Effects of Sulforaphane-Induced Cell Death upon Repeated Passage of Either P-Glycoprotein-Negative or P-Glycoprotein-Positive L1210 Cell Variants
Abstract
1. Introduction
2. Results
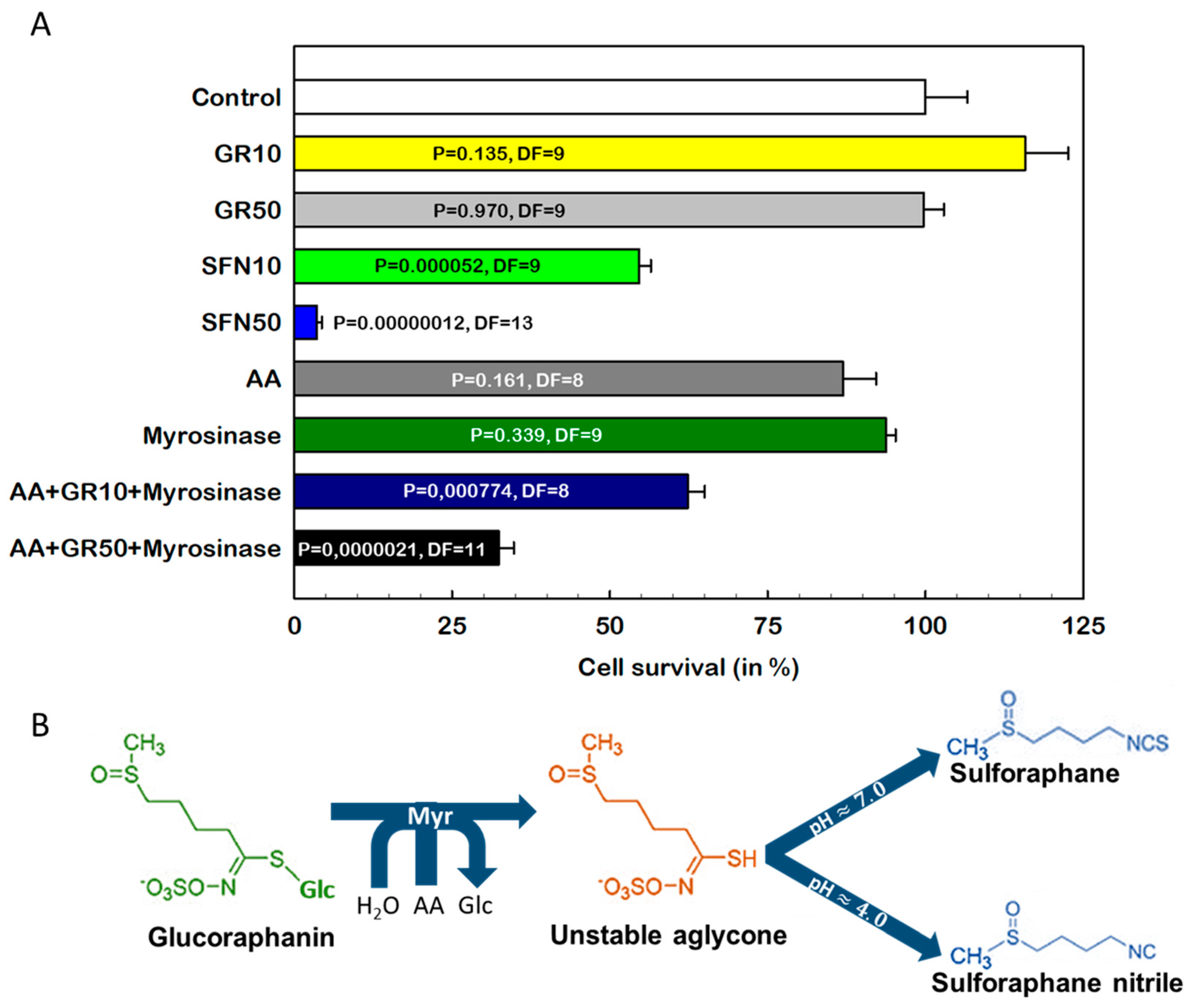
2.1. Comparison of the Effects of SFN and GR on Cell Death
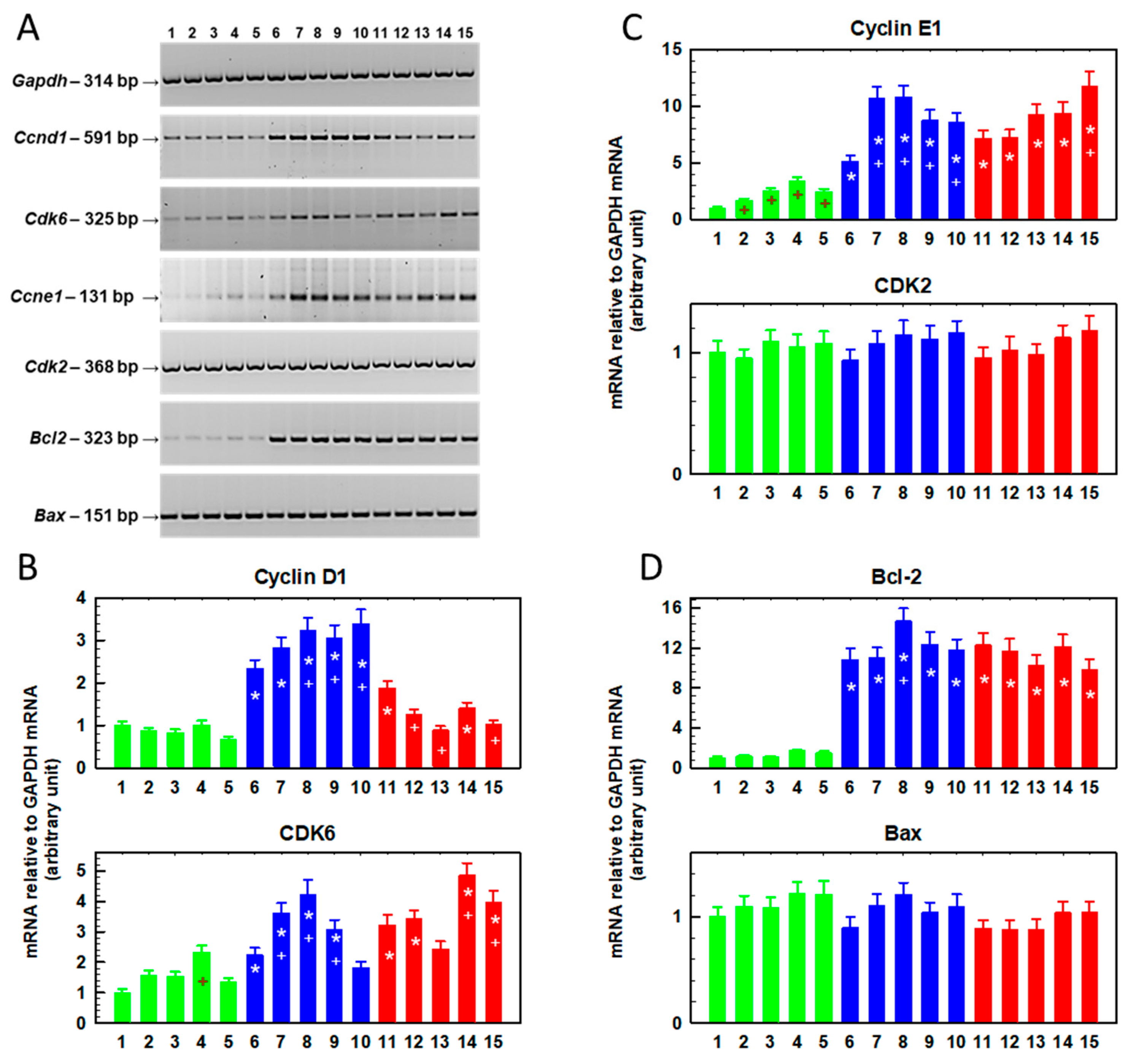
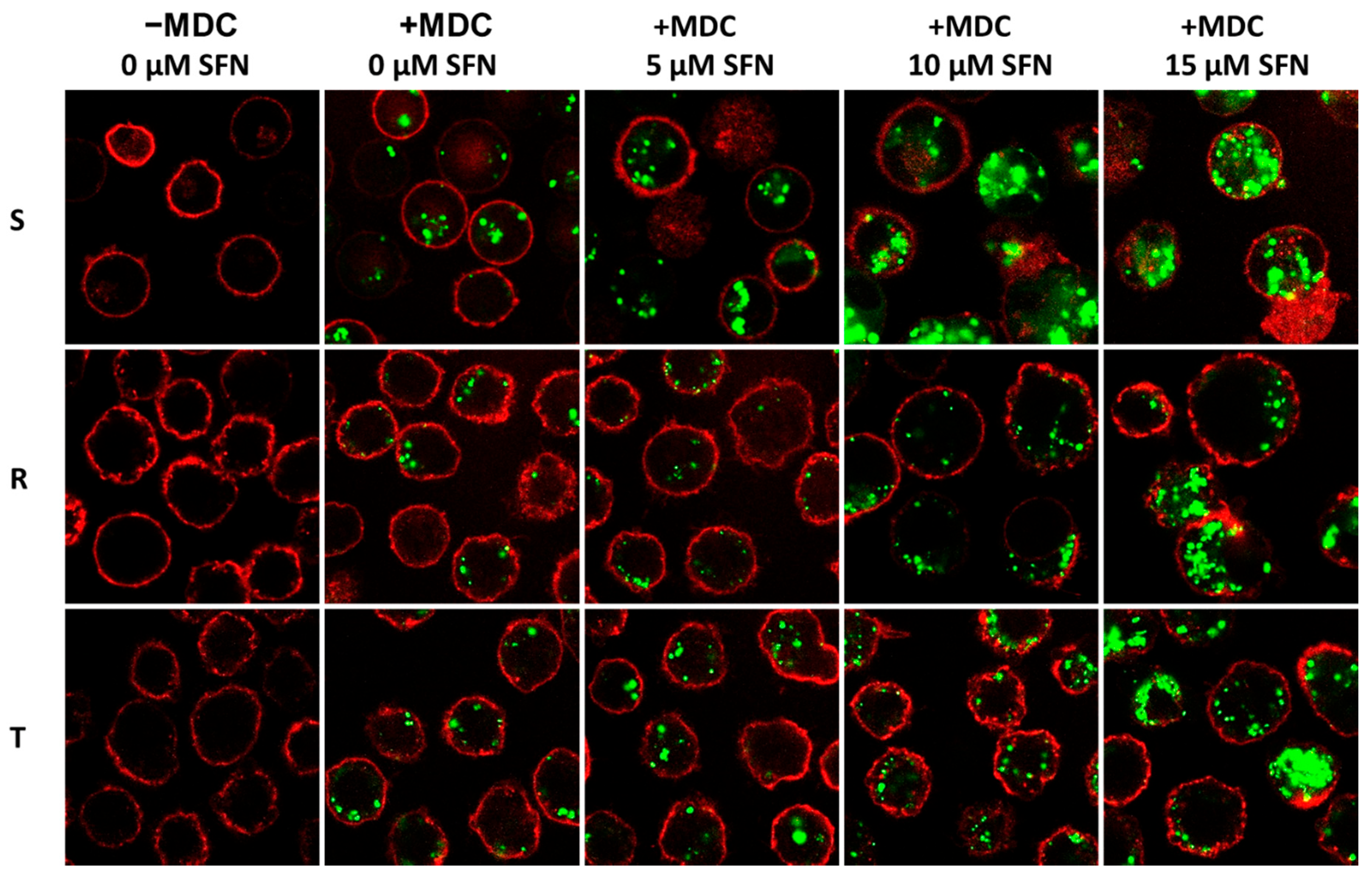
2.2. Cell Proliferation and Specific Gene Expression in S, R and T Cells Pretreated with SFN
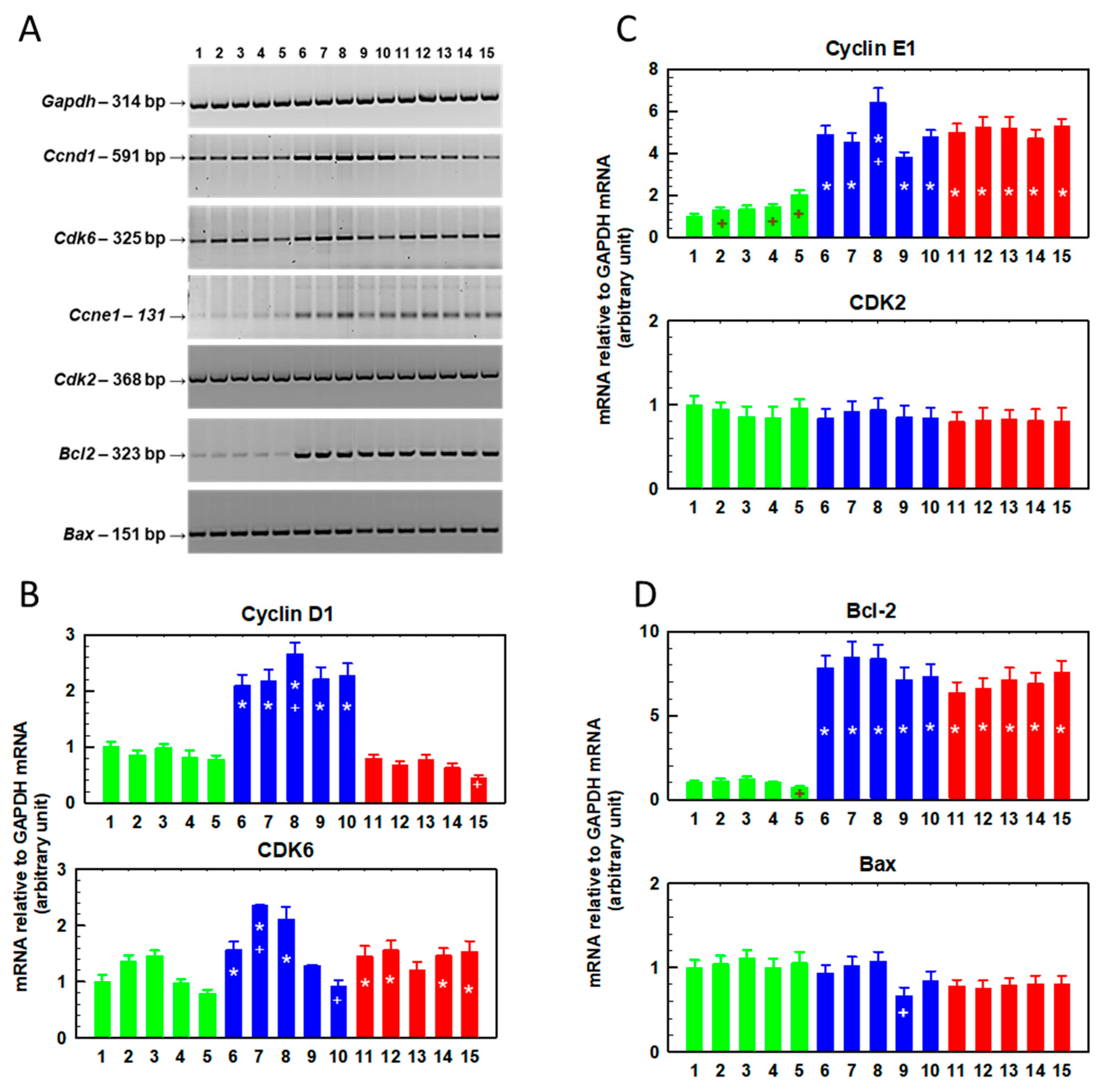
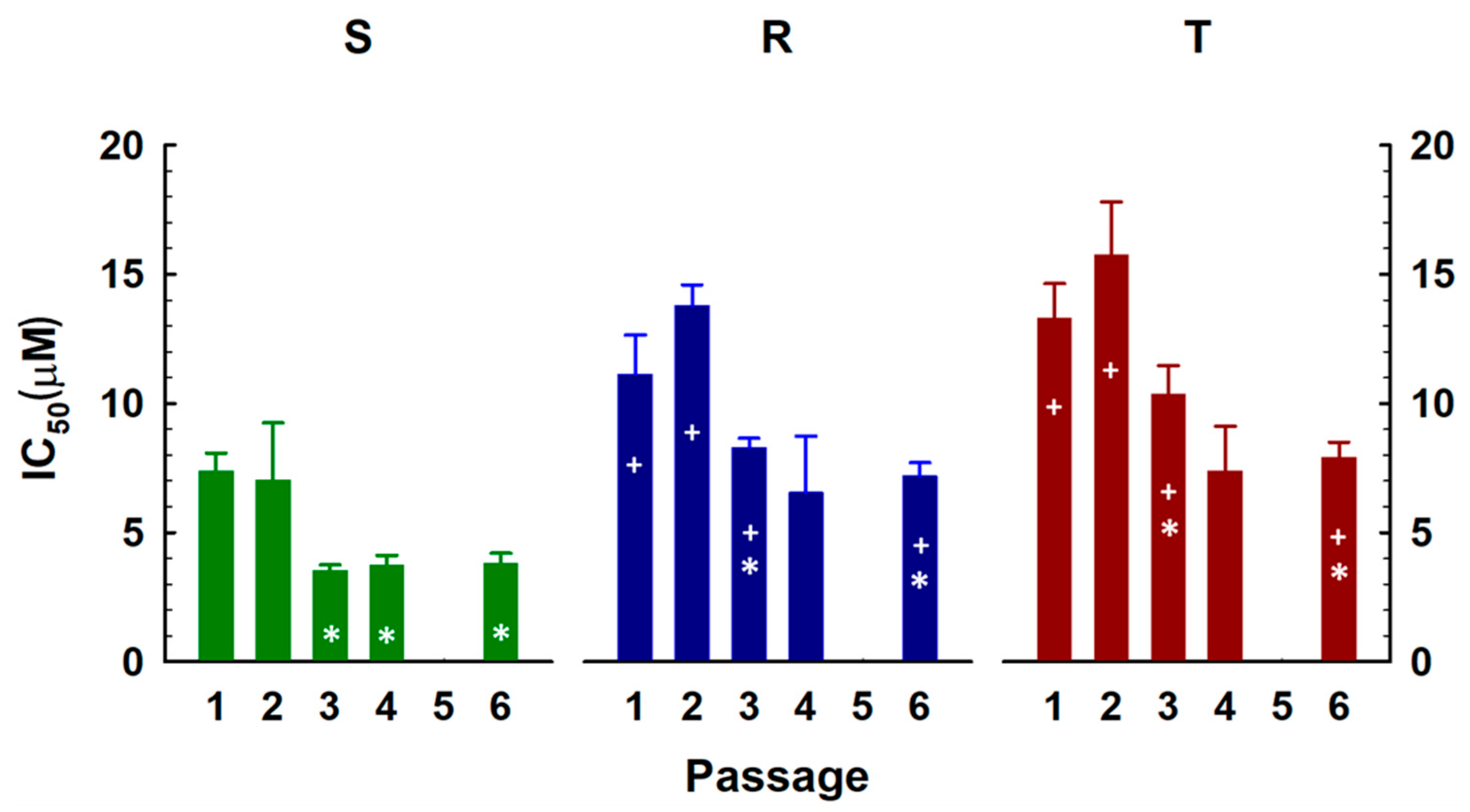
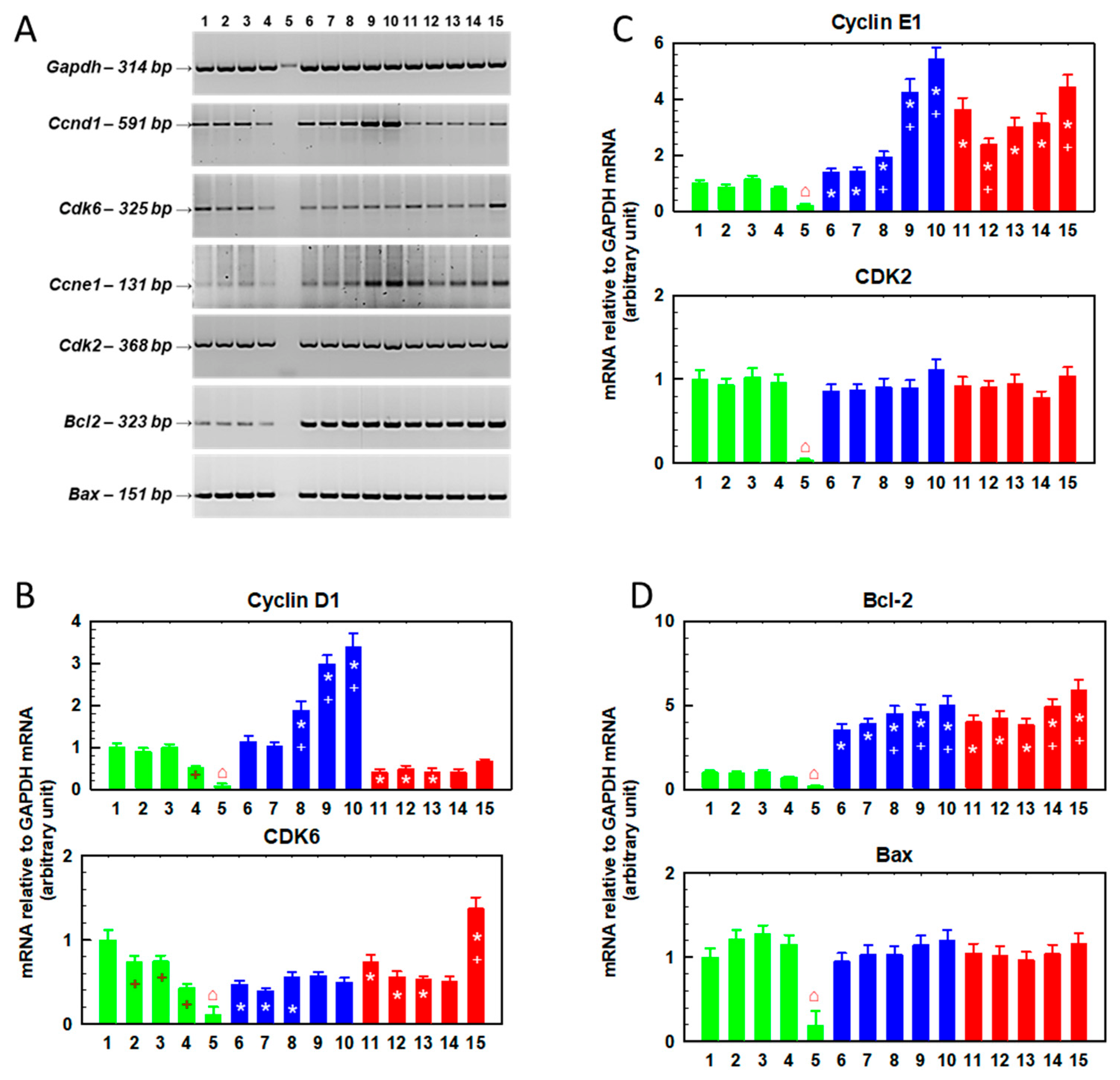
2.3. Effects of Repeated Cultivation of S, R and T Cells in SFN-Containing Medium on Cell Growth and Gene Expression
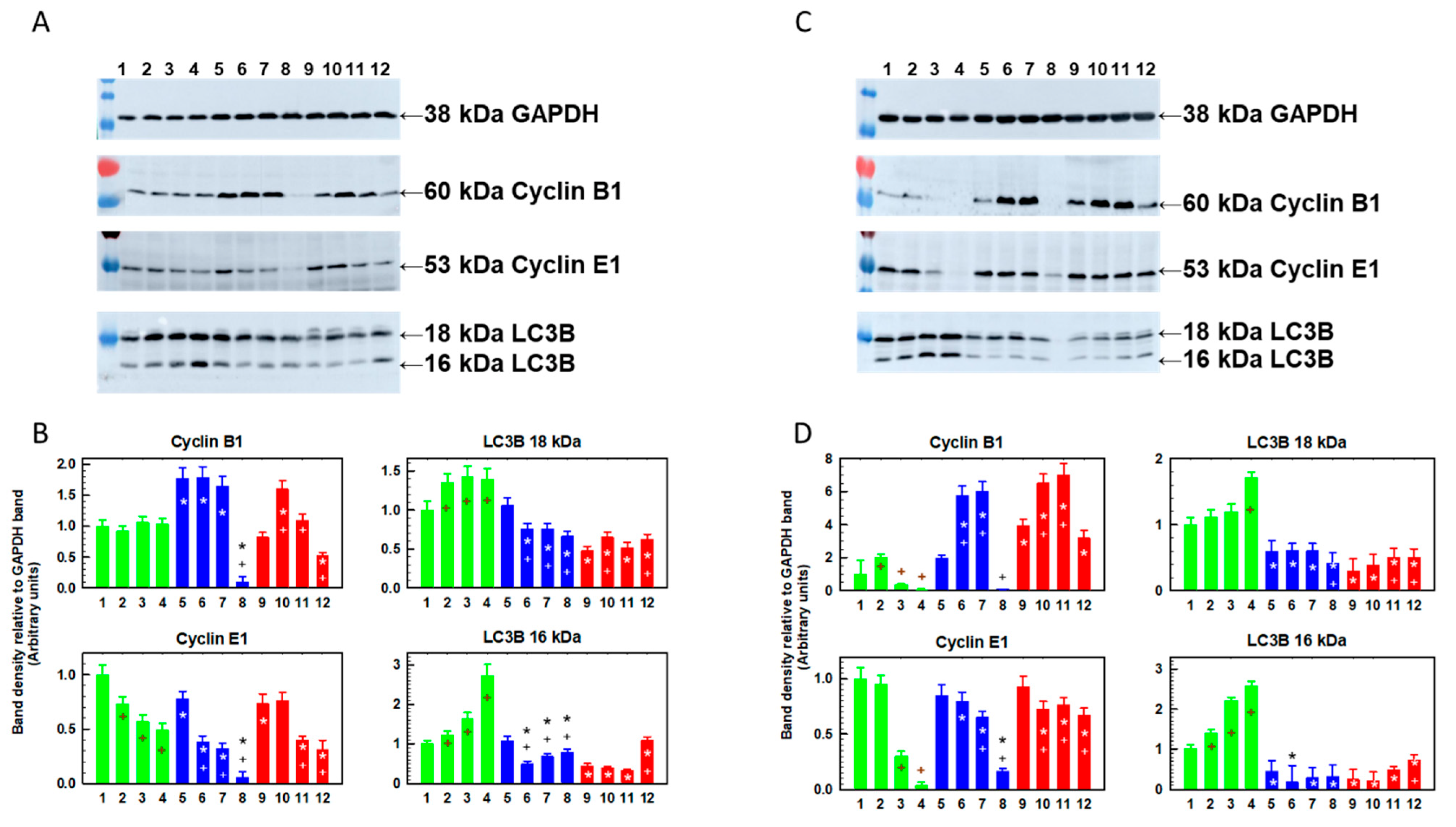
2.4. Changes in the Cellular Content of Cyclin B, Cyclin E and LC3B Protein Fragments Induced by Repeated Passaging in SFN-Containing Medium
3. Discussion
4. Materials and Methods
4.1. Chemicals
4.2. Cell Culture and Culture Conditions
4.3. Cell Passage in the Presence or Absence of SFN
4.4. Determination of IC50 Values for S, R and T Cells after Each Passage in the Presence of SFN
4.5. Cell Metabolic Activity Estimation Using the MTT Assay
4.6. Estimation of Gene Expression by RT-PCR
4.7. Western Blot Detection of Cyclin B1, E1 and LC3B Protein Fragments
4.8. Visualization of Autophagic Vacuoles by MDC
4.9. Statistical Analysis and Data Processing
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviation
| AA | ascorbic acid |
| ABCB1 | ATP Binding Cassette Subfamily B Member 1 |
| ABCC1 | ATP Binding Cassette Subfamily C Member 1 |
| ABCG2 | ATP Binding Cassette Subfamily G Member 2 |
| AITC | allyl isothiocyanate |
| Bax | Bcl-2-associated X protein |
| Bcl-2 | B-cell lymphoma-2 protein |
| Ccnb1 | gene for Cyclin B1 |
| Ccnd1 | gene for Cyclin D1 |
| Ccne1 | gene for Cyclin E1 |
| Cdk2 | cyclin dependent kinase 2 |
| Cdk6 | cyclin dependent kinase 6 |
| DMSO | dimethylsulfoxide |
| FBS | fetal bovine serum |
| GAPDH | glyceraldehyde-3-phosphate dehydrogenase |
| GR | glucoraphanin |
| GRP78/BiP | Endoplasmic reticulum chaperone (glucose reactive protein 78, also known as binding immunoglobulin protein) |
| HRP | horseradish peroxidase |
| ITC | isothiocyanate |
| LC3B | microtubule-associated protein 1 light chain 3 beta |
| MDC | monodansylcadaverine |
| MDR | multidrug resistance |
| MTT | 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide |
| Myr | myrosinase |
| NRF2 | nuclear factor erythroid 2-related factor 2 |
| PBS | phosphate buffered saline |
| R | drug-resistant ABCB1-positive L1210 cells obtained by selection with vincristine |
| S | drug-sensitive ABCB1-negative L1210 cells |
| SD | standard deviation |
| SDS | sodium dodecyl sulfate |
| SDS-PAGE | sodium dodecyl sulfate-polyacrylamide gel electrophoresis |
| SFN | sulforaphane |
| T | drug-resistant ABCB1-positive L1210 cells obtained by transfection with human ABCB1 gene |
| TQR | tariquidar |
| WGA | wheat-germ agglutinin |
| β-STAT3 | phosphorylated signal transducer and activator of transcription 3 |
References
- Cancer, Key Fact. Published on 3 February 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 1 May 2022).
- Wang, X.; Zhang, H.; Chen, X. Drug resistance and combating drug resistance in cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef] [PubMed]
- Pavlikova, L.; Seres, M.; Breier, A.; Sulova, Z. The roles of micrornas in cancer multidrug resistance. Cancers 2022, 14, 1090. [Google Scholar] [CrossRef] [PubMed]
- Deng, F.; Ghemtio, L.; Grazhdankin, E.; Wipf, P.; Xhaard, H.; Kidron, H. Binding site interactions of modulators of breast cancer resistance protein, multidrug resistance-associated protein 2, and p-glycoprotein activity. Mol. Pharm. 2020, 17, 2398–2410. [Google Scholar] [CrossRef] [PubMed]
- Gottesman, M.M.; Fojo, T.; Bates, S.E. Multidrug resistance in cancer: Role of atp-dependent transporters. Nature reviews. Cancer 2002, 2, 48–58. [Google Scholar] [PubMed]
- Breier, A.; Gibalova, L.; Seres, M.; Barancik, M.; Sulova, Z. New insight into p-glycoprotein as a drug target. Anticancer Agents Med. Chem. 2013, 13, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Mitsiogianni, M.; Koutsidis, G.; Mavroudis, N.; Trafalis, D.T.; Botaitis, S.; Franco, R.; Zoumpourlis, V.; Amery, T.; Galanis, A.; Pappa, A.; et al. The role of isothiocyanates as cancer chemo-preventive, chemo-therapeutic and anti-melanoma agents. Antioxidants 2019, 8, 106. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Bao, Y. Nanodelivery of natural isothiocyanates as a cancer therapeutic. Free Radic. Biol. Med. 2021, 167, 125–140. [Google Scholar] [CrossRef]
- Horakova, K.; Drobnica, L.; Nemec, P.; Antos, A.; Kristian, P. Cytotoxic and cancerostatis activity of isothiocyanates and related compounds. I. Activity of some naturally occurring isothiocyanates and their synthetic analogues on hela-cells. Neoplasma 1968, 15, 169–176. [Google Scholar]
- Bai, Y.; Wang, X.; Zhao, S.; Ma, C.; Cui, J.; Zheng, Y. Sulforaphane protects against cardiovascular disease via nrf2 activation. Oxid. Med. Cell Longev. 2015, 2015, 407580. [Google Scholar] [CrossRef]
- Esteve, M. Mechanisms underlying biological effects of cruciferous glucosinolate-derived isothiocyanates/indoles: A focus on metabolic syndrome. Front. Nutr. 2020, 7, 111. [Google Scholar] [CrossRef]
- Fimognari, C.; Turrini, E.; Ferruzzi, L.; Lenzi, M.; Hrelia, P. Natural isothiocyanates: Genotoxic potential versus chemoprevention. Mutat. Res. 2012, 750, 107–131. [Google Scholar] [CrossRef]
- Yeger, H.; Mokhtari, R.B. Perspective on dietary isothiocyanates in the prevention, development and treatment of cancer. J. Cancer Metastasis Treat. 2020, 6, 26. [Google Scholar] [CrossRef]
- Drobnica, Ľ.; Kristián, P.; Augustín, J. The chemistry of the—Ncs group. In Cyanates and Their Thio Derivatives (1977); John Wiley & Sons: Chichester, UK, 1977; pp. 1003–1221. [Google Scholar]
- Miyoshi, N. Chemical alterations and regulations of biomolecules in lifestyle-related diseases. Biosci. Biotechnol. Biochem. 2016, 80, 1046–1053. [Google Scholar] [CrossRef]
- de Figueiredo, S.M.; Filho, S.A.; Nogueira-Machado, J.A.; Caligiorne, R.B. The anti-oxidant properties of isothiocyanates: A review. Recent Pat Endocr. Metab. Immune Drug Discov. 2013, 7, 213–225. [Google Scholar] [CrossRef]
- Botanska, B.; Dovinova, I.; Barancik, M. The interplay between autophagy and redox signaling in cardiovascular diseases. Cells 2022, 11, 1203. [Google Scholar] [CrossRef]
- Wu, X.; Zhou, Q.H.; Xu, K. Are isothiocyanates potential anti-cancer drugs? Acta Pharmacol. Sin. 2009, 30, 501–512. [Google Scholar] [CrossRef]
- Kumar, S.; Sharma, S.; Kumar, V.; Sharma, R.; Minhas, A.; Boddu, R. Chapter 20—Cruciferous vegetables: A mine of phytonutrients for functional and nutraceutical enrichment. In Current Advances for Development of Functional Foods Modulating Inflammation and Oxidative Stress; Hernández-Ledesma, B., Martínez-Villaluenga, C., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 401–426. [Google Scholar]
- Hall, M.; Jobling, J.; Rogers, G. Some perspectives on rocket as a vegetable crop: A review. J. Fruit Ornam. Plant Res. 2012, 76, 21–41. [Google Scholar] [CrossRef]
- Rhaman, M.S.; Nakamura, T.; Nakamura, Y.; Munemasa, S.; Murata, Y. The myrosinases tgg1 and tgg2 function redundantly in reactive carbonyl species signaling in arabidopsis guard cells. Plant Cell Physiol. 2020, 61, 967–977. [Google Scholar] [CrossRef]
- Bauman, J.E.; Zang, Y.; Sen, M.; Li, C.; Wang, L.; Egner, P.A.; Fahey, J.W.; Normolle, D.P.; Grandis, J.R.; Kensler, T.W.; et al. Prevention of carcinogen-induced oral cancer by sulforaphane. Cancer Prev. Res. 2016, 9, 547–557. [Google Scholar] [CrossRef]
- Kontar, S.; Imrichova, D.; Bertova, A.; Mackova, K.; Poturnayova, A.; Sulova, Z.; Breier, A. Cell death effects induced by sulforaphane and allyl isothiocyanate on p-glycoprotein positive and negative variants in l1210 cells. Molecules 2020, 25, 2093. [Google Scholar] [CrossRef]
- Polekova, L.; Barancik, M.; Mrazova, T.; Pirker, R.; Wallner, J.; Sulova, Z.; Breier, A. Adaptation of mouse leukemia cells l1210 to vincristine. Evidence for expression of p-glycoprotein. Neoplasma 1992, 39, 73–77. [Google Scholar]
- Sulova, Z.; Ditte, P.; Kurucova, T.; Polakova, E.; Rogozanova, K.; Gibalova, L.; Seres, M.; Skvarkova, L.; Sedlak, J.; Pastorek, J.; et al. The presence of p-glycoprotein in l1210 cells directly induces down-regulation of cell surface saccharide targets of concanavalin a. Anticancer Res. 2010, 30, 3661–3668. [Google Scholar]
- Shikita, M.; Fahey, J.W.; Golden, T.R.; Holtzclaw, W.D.; Talalay, P. An unusual case of ‘uncompetitive activation’ by ascorbic acid: Purification and kinetic properties of a myrosinase from raphanus sativus seedlings. Biochem. J. 1999, 341 Pt 3, 725–732. [Google Scholar] [CrossRef]
- Rosenbergova, Z.; Kantorova, K.; Simkovic, M.; Breier, A.; Rebros, M. Optimisation of recombinant myrosinase production in pichia pastoris. Int. J. Mol. Sci. 2021, 22, 3677. [Google Scholar] [CrossRef]
- Deng, Q.; Zinoviadou, K.G.; Galanakis, C.M.; Orlien, V.; Grimi, N.; Vorobiev, E.; Lebovka, N.; Barba, F.J. The effects of conventional and non-conventional processing on glucosinolates and its derived forms, isothiocyanates: Extraction, degradation, and applications. Food Eng. Rev. 2015, 7, 357–381. [Google Scholar] [CrossRef]
- Shekarri, Q.; Dekker, M. A physiological-based model for simulating the bioavailability and kinetics of sulforaphane from broccoli products. Foods 2021, 10, 2761. [Google Scholar] [CrossRef] [PubMed]
- Kyca, T.; Pavlikova, L.; Bohacova, V.; Misak, A.; Poturnayova, A.; Breier, A.; Sulova, Z.; Seres, M. Insight into bortezomib focusing on its efficacy against p-gp-positive mdr leukemia cells. Int. J. Mol. Sci. 2021, 22, 5504. [Google Scholar] [CrossRef]
- Gao, X.; Leone, G.W.; Wang, H. Chapter four-cyclin d-cdk4/6 functions in cancer. In Advances in Cancer Research; Tew, K.D., Fisher, P.B., Eds.; Academic Press: Cambridge, MA, USA, 2020; Volume 148, pp. 147–169. [Google Scholar]
- Sonntag, R.; Giebeler, N.; Nevzorova, Y.A.; Bangen, J.-M.; Fahrenkamp, D.; Lambertz, D.; Haas, U.; Hu, W.; Gassler, N.; Cubero, F.J.; et al. Cyclin e1 and cyclin-dependent kinase 2 are critical for initiation, but not for progression of hepatocellular carcinoma. Proc. Natl. Acad. Sci. USA 2018, 115, 9282–9287. [Google Scholar] [CrossRef]
- García-Aranda, M.; Pérez-Ruiz, E.; Redondo, M. Bcl-2 inhibition to overcome resistance to chemo- and immunotherapy. Int. J. Mol. Sci. 2018, 19, 3950. [Google Scholar] [CrossRef] [PubMed]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. Bcl-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef]
- Seres, M.; Pavlikova, L.; Bohacova, V.; Kyca, T.; Borovska, I.; Lakatos, B.; Breier, A.; Sulova, Z. Overexpression of grp78/bip in p-glycoprotein-positive l1210 cells is responsible for altered response of cells to tunicamycin as a stressor of the endoplasmic reticulum. Cells 2020, 9, 890. [Google Scholar] [CrossRef] [PubMed]
- Suppipat, K.; Park, C.S.; Shen, Y.; Zhu, X.; Lacorazza, H.D. Sulforaphane induces cell cycle arrest and apoptosis in acute lymphoblastic leukemia cells. PLoS ONE 2012, 7, e51251. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Li, P.; Wang, S.; Cheng, G.; Wang, L.; Mi, X.; Su, X.; Wang, Y.; Zan, L. Tp53inp2 promotes bovine adipocytes differentiation through autophagy activation. Animals 2019, 9, 1060. [Google Scholar] [CrossRef]
- Zschemisch, N.H.; Liedtke, C.; Dierssen, U.; Nevzorova, Y.A.; Wustefeld, T.; Borlak, J.; Manns, M.P.; Trautwein, C. Expression of a cyclin e1 isoform in mice is correlated with the quiescent cell cycle status of hepatocytes in vivo. Hepatology 2006, 44, 164–173. [Google Scholar] [CrossRef]
- Huang, Y.; Sramkoski, R.M.; Jacobberger, J.W. The kinetics of g2 and m transitions regulated by b cyclins. PLoS ONE 2013, 8, e80861. [Google Scholar] [CrossRef]
- Peppard, J.V.; Rugg, C.; Smicker, M.; Dureuil, C.; Ronan, B.; Flamand, O.; Durand, L.; Pasquier, B. Identifying small molecules which inhibit autophagy: A phenotypic screen using image-based high-content cell analysis. Curr. Chem. Genom. Transl. Med. 2014, 8, 3–15. [Google Scholar] [CrossRef]
- Vázquez, C.L.; Colombo, M.I. Assays to assess autophagy induction and fusion of autophagic vacuoles with a degradative compartment, using monodansylcadaverine (mdc) and dq-bsa. Methods Enzymol. 2009, 452, 85–95. [Google Scholar]
- Elefantova, K.; Lakatos, B.; Kubickova, J.; Sulova, Z.; Breier, A. Detection of the mitochondrial membrane potential by the cationic dye jc-1 in l1210 cells with massive overexpression of the plasma membrane abcb1 drug transporter. Int. J. Mol. Sci. 2018, 19, 1985. [Google Scholar] [CrossRef]
- Bubencikova, T.; Cholujova, D.; Messingerova, L.; Mislovicova, D.; Seres, M.; Breier, A.; Sulova, Z. Detection of glycomic alterations induced by overexpression of p-glycoprotein on the surfaces of l1210 cells using sialic acid binding lectins. Int. J. Mol. Sci. 2012, 13, 15177–15192. [Google Scholar] [CrossRef]
- Pavlikova, L.; Seres, M.; Imrichova, D.; Hano, M.; Rusnak, A.; Zamorova, M.; Katrlik, J.; Breier, A.; Sulova, Z. The expression of p-gp in leukemia cells is associated with cross-resistance to protein n-glycosylation inhibitor tunicamycin. Gen. Physiol. Biophys. 2016, 35, 497–510. [Google Scholar] [CrossRef]
- Bhat, R.; Vyas, D. Myrosinase: Insights on structural, catalytic, regulatory, and environmental interactions. Crit. Rev. Biotechnol. 2019, 39, 508–523. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejski, D.; Koss-Mikolajczyk, I.; Glatt, H.; Bartoszek, A. The comparison of cytotoxic and genotoxic activities of glucosinolates, isothiocyanates, and indoles. Sci. Rep. 2022, 12, 4875. [Google Scholar] [CrossRef] [PubMed]
- Rosenbergova, Z.; Hegyi, Z.; Ferko, M.; Andelova, N.; Rebros, M. Improved production of recombinant myrosinase in pichia pastoris. Int. J. Mol. Sci. 2021, 22, 11889. [Google Scholar] [CrossRef] [PubMed]
- Galadova, H.; Polozsanyi, Z.; Breier, A.; Simkovic, M. Sulphoraphane affinity-based chromatography for the purification of myrosinase from lepidium sativum seeds. Biomolecules 2022, 12, 406. [Google Scholar] [CrossRef]
- Rai, Y.; Pathak, R.; Kumari, N.; Sah, D.K.; Pandey, S.; Kalra, N.; Soni, R.; Dwarakanath, B.S.; Bhatt, A.N. Mitochondrial biogenesis and metabolic hyperactivation limits the application of mtt assay in the estimation of radiation induced growth inhibition. Sci. Rep. 2018, 8, 1531. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Kozumbo, W.J. The phytoprotective agent sulforaphane prevents inflammatory degenerative diseases and age-related pathologies via nrf2-mediated hormesis. Pharmacol. Res. 2021, 163, 105283. [Google Scholar] [CrossRef]
- Semeniuk, M.; Ceré, L.I.; Ciriaci, N.; Bucci-Muñoz, M.; Villanueva, S.S.M.; Mottino, A.D.; Catania, V.A.; Rigalli, J.P.; Ruiz, M.L. Regulation of hepatic p-gp expression and activity by genistein in rats. Arch. Toxicol. 2020, 94, 1625–1635. [Google Scholar] [CrossRef]
- Ji, L.; Liu, X.; Zhang, S.; Tang, S.; Yang, S.; Li, S.; Qi, X.; Yu, S.; Lu, L.; Meng, X.; et al. The novel triazolonaphthalimide derivative lss-11 synergizes the anti-proliferative effect of paclitaxel via stat3-dependent mdr1 and mrp1 downregulation in chemoresistant lung cancer cells. Molecules 2017, 22, 1822. [Google Scholar] [CrossRef]
- Wang, X.; Campos, C.R.; Peart, J.C.; Smith, L.K.; Boni, J.L.; Cannon, R.E.; Miller, D.S. Nrf2 upregulates atp binding cassette transporter expression and activity at the blood–brain and blood–spinal cord barriers. J. Neurosci. 2014, 34, 8585–8593. [Google Scholar] [CrossRef]
- Harris, K.E.; Jeffery, E.H. Sulforaphane and erucin increase mrp1 and mrp2 in human carcinoma cell lines. J. Nutr. Biochem. 2008, 19, 246–254. [Google Scholar] [CrossRef]
- Hsu, C.; Lin, L.-I.; Cheng, Y.-C.; Feng, Z.-R.; Shao, Y.-Y.; Cheng, A.-L.; Ou, D.-L. Cyclin e1 inhibition can overcome sorafenib resistance in hepatocellular carcinoma cells through mcl-1 suppression. Clin. Cancer Res. 2016, 22, 2555–2564. [Google Scholar] [CrossRef]
- Liang, H.; Chen, Z.; Sun, L. Inhibition of cyclin e1 overcomes temozolomide resistance in glioblastoma by mcl-1 degradation. Mol. Carcinog. 2019, 58, 1502–1511. [Google Scholar] [CrossRef]
- Wegler, C.; Olander, M.; Wisniewski, J.R.; Lundquist, P.; Zettl, K.; Asberg, A.; Hjelmesaeth, J.; Andersson, T.B.; Artursson, P. Global variability analysis of mrna and protein concentrations across and within human tissues. NAR Genom. Bioinform. 2020, 2, lqz010. [Google Scholar] [CrossRef]
- Wang, D. Discrepancy between mrna and protein abundance: Insight from information retrieval process in computers. Comput. Biol. Chem. 2008, 32, 462–468. [Google Scholar] [CrossRef]
- O’Leary, B.; Finn, R.S.; Turner, N.C. Treating cancer with selective cdk4/6 inhibitors. Nature reviews. Clin. Oncol. 2016, 13, 417–430. [Google Scholar]
- Álvarez-Fernández, M.; Malumbres, M. Mechanisms of sensitivity and resistance to cdk4/6 inhibition. Cancer Cell 2020, 37, 514–529. [Google Scholar] [CrossRef]
- Watt, A.C.; Goel, S. Cellular mechanisms underlying response and resistance to cdk4/6 inhibitors in the treatment of hormone receptor-positive breast cancer. Breast Cancer Res. 2022, 24, 17. [Google Scholar] [CrossRef]
- Freeman-Cook, K.; Hoffman, R.L.; Miller, N.; Almaden, J.; Chionis, J.; Zhang, Q.; Eisele, K.; Liu, C.; Zhang, C.; Huser, N.; et al. Expanding control of the tumor cell cycle with a cdk2/4/6 inhibitor. Cancer Cell 2021, 39, 1404–1421.e1411. [Google Scholar] [CrossRef]
- Kasimir-Bauer, S.; Beelen, D.; Flasshove, M.; Noppeney, R.; Seeber, S.; Scheulen, M.E. Impact of the expression of p glycoprotein, the multidrug resistance-related protein, bcl-2, mutant p53, and heat shock protein 27 on response to induction therapy and long-term survival in patients with de novo acute myeloid leukemia. Exp. Hematol. 2002, 30, 1302–1308. [Google Scholar] [CrossRef]
- Campbell, K.J.; Tait, S.W.G. Targeting bcl-2 regulated apoptosis in cancer. Open. Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. Lc3 and autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar]
- Migneault, F.; Hébert, M.-J. Autophagy, tissue repair, and fibrosis: A delicate balance. Matrix Biol. 2021, 100–101, 182–196. [Google Scholar] [CrossRef]
- Herman-Antosiewicz, A.; Johnson, D.E.; Singh, S.V. Sulforaphane causes autophagy to inhibit release of cytochrome c and apoptosis in human prostate cancer cells. Cancer Res. 2006, 66, 5828–5835. [Google Scholar] [CrossRef]
- Lu, Z.; Ren, Y.; Yang, L.; Jia, A.; Hu, Y.; Zhao, Y.; Zhao, W.; Yu, B.; Zhao, W.; Zhang, J.; et al. Inhibiting autophagy enhances sulforaphane-induced apoptosis via targeting nrf2 in esophageal squamous cell carcinoma. Acta Pharm. Sin. B 2021, 11, 1246–1260. [Google Scholar] [CrossRef] [PubMed]
- Tainton, K.M.; Smyth, M.J.; Jackson, J.T.; Tanner, J.E.; Cerruti, L.; Jane, S.M.; Darcy, P.K.; Johnstone, R.W. Mutational analysis of p-glycoprotein: Suppression of caspase activation in the absence of atp-dependent drug efflux. Cell Death Differ. 2004, 11, 1028–1037. [Google Scholar] [CrossRef]
- Cagala, M.; Pavlikova, L.; Seres, M.; Kadlecikova, K.; Breier, A.; Sulova, Z. Development of resistance to endoplasmic reticulum stress-inducing agents in mouse leukemic l1210 cells. Molecules 2020, 25, 2517. [Google Scholar] [CrossRef]
- Sulova, Z.; Mislovicova, D.; Gibalova, L.; Vajcnerova, Z.; Polakova, E.; Uhrik, B.; Tylkova, L.; Kovarova, A.; Sedlak, J.; Breier, A. Vincristine-induced overexpression of p-glycoprotein in l1210 cells is associated with remodeling of cell surface saccharides. J. Proteome Res. 2009, 8, 513–520. [Google Scholar] [CrossRef]
- Pastan, I.; Gottesman, M.M.; Ueda, K.; Lovelace, E.; Rutherford, A.V.; Willingham, M.C. A retrovirus carrying an mdr1 cdna confers multidrug resistance and polarized expression of p-glycoprotein in mdck cells. Proc. Natl. Acad. Sci. USA 1988, 85, 4486–4490. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage t4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. Proc. Natl. Acad. Sci. USA 1979, 76, 4350–4354. [Google Scholar] [CrossRef] [PubMed]
- Towbin, H.; Staehelin, T.; Gordon, J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: Procedure and some applications. 1979. Biotechnology 1992, 24, 145–149. [Google Scholar] [PubMed]
| One passage in the presence of SFN | ||||||||||||||||||
| Gene | Ccnd1 | Cdk6 | Ccne1 | Cdk2 | Bcl2 | Bax | ||||||||||||
| Cell variants | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T |
| Compared to S | ↑ | 0 | ↑ | ↑ | ↑ | ↑ | 0 | 0 | ↑ | ↑ | 0 | 0 | ||||||
| SFN (μM) | Compared to cells passaged in the absence of SFN | |||||||||||||||||
| 2.5 | ||||||||||||||||||
| 5.0 | ||||||||||||||||||
| 7.5 | ||||||||||||||||||
| 10.0 | ||||||||||||||||||
| One passage in the presence of SFN followed by passage in the absence of SFN | ||||||||||||||||||
| Gene | Ccnd1 | Cdk6 | Ccne1 | Cdk2 | Bcl2 | Bax | ||||||||||||
| Cell variants | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T |
| Compared to S | ↑ | 0 | ↑ | ↑ | ↑ | ↑ | 0 | 0 | ↑ | ↑ | 0 | 0 | ||||||
| SFN (μM) | Compared to cells passaged in the absence of SFN | |||||||||||||||||
| 2.5 | ||||||||||||||||||
| 5.0 | ||||||||||||||||||
| 7.5 | ||||||||||||||||||
| 10.0 | ||||||||||||||||||
| Six consecutive passages in the presence of SFN | ||||||||||||||||||
| Gene | Ccnd1 | Cdk6 | Ccne1 | Cdk2 | Bcl2 | Bax | ||||||||||||
| Cell variants | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T | S | R | T |
| Compared to S | 0 | ↓ | ↓ | ↓ | ↑ | ↑ | 0 | 0 | ↑ | ↑ | 0 | 0 | ||||||
| SFN (μM) | Compared to cells passaged in the absence of SFN | |||||||||||||||||
| 2.5 | ||||||||||||||||||
| 5.0 | ||||||||||||||||||
| 7.5 | ||||||||||||||||||
| 10.0 | ||||||||||||||||||
| One passage in the presence of SFN | ||||||||||||
| Proteins | Cyclin B1 | Cyclin E1 | LC3B | |||||||||
| 16 kDa | 18 kDa | |||||||||||
| Cell variants | S | R | T | S | R | T | S | R | T | S | R | T |
| Compared to S | ↑ | 0 | ↓ | ↓ | 0 | ↓ | 0 | ↓ | ||||
| SFN (μM) | Compared to cells passaged in the absence of SFN | |||||||||||
| 5 | ||||||||||||
| 10 | ||||||||||||
| 15 | ||||||||||||
| Four passages in the presence of SFN | ||||||||||||
| Proteins | Cyclin B1 | Cyclin E1 | LC3B | |||||||||
| 16 kDa | 18 kDa | |||||||||||
| Cell variants | S | R | T | S | R | T | S | R | T | S | R | T |
| Compared to S | 0 | ↑ | 0 | 0 | ↓ | ↓ | ↓ | ↓ | ||||
| SFN (μM) | Compared to cells passaged in the absence of SFN | |||||||||||
| 5 | ||||||||||||
| 10 | ||||||||||||
| 15 | ||||||||||||
| Gene | Primer Sequences | TA (°C) | PCR Product (bp) |
|---|---|---|---|
| ABCB1 * | F: 5′-GCAATGGAGGAGCAAAGAAG-3′ | 59 | 150 |
| R: 5′-CCAAAGTTCCCACCACCATA-3′ | |||
| Abcb1 | F: 5′-AGGTAGAGACACGTGAGGTCG-3′ | 57 | 158 |
| R: 5′-CAGCCAACCTGCATAACG-3′ | |||
| Bax | F: 5′-CTAGCAAAGTAGAAGAGGGCAACC-3′ | 58 | 151 |
| R: 5′-ATGAACTGGACAGCAATATGGAG-3′ | |||
| Bcl2 | F: 5′-GCATGCTGGGGCCATATAGTT-3′ | 58 | 323 |
| R: 5′-GGCTGGGGATGACTTCTCTC-3′ | |||
| Ccnb1 | F: 5′-GGTGACTTCGCCTTTGTGAC-3′ | 58 | 125 |
| R: 5′-CTACGGAGGAAGTGCAGAGG-3′ | |||
| Ccnd1 | F: 5′-GGCCTTCAGGCAAAAACCAG-3′ | 58 | 591 |
| R: 5′-TCACCCTGAGAGTAGGGAGC-3′ | |||
| Ccne1 | F: 5′-AGGATGACGCTGCAGAAAGT-3′ | 58 | 131 |
| R: 5′-GGAAAATCAGACCACCCAGA-3′ | |||
| Cdk2 | F: 5′-CTTTGCTGAAATGGTGACCCG-3′ | 57 | 368 |
| R: 5′-CCAGGGCCAAGTCAGACCAC-3′ | |||
| Cdk6 | F: 5′-GCCGGGCTCTGGAACTTTAT-3′ | 57 | 325 |
| R: 5′-CGCCGATCAGCAGTATGAGT-3′ | |||
| Gapdh | F: 5′-CAATGTGTCCGTCGTGGAT-3′ | 57 | 314 |
| R: 5′-GTGGGTGGTCCAGGGTTT-3′ |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bertova, A.; Kontar, S.; Polozsanyi, Z.; Simkovic, M.; Rosenbergova, Z.; Rebros, M.; Sulova, Z.; Breier, A.; Imrichova, D. Effects of Sulforaphane-Induced Cell Death upon Repeated Passage of Either P-Glycoprotein-Negative or P-Glycoprotein-Positive L1210 Cell Variants. Int. J. Mol. Sci. 2022, 23, 10818. https://doi.org/10.3390/ijms231810818
Bertova A, Kontar S, Polozsanyi Z, Simkovic M, Rosenbergova Z, Rebros M, Sulova Z, Breier A, Imrichova D. Effects of Sulforaphane-Induced Cell Death upon Repeated Passage of Either P-Glycoprotein-Negative or P-Glycoprotein-Positive L1210 Cell Variants. International Journal of Molecular Sciences. 2022; 23(18):10818. https://doi.org/10.3390/ijms231810818
Chicago/Turabian StyleBertova, Anna, Szilvia Kontar, Zoltan Polozsanyi, Martin Simkovic, Zuzana Rosenbergova, Martin Rebros, Zdena Sulova, Albert Breier, and Denisa Imrichova. 2022. "Effects of Sulforaphane-Induced Cell Death upon Repeated Passage of Either P-Glycoprotein-Negative or P-Glycoprotein-Positive L1210 Cell Variants" International Journal of Molecular Sciences 23, no. 18: 10818. https://doi.org/10.3390/ijms231810818
APA StyleBertova, A., Kontar, S., Polozsanyi, Z., Simkovic, M., Rosenbergova, Z., Rebros, M., Sulova, Z., Breier, A., & Imrichova, D. (2022). Effects of Sulforaphane-Induced Cell Death upon Repeated Passage of Either P-Glycoprotein-Negative or P-Glycoprotein-Positive L1210 Cell Variants. International Journal of Molecular Sciences, 23(18), 10818. https://doi.org/10.3390/ijms231810818






