Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry
Abstract
:1. Introduction
2. Results
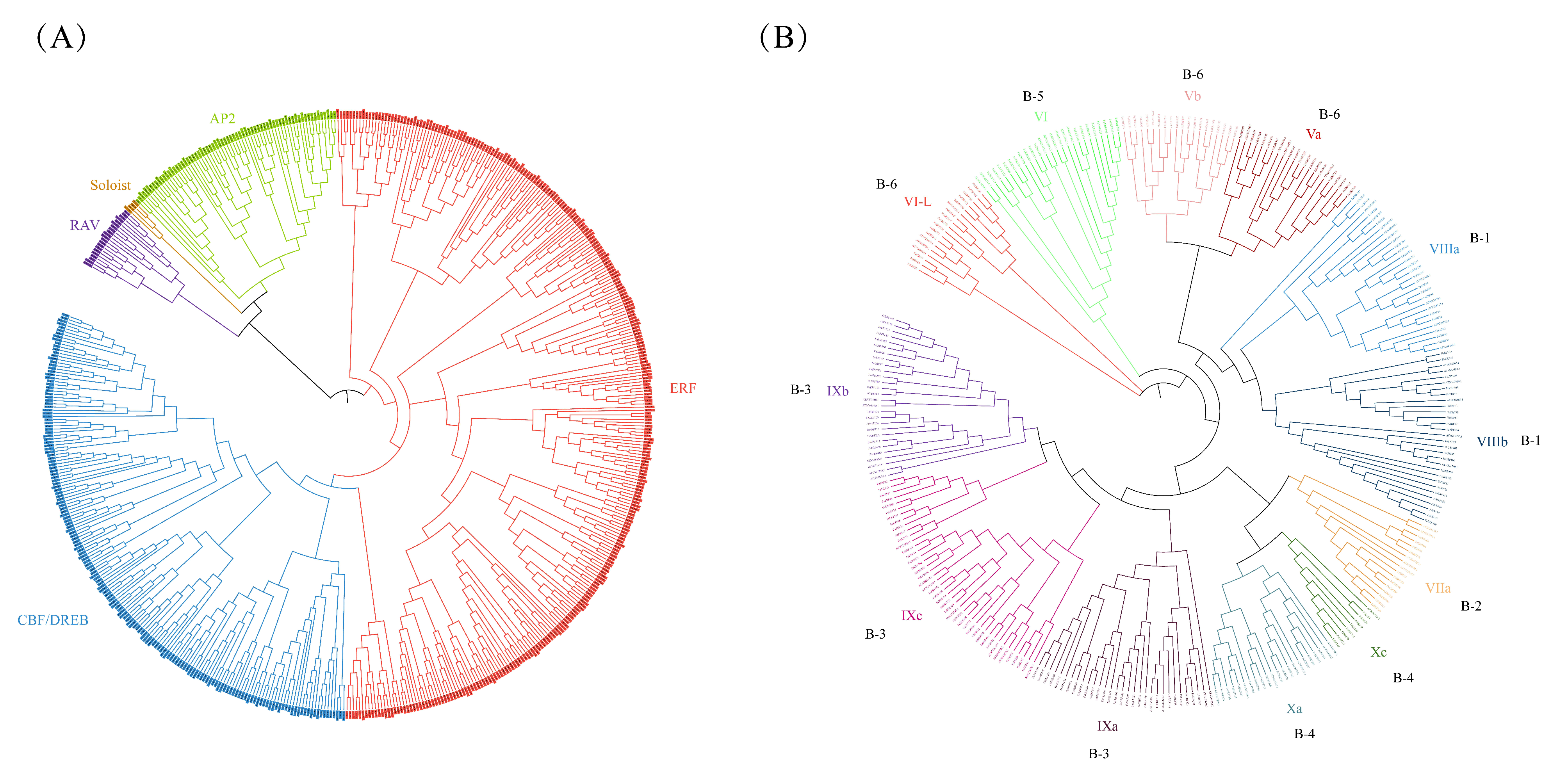
2.1. Identification and Characterization of FaERFs
2.2. Gene Structure and Conserved Motif of FaERFs
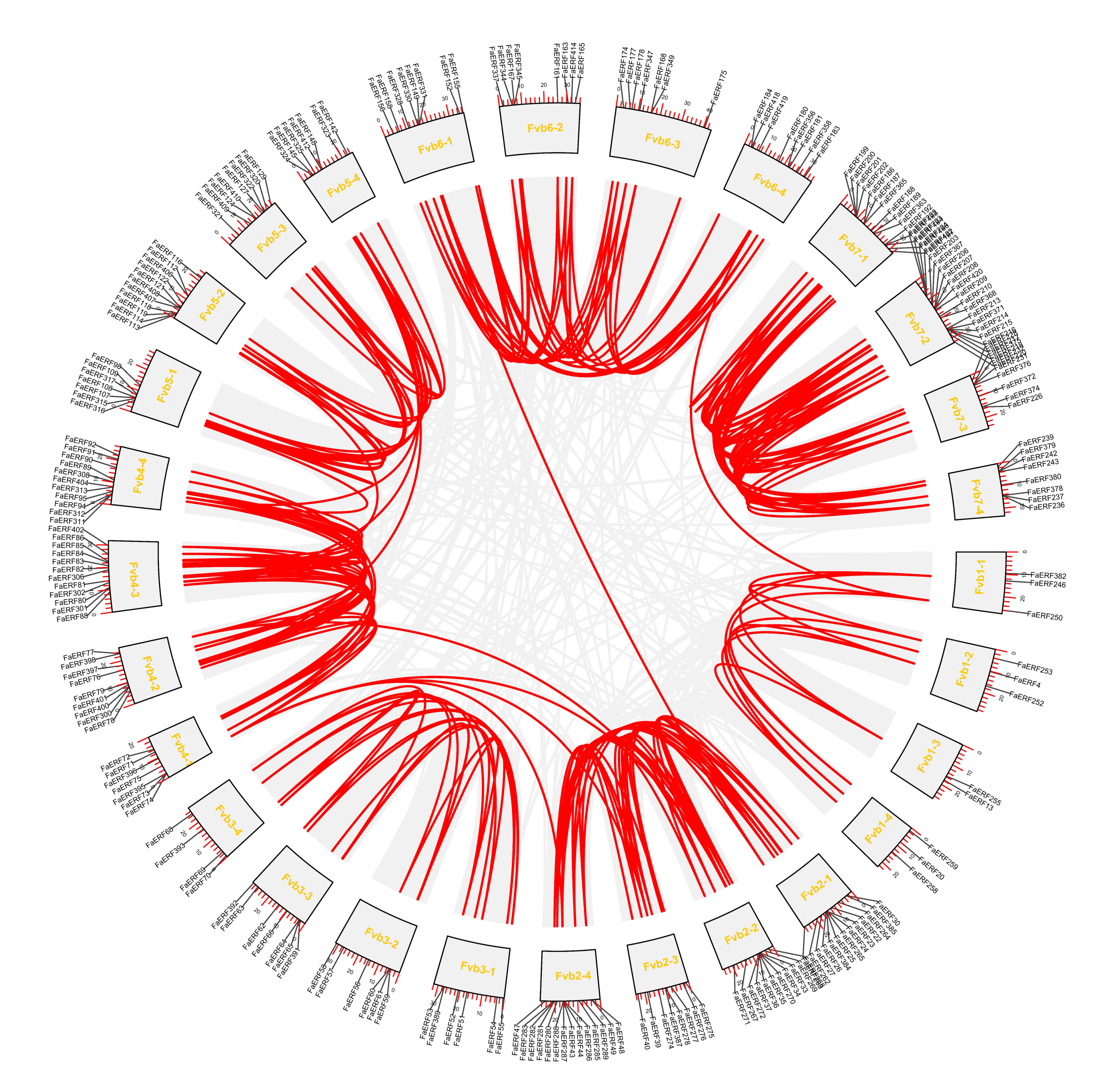
2.3. Chromosomal Location and Synteny Analysis of FaERFs
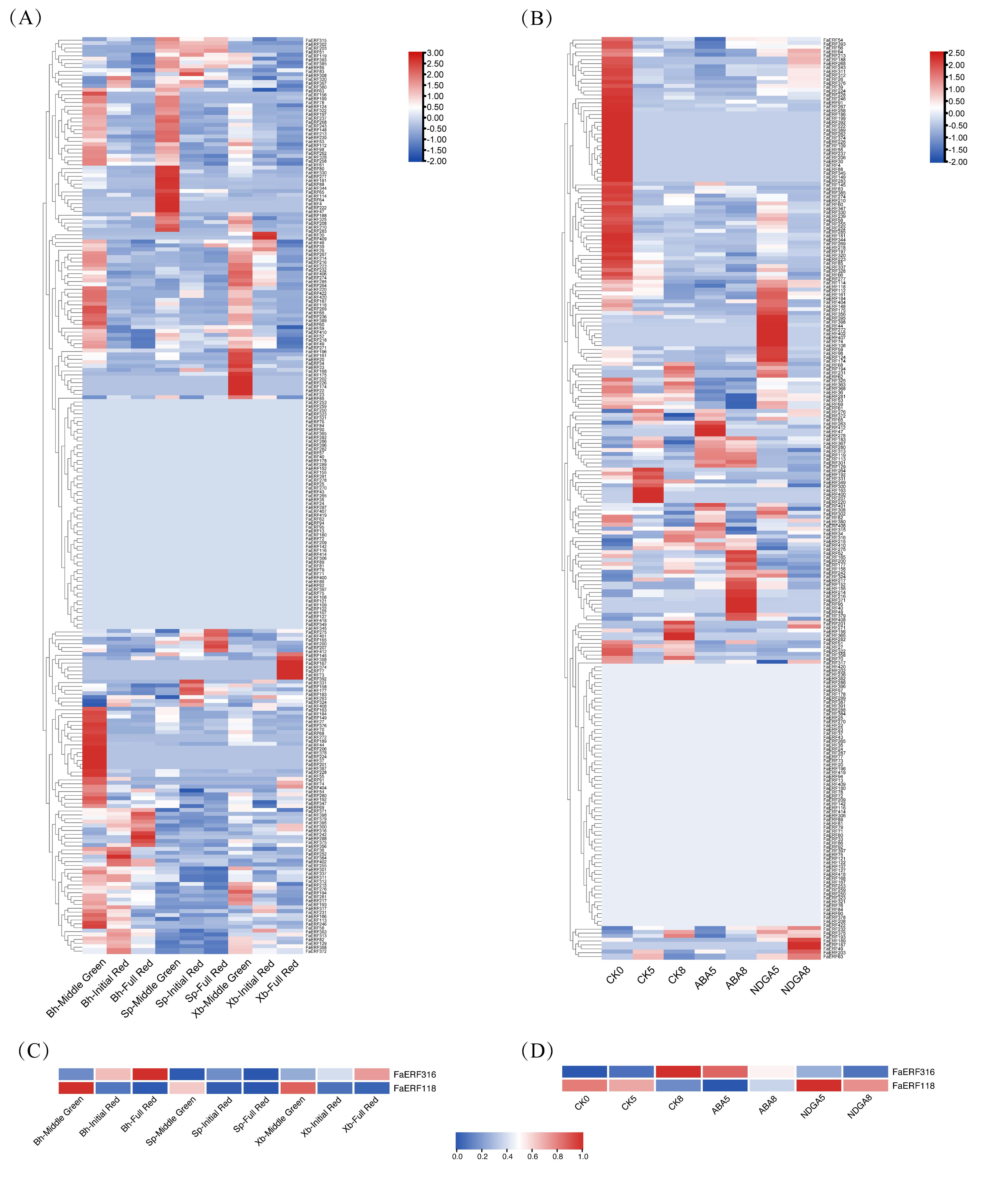
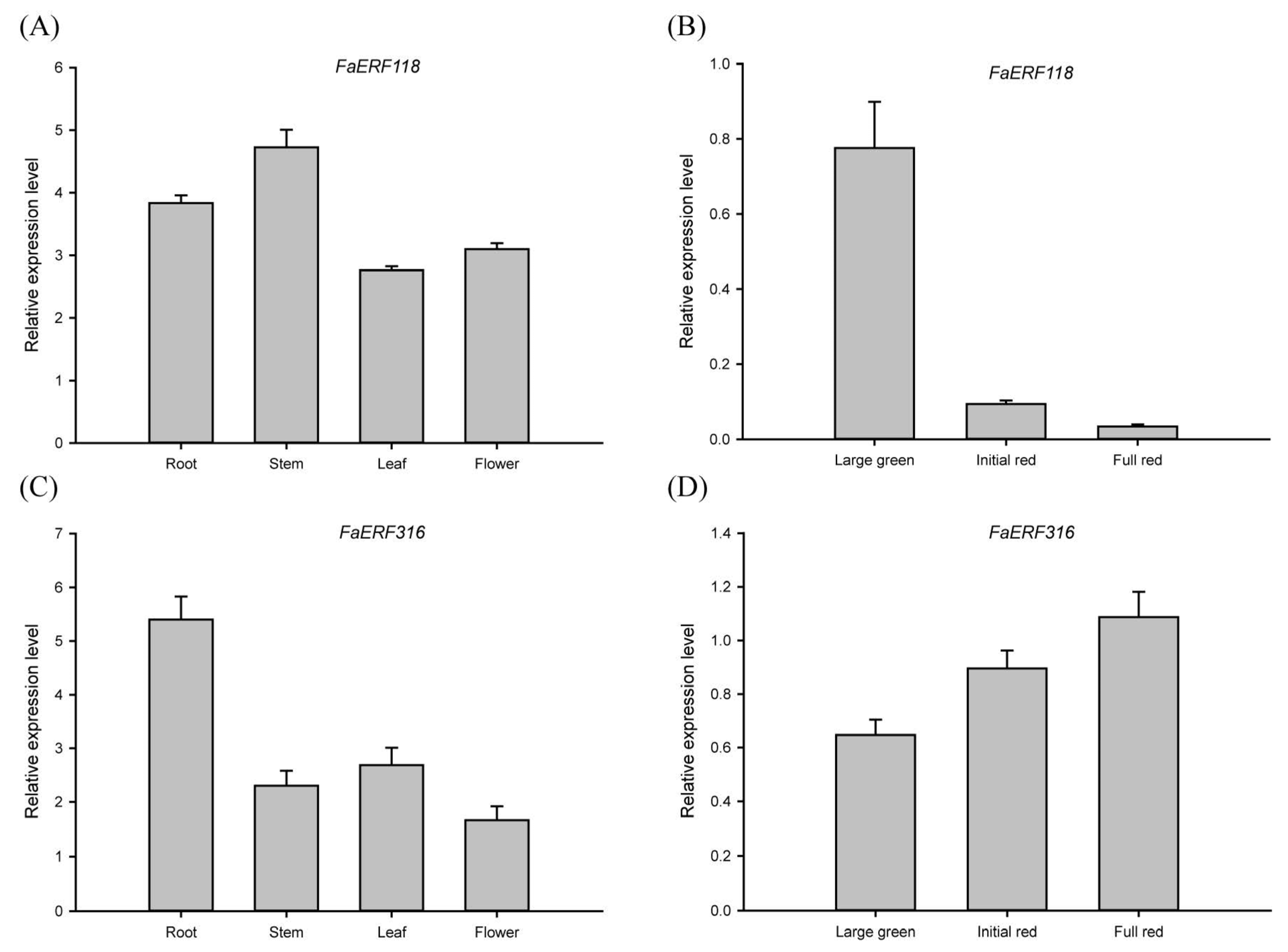
2.4. Expression Profiles Analysis of FaERF Genes
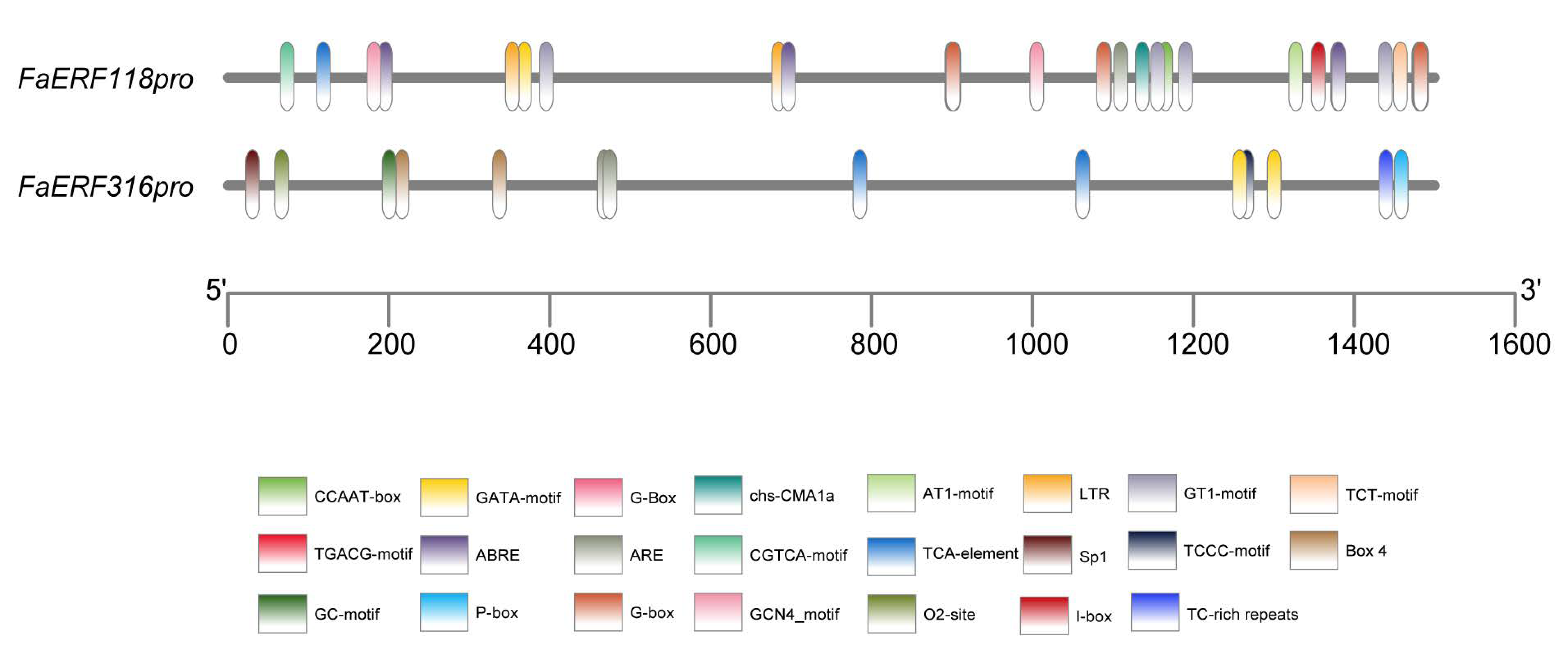
2.5. Regulatory Mechanism Analysis in the Promoter Regions of FaERF118 and FaERF316
2.6. Protein–Protein Network Analysis of FaERF118 and FaERF316
3. Discussion
4. Materials and Methods
4.1. Plant Materials, RNA Extraction, cDNA Synthesis
4.2. Identification of FaERF Genes in Strawberry
4.3. Sequence Analysis of FaERF Genes
4.4. Gene Structure and Conserved Motif Analysis of FaERF Genes
4.5. Chromosomal Mapping and Synteny Analyses of FaERF Genes
4.6. Expression Profile Analysis of FaERF Genes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.-Z.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.; Samaha, R. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. Planttfdb 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Feng, K.; Hou, X.-L.; Xing, G.-M.; Liu, J.-X.; Duan, A.-Q.; Xu, Z.-S.; Li, M.-Y.; Zhuang, J.; Xiong, A.-S. Advances in AP2/ERF super-family transcription factors in plant. Crit. Rev. Biotechnol. 2020, 40, 750–776. [Google Scholar] [CrossRef]
- Okamuro, J.K.; Caster, B.; Villarroel, R.; Van Montagu, M.; Jofuku, K.D. The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc. Natl. Acad. Sci. USA 1997, 94, 7076–7081. [Google Scholar] [CrossRef]
- Riechmann, J.L.; Meyerowitz, E.M. The AP2/EREBP family of plant transcription factors. Biol. Chem. 1998, 379, 633–646. [Google Scholar]
- Allen, M.D.; Yamasaki, K.; Ohme-Takagi, M.; Tateno, M.; Suzuki, M. A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 1998, 17, 5484–5496. [Google Scholar] [CrossRef]
- Sakuma, Y.; Liu, Q.; Dubouzet, J.G.; Abe, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef]
- Stockinger, E.J.; Gilmour, S.J.; Thomashow, M.F. Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. USA 1997, 94, 1035–1040. [Google Scholar] [CrossRef]
- Ohme-Takagi, M.; Shinshi, H. Ethylene-inducible DNA binding proteins that interact with an ethylene-responsive element. Plant Cell 1995, 7, 173–182. [Google Scholar] [PubMed] [Green Version]
- Srivastava, R.; Kumar, R. The expanding roles of APETALA2/Ethylene Responsive Factors and their potential applications in crop improvement. Brief. Funct. Genom. 2019, 18, 240–254. [Google Scholar] [CrossRef] [PubMed]
- Li, B.-J.; Grierson, D.; Shi, Y.; Chen, K.-S. Roles of abscisic acid in regulating ripening and quality of strawberry, a model non-climacteric fruit. Hortic. Res. 2022, 9, uhac089. [Google Scholar] [CrossRef]
- Bai, Q.; Huang, Y.; Shen, Y. The physiological and molecular mechanism of abscisic acid in regulation of fleshy fruit ripening. Front. Plant Sci. 2021, 11, 619953. [Google Scholar] [CrossRef]
- Kou, X.; Feng, Y.; Yuan, S.; Zhao, X.; Wu, C.; Wang, C.; Xue, Z. Different regulatory mechanisms of plant hormones in the ripening of climacteric and non-climacteric fruits: A review. Plant Mol. Biol. 2021, 107, 477–497. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Li, Z.; Liu, M. Role of ethylene response factors (ERFs) in fruit ripening. Food Qual. Saf. 2020, 4, 15–20. [Google Scholar] [CrossRef]
- Xie, X.-L.; Shen, S.-L.; Yin, X.-R.; Xu, Q.; Sun, C.-D.; Grierson, D.; Ferguson, I.; Chen, K.-S. Isolation, classification and transcription profiles of the AP2/ERF transcription factor superfamily in citrus. Mol. Biol. Rep. 2014, 41, 4261–4271. [Google Scholar] [CrossRef] [PubMed]
- Kuang, J.-F.; Chen, J.-Y.; Luo, M.; Wu, K.-Q.; Sun, W.; Jiang, Y.-M.; Lu, W.-J. Histone deacetylase HD2 interacts with ERF1 and is involved in longan fruit senescence. J. Exp. Bot. 2012, 63, 441–454. [Google Scholar] [CrossRef]
- Viola, I.L.; Camoirano, A.; Gonzalez, D.H. Redox-dependent modulation of anthocyanin biosynthesis by the TCP transcription factor TCP15 during exposure to high light intensity conditions in Arabidopsis. Plant Physiol. 2016, 170, 74–85. [Google Scholar] [CrossRef]
- Liu, L.; Lin, N.; Liu, X.; Yang, S.; Wang, W.; Wan, X. From chloroplast biogenesis to chlorophyll accumulation: The interplay of light and hormones on gene expression in camellia sinensis cv. Shuchazao leaves. Front. Plant Sci. 2020, 11, 256. [Google Scholar] [CrossRef]
- Sirko, A.; Wawrzyńska, A.; Brzywczy, J.; Sieńko, M. Control of ABA signaling and crosstalk with other hormones by the selective degradation of pathway components. Int. J. Mol. Sci. 2021, 22, 4638. [Google Scholar] [CrossRef] [PubMed]
- Chai, L.; Shen, Y.-Y. FaABI4 is involved in strawberry fruit ripening. Sci. Hortic. 2016, 210, 34–40. [Google Scholar] [CrossRef]
- Wang, H.; Zhang, Y.; Norris, A.; Jiang, C.-Z. S1-bZIP transcription factors play important roles in the regulation of fruit quality and stress response. Front. Plant Sci. 2022, 12, 802802. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Tang, Y.-M.; Wang, Y.; Sun, B.; Chen, T.; Lei, D.-Y.; Zhang, F.; Luo, Y.; Zhang, Y.; Wang, X.-R.; et al. Enhance sucrose accumulation in strawberry fruits by eliminating the translational repression of FabZIPs1.1. Sci. Hortic. 2020, 259, 108850. [Google Scholar] [CrossRef]
- Shoji, T.; Yuan, L. ERF gene clusters: Working together to regulate metabolism. Trends Plant Sci. 2021, 26, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Klay, I.; Gouia, S.; Liu, M.; Mila, I.; Khoudi, H.; Bernadac, A.; Bouzayen, M.; Pirrello, J. Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018, 274, 137–145. [Google Scholar] [CrossRef]
- Khaksar, G.; Sirikantaramas, S. Transcriptome-wide identification and expression profiling of the ERF gene family suggest roles as transcriptional activators and repressors of fruit ripening in durian. PLoS ONE 2021, 16, e0252367. [Google Scholar] [CrossRef]
- Brumos, J. Gene regulation in climacteric fruit ripening. Curr. Opin. Plant Biol. 2021, 63, 102042. [Google Scholar] [CrossRef]
- Edger, P.P.; Poorten, T.J.; VanBuren, R.; Hardigan, M.A.; Colle, M.; McKain, M.R.; Smith, R.D.; Teresi, S.J.; Nelson, A.D.; Wai, C.M. Origin and evolution of the octoploid strawberry genome. Nat. Genet. 2019, 51, 541–547. [Google Scholar] [CrossRef]
- Su, Y.; Ma, Z.; Mao, J.; Li, W.; Cao, X.; Chen, B. Genome-wide identification and characterization of the strawberry (Fragaria vesca) FvAP2/ERF gene family in abiotic stress. In Plant Molecular Biology Reporter; Springer: Berlin/Heidelberg, Germany, 2022; pp. 1–15. [Google Scholar]
- Zhang, Z.; Li, X. Genome-wide identification of AP2/ERF superfamily genes and their expression during fruit ripening of Chinese jujube. Sci. Rep. 2018, 8, 15612. [Google Scholar] [CrossRef]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, R.; Song, J.; Lv, Z.; Qi, X.; Han, X.; Guo, Q.; Wang, S.; Shi, J.; Jian, Z.; Hu, Q. Genome-wide identification and comprehensive analysis of the AP2/ERF gene family in pomegranate fruit development and postharvest preservation. Genes 2022, 13, 895. [Google Scholar] [CrossRef] [PubMed]
- Girardi, C.L.; Rombaldi, C.V.; Dal Cero, J.; Nobile, P.M.; Laurens, F.; Bouzayen, M.; Quecini, V. Genome-wide analysis of the AP2/ERF superfamily in apple and transcriptional evidence of ERF involvement in scab pathogenesis. Sci. Hortic. 2013, 151, 112–121. [Google Scholar] [CrossRef]
- Li, X.; Tao, S.; Wei, S.; Ming, M.; Huang, X.; Zhang, S.; Wu, J. The mining and evolutionary investigation of AP2/ERF genes in pear (Pyrus). BMC Plant Biol. 2018, 18, 46. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Pan, X.; Liu, S.; Lin, W.; Li, Y.; Zhang, X. Genome-wide analysis of AP2/ERF transcription factors in pineapple reveals functional divergence during flowering induction mediated by ethylene and floral organ development. Genomics 2021, 113, 474–489. [Google Scholar] [CrossRef]
- Li, M.; Li, B.; Yang, M.; Wang, L.; Hou, G.; Lin, Y.; Zhang, Y.; Zhang, Y.; Chen, Q.; Wang, Y. Genome-wide identification and expression of MAPK gene family in cultivated strawberry and their involvement in fruit developing and ripening. Int. J. Mol. Sci. 2022, 23, 5201. [Google Scholar] [CrossRef]
- Li, M.; Wang, L.; Liu, Y.; Lin, Y.; Zhang, Y.; Long, Y.; Luo, C.; Zhang, Y.; Chen, Q.; Chen, P. Characterization and regulation mechanism analysis of ubiquitin-conjugating family genes in strawberry reveals a potential role in fruit ripening. BMC Plant Biol. 2022, 22, 39. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, J.; Wu, L.; Zhang, Y.; Chen, Q.; Li, M.; Zhang, Y.; Luo, Y.; Wang, Y.; Wang, X. Genome-wide identification of GMP genes in rosaceae and functional characterization of FaGMP4 in strawberry (Fragaria× ananassa). Genes Genom. 2021, 43, 587–599. [Google Scholar] [CrossRef]
- Lin, Y.; Zhang, L.; Zhang, J.; Zhang, Y.; Wang, Y.; Chen, Q.; Luo, Y.; Zhang, Y.; Li, M.; Wang, X. Identification of anthocyanins-related glutathione s-transferase (GST) genes in the genome of cultivated strawberry (Fragaria× ananassa). Int. J. Mol. Sci. 2020, 21, 8708. [Google Scholar] [CrossRef]
- Cramer, G.R.; Ghan, R.; Schlauch, K.A.; Tillett, R.L.; Heymann, H.; Ferrarini, A.; Delledonne, M.; Zenoni, S.; Fasoli, M.; Pezzotti, M. Transcriptomic analysis of the late stages of grapevine (Vitis vinifera cv. Cabernet sauvignon) berry ripening reveals significant induction of ethylene signaling and flavor pathways in the skin. BMC Plant Biol. 2014, 14, 370. [Google Scholar] [CrossRef]
- Xing, S.N.; Chen, K.L.; Zhu, H.C.; Zhang, R.; Zhang, H.W.; Li, B.B.; Gao, C.X. Fine-tuning sugar content in strawberry. Genome Biol. 2020, 21, 230. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Yu, H.; Wang, X.; Xie, X.; Yue, X.; Tang, H. An alternative cetyltrimethylammonium bromide-based protocol for RNA isolation from blackberry (Rubus L.). Genet. Mol. Res. GMR 2012, 11, 1773–1782. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-h.; Jin, H.; Marler, B.; Guo, H. Mcscanx: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. Tbtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Zhao, F.; Li, G.; Hu, P.; Zhao, X.; Li, L.; Wei, W.; Feng, J.; Zhou, H. Identification of basic/helix-loop-helix transcription factors reveals candidate genes involved in anthocyanin biosynthesis from the strawberry white-flesh mutant. Sci. Rep. 2018, 8, 2721. [Google Scholar] [CrossRef]
- Li, D.D.; Li, L.; Luo, Z.S.; Mou, W.S.; Mao, L.C.; Ying, T.J. Comparative transcriptome analysis reveals the influence of abscisic acid on the metabolism of pigments, ascorbic acid and folic acid during strawberry fruit ripening. PLoS ONE 2015, 10, e0130037. [Google Scholar] [CrossRef]



| Promoter | Transcription Factor Gene Family |
| FaERF118-pro | bHLH (BIM1, PIF3), Trihelix (ASIL2), NAC, TCP (TCP19, TCP20, TCP15), C2H2 (ZAT10, TFIIIA), ARF (ARF6, ARF5), MIKC_MADS (AGL22), GRAS (RGA1), CPP (SOL2), BBR-BPC (BPC6, BPC2), CAMTA (CMTA5), bZIP (AREB1, AREB3), DOF (DOF2.4, DOF1.5, DOF 5.1, DOF 1), MYB (MYB50, MYB60), G2-like (GLK1) |
| FaERF316-pro | bZIP (bZIP53, GBF3, bZIP42, bZIP44), Trihelix (ASR3), GRAS (RGA 1), ERF (ERF005, ABI4), BBR-BPC (BPC6, BPC2), MYB (MYB121, MYB50, MYB27, MYB83, MYB84), C2H2 (JKD, TFIIIA) DOF (DOF2.4, DOF 5.1, DOF 5.4), LBD (ASL4, AS2) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Guo, C.; Deng, M.; Li, S.; Chen, Y.; Gu, X.; Tang, G.; Lin, Y.; Wang, Y.; He, W.; et al. Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry. Int. J. Mol. Sci. 2022, 23, 10550. https://doi.org/10.3390/ijms231810550
Zhang Y, Guo C, Deng M, Li S, Chen Y, Gu X, Tang G, Lin Y, Wang Y, He W, et al. Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry. International Journal of Molecular Sciences. 2022; 23(18):10550. https://doi.org/10.3390/ijms231810550
Chicago/Turabian StyleZhang, Yunting, Chenhui Guo, Meiyi Deng, Shanlin Li, Yingying Chen, Xianjie Gu, Guohao Tang, Yuanxiu Lin, Yan Wang, Wen He, and et al. 2022. "Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry" International Journal of Molecular Sciences 23, no. 18: 10550. https://doi.org/10.3390/ijms231810550
APA StyleZhang, Y., Guo, C., Deng, M., Li, S., Chen, Y., Gu, X., Tang, G., Lin, Y., Wang, Y., He, W., Li, M., Zhang, Y., Luo, Y., Wang, X., Chen, Q., & Tang, H. (2022). Genome-Wide Analysis of the ERF Family and Identification of Potential Genes Involved in Fruit Ripening in Octoploid Strawberry. International Journal of Molecular Sciences, 23(18), 10550. https://doi.org/10.3390/ijms231810550








