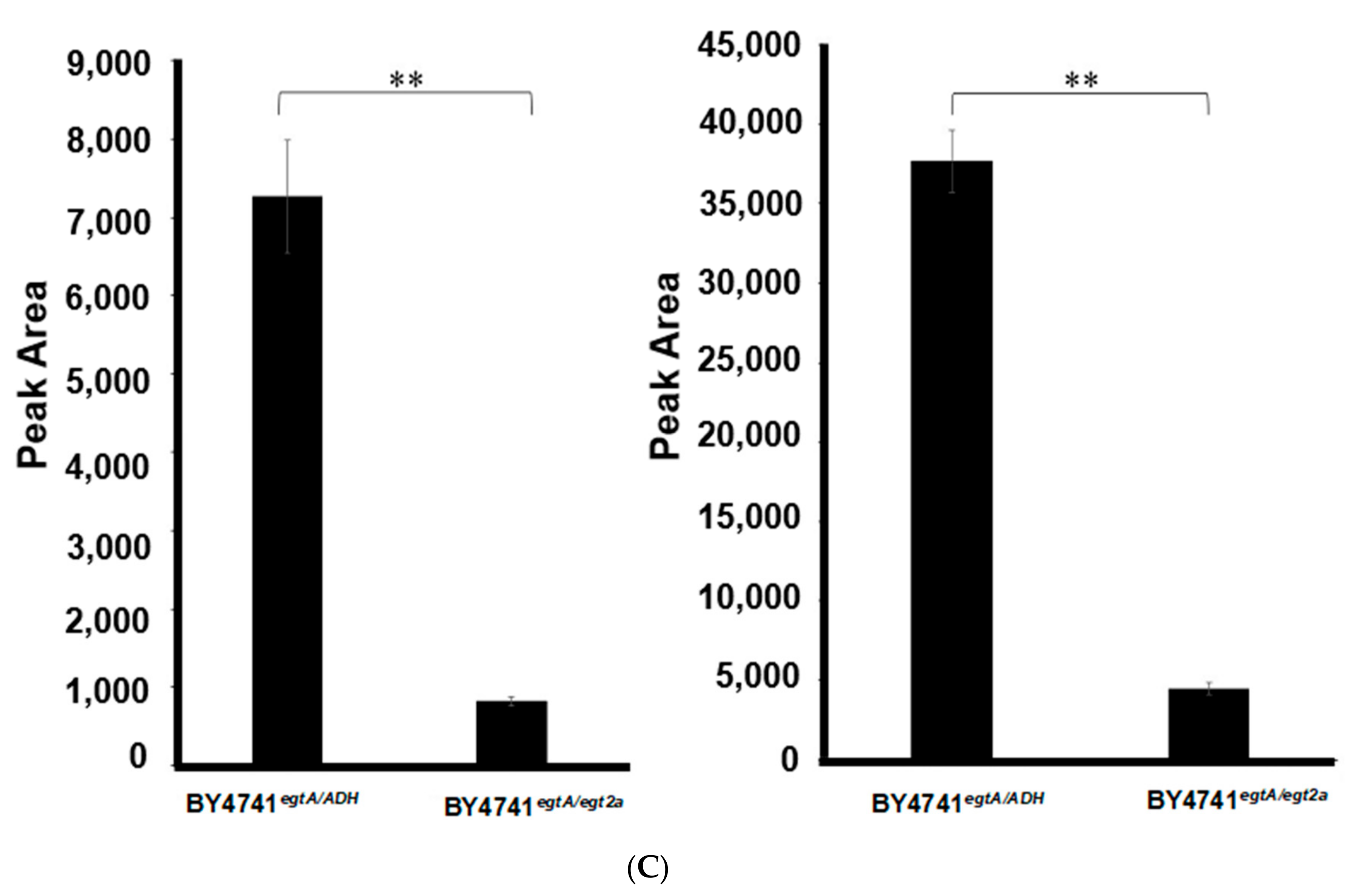
2.5. Co-Expression of egtA with the egtB Candidate AFUA_2G13295 Decreases the Level of EGT in S. cerevisiae
2.6. In Silico Modelling of A. fumigatus EgtA Structure Predicts a Didomain Structure with Opposed Active Sites
3. Discussion
4. Methods
4.1. Cloning and Expression of A. fumigatus egtA and Related Genes in S. cerevisiae
4.2. Protein and EGT Extraction from S. cerevisiae
4.3. Immunoblot Analysis of Yeast Cell Lysates
4.4. Alkylation of Intracellular and Extracellular EGT
4.5. RP–HPLC and LC–MS Analyses of Alkylated Ergothioneine
4.6. Yeast Culture and Immobilised Yeast (IY) Preparation in Alginate Beads
4.7. AlphaFold Prediction of Fungal EGT Biosynthetic Enzyme Structure
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seebeck, F.P. In vitro reconstitution of Mycobacterial ergothioneine biosynthesis. J. Am. Chem. Soc. 2010, 132, 6632–6633. [Google Scholar] [CrossRef] [PubMed]
- Sheridan, K.J.; Lechner, B.E.; Keeffe, G.O.; Keller, M.A.; Werner, E.R.; Lindner, H.; Jones, G.W.; Haas, H.; Doyle, S. Ergothioneine Biosynthesis and Functionality in the Opportunistic Fungal Pathogen, Aspergillus fumigatus. Sci. Rep. 2016, 6, 35306. [Google Scholar] [CrossRef] [PubMed]
- Bello, M.H.; Barrera-Perez, V.; Morin, D.; Epstein, L. The Neurospora crassa mutant NcDeltaEgt-1 identifies an ergothioneine biosynthetic gene and demonstrates that ergothioneine enhances conidial survival and protects against peroxide toxicity during conidial germination. Fungal Genet. Biol. 2012, 49, 160–172. [Google Scholar] [CrossRef] [PubMed]
- Cumming, B.M.; Chinta, K.C.; Reddy, V.P.; Steyn, A.J.C. Role of Ergothioneine in Microbial Physiology and Pathogenesis. Antioxid Redox Signal. 2018, 28, 431–444. [Google Scholar] [CrossRef] [PubMed]
- Gamage, A.M.; Liao, C.; Cheah, I.K.; Chen, Y.; Lim, D.R.X.; Ku, J.W.K.; Chee, R.S.L.; Gengenbacher, M.; Seebeck, F.P.; Halliwell, B.; et al. The proteobacterial species Burkholderia pseudomallei produces ergothioneine, which enhances virulence in mammalian infection. FASEB J. 2018, 32, 6395–6409. [Google Scholar] [CrossRef]
- Gallagher, L.; Owens, R.A.; Dolan, S.K.; O’Keeffe, G.; Schrettl, M.; Kavanagh, K.; Jones, G.W.; Doyle, S. The Aspergillus fumigatus protein gliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot. Cell 2012, 11, 1226–1238. [Google Scholar] [CrossRef]
- Saini, V.; Cumming, B.M.; Guidry, L.; Lamprecht, D.A.; Adamson, J.H.; Reddy, V.P.; Chinta, K.C.; Mazorodze, J.H.; Glasgow, J.N.; Richard-Greenblatt, M.; et al. Ergothioneine Maintains Redox and Bioenergetic Homeostasis Essential for Drug Susceptibility and Virulence of Mycobacterium tuberculosis. Cell Rep. 2016, 14, 572–585. [Google Scholar] [CrossRef]
- Traynor, A.M.; Sheridan, K.J.; Jones, G.W.; Calera, J.A.; Doyle, S. Involvement of Sulfur in the Biosynthesis of Essential Metabolites in Pathogenic Fungi of Animals, Particularly Aspergillus spp.: Molecular and Therapeutic Implications. Front. Microbiol. 2019, 10, 2859. [Google Scholar] [CrossRef]
- Peckelsen, K.; Martens, J.; Czympiel, L.; Oomens, J.; Berden, G.; Gründemann, D.; Meijer, A.; Schäfer, M. Ergothioneine and related histidine derivatives in the gas phase: Tautomer structures determined by IRMPD spectroscopy and theory. Phys. Chem. Chem. Phys. 2017, 19, 23362–23372. [Google Scholar] [CrossRef]
- Ames, B.N. Prolonging healthy aging: Longevity vitamins and proteins. Proc. Natl. Acad. Sci. USA 2018, 115, 10836–10844. [Google Scholar] [CrossRef]
- Nakamichi, N.; Kato, Y. Physiological Roles of Carnitine/Organic Cation Transporter OCTN1/SLC22A4 in Neural Cells. Biol. Pharm. Bull. 2017, 40, 1146–1152. [Google Scholar] [CrossRef] [PubMed]
- Halliwell, B.; Cheah, I.K.; Tang, R.M.Y. Ergothioneine—A diet-derived antioxidant with therapeutic potential. FEBS Lett. 2018, 592, 3357–3366. [Google Scholar] [CrossRef] [PubMed]
- Turner, E.; Brewster, J.A.; Simpson, N.A.; Walker, J.J.; Fisher, J. Imidazole-based erythrocyte markers of oxidative stress in preeclampsia--an NMR investigation. Reprod. Sci. 2009, 16, 1040–1051. [Google Scholar] [CrossRef]
- Kerley, R.N.; McCarthy, C.; Kell, D.B.; Kenny, L.C. The potential therapeutic effects of ergothioneine in pre-eclampsia. Free Radic. Biol. Med. 2018, 117, 145–157. [Google Scholar] [CrossRef] [PubMed]
- Williamson, R.D.; McCarthy, F.P.; Manna, S.; Groarke, E.; Kell, D.B.; Kenny, L.C.; McCarthy, C.M. L-(+)-Ergothioneine Significantly Improves the Clinical Characteristics of Preeclampsia in the Reduced Uterine Perfusion Pressure Rat Model. Hypertension 2020, 75, 561–568. [Google Scholar] [CrossRef] [PubMed]
- Morillon, A.C.; Williamson, R.D.; Baker, P.N.; Kell, D.B.; Kenny, L.C.; English, J.A.; McCarthy, F.P.; McCarthy, C. Effect of L-Ergothioneine on the metabolic plasma profile of the RUPP rat model of pre-eclampsia. PLoS ONE 2020, 15, e0230977. [Google Scholar] [CrossRef]
- Pluskal, T.; Ueno, M.; Yanagida, M. Genetic and metabolomic dissection of the ergothioneine and selenoneine biosynthetic pathway in the fission yeast, S. pombe, and construction of an overproduction system. PLoS ONE 2014, 9, e97774. [Google Scholar] [CrossRef]
- Jones, G.W.; Doyle, S.; Fitzpatrick, D.A. The evolutionary history of the genes involved in the biosynthesis of the antioxidant ergothioneine. Gene 2014, 549, 161–170. [Google Scholar] [CrossRef]
- Bello, M.H.; Mogannam, J.C.; Morin, D.; Epstein, L. Endogenous ergothioneine is required for wild type levels of conidiogenesis and conidial survival but does not protect against 254 nm UV-induced mutagenesis or kill. Fungal Genet. Biol. 2014, 73, 120–127. [Google Scholar] [CrossRef][Green Version]
- Burn, R.; Misson, L.; Meury, M.; Seebeck, F.P. Anaerobic Origin of Ergothioneine. Angew. Chem. Int. Ed. Engl. 2017, 56, 12508–12511. [Google Scholar] [CrossRef]
- Leisinger, F.; Burn, R.; Meury, M.; Lukat, P.; Seebeck, F.P. Structural and Mechanistic Basis for Anaerobic Ergothioneine Biosynthesis. J. Am. Chem. Soc. 2019, 141, 6906–6914. [Google Scholar] [CrossRef] [PubMed]
- Stampfli, A.R.; Seebeck, F.P. The catalytic mechanism of sulfoxide synthases. Curr. Opin. Chem. Biol. 2020, 59, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Stampfli, A.R.; Blankenfeldt, W.; Seebeck, F.P. Structural basis of ergothioneine biosynthesis. Curr. Opin. Struct. Biol. 2020, 65, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Kalaras, M.D.; Richie, J.P.; Calcagnotto, A.; Beelman, R.B. Mushrooms: A rich source of the antioxidants ergothioneine and glutathione. Food Chem. 2017, 233, 429–433. [Google Scholar] [CrossRef]
- Bao, H.N.; Osako, K.; Ohshima, T. Value-added use of mushroom ergothioneine as a colour stabilizer in processed fish meats. J. Sci. Food Agric. 2010, 90, 1634–1641. [Google Scholar] [CrossRef]
- Feeney, M.J.; Dwyer, J.; Hasler-Lewis, C.M.; Milner, J.A.; Noakes, M.; Rowe, S.; Wach, M.; Beelman, R.B.; Caldwell, J.; Cantorna, M.T.; et al. Mushrooms and Health Summit proceedings. J. Nutr. 2014, 144, 1128s–1136s. [Google Scholar] [CrossRef]
- Fujitani, Y.; Alamgir, K.M.; Tani, A. Ergothioneine production using Methylobacterium species, yeast, and fungi. J. Biosci. Bioeng. 2018, 126, 715–722. [Google Scholar] [CrossRef]
- Tanaka, N.; Kawano, Y.; Satoh, Y.; Dairi, T.; Ohtsu, I. Gram-scale fermentative production of ergothioneine driven by overproduction of cysteine in Escherichia coli. Sci. Rep. 2019, 9, 1895. [Google Scholar] [CrossRef]
- van der Hoek, S.A.; Darbani, B.; Zugaj, K.E.; Prabhala, B.K.; Biron, M.B.; Randelovic, M.; Medina, J.B.; Kell, D.B.; Borodina, I. Engineering the Yeast Saccharomyces cerevisiae for the Production of L-(+)-Ergothioneine. Front. Bioeng. Biotechnol. 2019, 7, 262. [Google Scholar] [CrossRef]
- Osawa, R.; Kamide, T.; Satoh, Y.; Kawano, Y.; Ohtsu, I.; Dairi, T. Heterologous and High Production of Ergothioneine in Escherichia coli. J. Agric. Food Chem. 2018, 66, 1191–1196. [Google Scholar] [CrossRef]
- Takusagawa, S.; Satoh, Y.; Ohtsu, I.; Dairi, T. Ergothioneine production with Aspergillus oryzae. Biosci. Biotechnol. Biochem. 2019, 83, 181–184. [Google Scholar] [CrossRef] [PubMed]
- Kamide, T.; Takusagawa, S.; Tanaka, N.; Ogasawara, Y.; Kawano, Y.; Ohtsu, I.; Satoh, Y.; Dairi, T. High Production of Ergothioneine in Escherichia coli using the Sulfoxide Synthase from Methylobacterium strains. J. Agric. Food Chem. 2020, 68, 6390–6394. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; He, Y.; Wu, X.; Wang, L.; Dong, Z.; Chen, X. Toward more efficient ergothioneine production using the fungal ergothioneine biosynthetic pathway. Microb. Cell Fact. 2022, 21, 76. [Google Scholar] [CrossRef]
- Kim, M.; Jeong, D.W.; Oh, J.W.; Jeong, H.J.; Ko, Y.J.; Park, S.E.; Han, S.O. Efficient Synthesis of Food-Derived Antioxidant l-Ergothioneine by Engineered Corynebacterium glutamicum. J. Agric. Food Chem. 2022, 70, 1516–1524. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef] [PubMed]
- Sotgia, S.; Pisanu, E.; Pintus, G.; Erre, G.L.; Pinna, G.A.; Deiana, L.; Carru, C.; Zinellu, A. Plasma L-ergothioneine measurement by high-performance liquid chromatography and capillary electrophoresis after a pre-column derivatization with 5-iodoacetamidofluorescein (5-IAF) and fluorescence detection. PLoS ONE 2013, 8, e70374. [Google Scholar] [CrossRef]
- Yu, Y.H.; Pan, H.Y.; Guo, L.Q.; Lin, J.F.; Liao, H.L.; Li, H.Y. Successful biosynthesis of natural antioxidant ergothioneine in Saccharomyces cerevisiae required only two genes from Grifola frondosa. Microb. Cell Fact. 2020, 19, 164. [Google Scholar] [CrossRef]
- van der Hoek, S.A.; Rusnák, M.; Wang, G.; Stanchev, L.D.; de Fátima Alves, L.; Jessop-Fabre, M.M.; Paramasivan, K.; Jacobsen, I.H.; Sonnenschein, N.; Martínez, J.L.; et al. Engineering precursor supply for the high-level production of ergothioneine in Saccharomyces cerevisiae. Metab. Eng. 2022, 70, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Mumberg, D.; Müller, R.; Funk, M. Yeast vectors for the controlled expression of heterologous proteins in different genetic backgrounds. Gene 1995, 156, 119–122. [Google Scholar] [CrossRef]
- Sikorski, R.S.; Hieter, P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 1989, 122, 19–27. [Google Scholar] [CrossRef]
- Schwimmer, C.; Masison, D.C. Antagonistic interactions between yeast [PSI(+)] and [URE3] prions and curing of [URE3] by Hsp70 protein chaperone Ssa1p but not by Ssa2p. Mol. Cell. Biol. 2002, 22, 3590–3598. [Google Scholar] [CrossRef] [PubMed]
- Dolan, S.K.; Bock, T.; Hering, V.; Owens, R.A.; Jones, G.W.; Blankenfeldt, W.; Doyle, S. Structural, mechanistic and functional insight into gliotoxin bis-thiomethylation in Aspergillus fumigatus. Open Biol. 2017, 7, 160292. [Google Scholar] [CrossRef] [PubMed]
- Traynor, A.M.; Owens, R.A.; Coughlin, C.M.; Holton, M.C.; Jones, G.W.; Calera, J.A.; Doyle, S. At the metal-metabolite interface in Aspergillus fumigatus: Towards untangling the intersecting roles of zinc and gliotoxin. Microbiology 2021, 167, 001106. [Google Scholar] [CrossRef] [PubMed]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Jeong, J.H.; Cha, H.J.; Ha, S.C.; Rojviriya, C.; Kim, Y.G. Structural insights into the histidine trimethylation activity of EgtD from Mycobacterium smegmatis. Biochem. Biophys. Res. Commun. 2014, 452, 1098–1103. [Google Scholar] [CrossRef] [PubMed]
- Vit, A.; Misson, L.; Blankenfeldt, W.; Seebeck, F.P. Crystallization and preliminary X-ray analysis of the ergothioneine-biosynthetic methyltransferase EgtD. Acta Cryst. F Struct. Biol. Commun. 2014, 70, 676–680. [Google Scholar] [CrossRef]
- Vit, A.; Misson, L.; Blankenfeldt, W.; Seebeck, F.P. Ergothioneine biosynthetic methyltransferase EgtD reveals the structural basis of aromatic amino acid betaine biosynthesis. Chembiochem 2015, 16, 119–125. [Google Scholar] [CrossRef]
- Naowarojna, N.; Irani, S.; Hu, W.; Cheng, R.; Zhang, L.; Li, X.; Chen, J.; Zhang, Y.J.; Liu, P. Crystal Structure of the Ergothioneine Sulfoxide Synthase from Candidatus Chloracidobacterium thermophilum and Structure-Guided Engineering to Modulate Its Substrate Selectivity. ACS Catal. 2019, 9, 6955–6961. [Google Scholar] [CrossRef]
- Goncharenko, K.V.; Vit, A.; Blankenfeldt, W.; Seebeck, F.P. Structure of the sulfoxide synthase EgtB from the ergothioneine biosynthetic pathway. Angew. Chem. Int. Ed. Engl. 2015, 54, 2821–2824. [Google Scholar] [CrossRef]
- Zhang, Y.; Skolnick, J. TM-align: A protein structure alignment algorithm based on the TM-score. Nucleic Acids Res. 2005, 33, 2302–2309. [Google Scholar] [CrossRef]



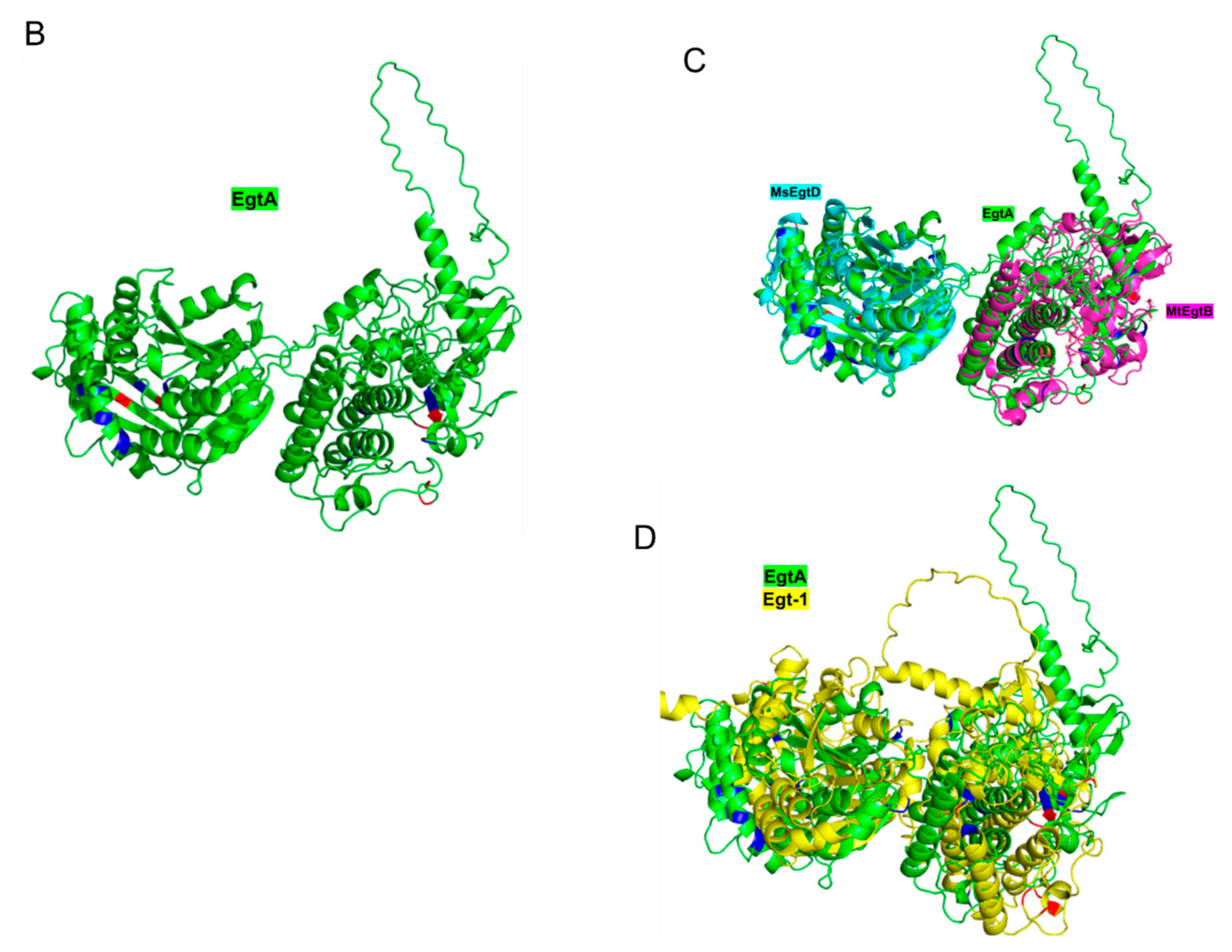
| Comparison | TM-score | SI% | SS% | Length |
|---|---|---|---|---|
| EgtA vs. Egt-1 | 0.47 | 29 | 41 | 787 |
| EgtA vs. MsEgtD | 0.84 | 25 | 46 | 315 |
| EgtA vs. MtEgtB | 0.67 | 24 | 35 | 388 |
| Egt-1 vs. MsEgtD | 0.8 | 25 | 44 | 312 |
| Egt-1 vs. MtEgtB | 0.66 | 23 | 37 | 380 |
| Strain | Genotype | Source |
|---|---|---|
| BY4741 | MATa his3Δ 1; leu2Δ; met15Δ 0; ura3Δ 0 | Euroscarf |
| R1158 | MATa his3Δ 1; leu2Δ; met15Δ; pNFS1::kanR-tet07-TATA; ura3::CMV-tTA | Dharmacon |
| R1158 | MATa his3Δ 1; leu2Δ; met15Δ; pSAH1::kanR-tet07-TATA; ura3::CMV-tTA | Dharmacon |
| Y258 | MATa pep4-3; his4-580; ura3-52; leu2-3 | Dharmacon |
| Top 10 | E. coli | Thermofisher |
| Xli-blue | E. coli | Thermofisher |
| Plasmid Name | Description | Source |
|---|---|---|
| pUC57 | E. coli expression vector | GenScript |
| pRS316 | Centromeric Saccharomyces cerevisiae shuttle vector—URA3 marker | Sikorski and Hieter [40] |
| pC210 | SSA1 under control of SSA2 promoter—LEU2 marker | Schwimmer and Masison [41] |
| p413-ADH | Low-copy centromeric plasmid with an ADH promoter based on the pRS vector series—HIS3 marker | Mumberg et al. [39] |
| p413-GPD | Low-copy centromeric plasmid with a GPD promoter based on the pRS vector series—HIS3 marker | Mumberg et al. [39] |
| p416-ADH | Low-copy centromeric plasmid with an ADH promoter based on the pRS vector series—URA3 marker | Mumberg et al. [39] |
| p416-GPD | Low-copy centromeric plasmid with a GPD promoter based on the pRS vector series—URA3 marker | Mumberg et al. [39] |
| p423-ADH | High-copy 2µ plasmid with an ADH promoter based on the pRS vector series—HIS3 marker | Mumberg et al. [39] |
| p423-GPD | High-copy 2µ plasmid with a GPD promoter based on the pRS vector series—HIS3 marker | Mumberg et al. [39] |
| p426-ADH | High-copy 2µ plasmid with an ADH promoter based on the pRS vector series—URA3 marker | Mumberg et al. [39] |
| p426-GPD | High-copy 2µ plasmid with a GPD promoter based on the pRS vector series—URA3 marker | Mumberg et al. [39] |
| p426-GPD-egtA | Aspergillus fumigatus egtA under control of GDP promoter—URA3 marker | This study |
| p423-ADH-egt2a | Aspergillus fumigatus egt2a under control of ADH promoter—HIS3 marker | This study |
| p423-ADH-egt2b | Aspergillus fumigatus egt2b under control of ADH promoter—HIS3 marker | This study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Doyle, S.; Cuskelly, D.D.; Conlon, N.; Fitzpatrick, D.A.; Gilmartin, C.B.; Dix, S.H.; Jones, G.W. A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae. Int. J. Mol. Sci. 2022, 23, 10832. https://doi.org/10.3390/ijms231810832
Doyle S, Cuskelly DD, Conlon N, Fitzpatrick DA, Gilmartin CB, Dix SH, Jones GW. A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae. International Journal of Molecular Sciences. 2022; 23(18):10832. https://doi.org/10.3390/ijms231810832
Chicago/Turabian StyleDoyle, Sean, Daragh D. Cuskelly, Niall Conlon, David A. Fitzpatrick, Ciara B. Gilmartin, Sophia H. Dix, and Gary W. Jones. 2022. "A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae" International Journal of Molecular Sciences 23, no. 18: 10832. https://doi.org/10.3390/ijms231810832
APA StyleDoyle, S., Cuskelly, D. D., Conlon, N., Fitzpatrick, D. A., Gilmartin, C. B., Dix, S. H., & Jones, G. W. (2022). A Single Aspergillus fumigatus Gene Enables Ergothioneine Biosynthesis and Secretion by Saccharomyces cerevisiae. International Journal of Molecular Sciences, 23(18), 10832. https://doi.org/10.3390/ijms231810832






