Perfluorobutanoic Acid (PFBA) Induces a Non-Enzymatic Oxidative Stress Response in Soybean (Glycine max L. Merr.)
Abstract
:1. Introduction
2. Results
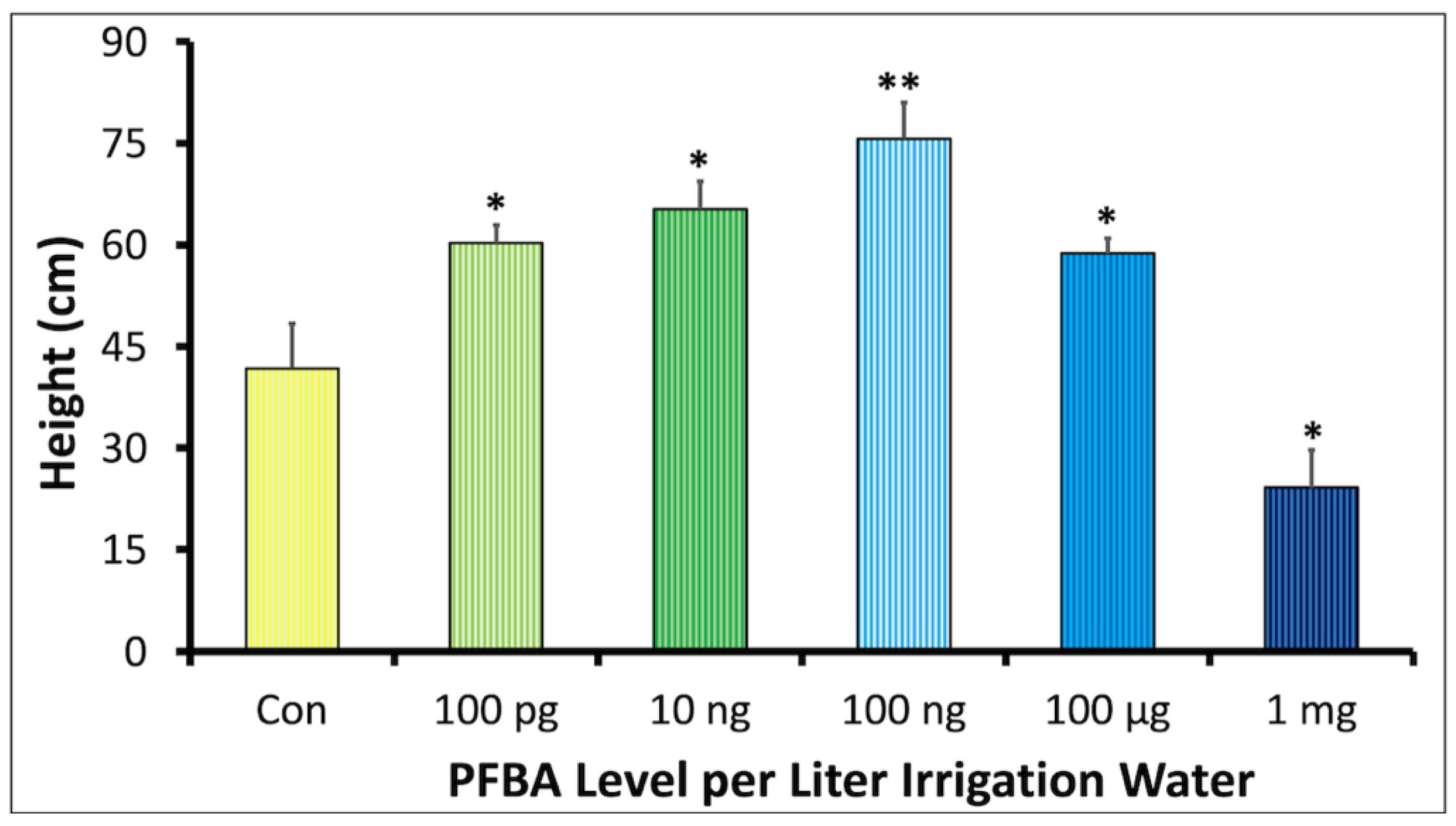
2.1. Effect of PFBA Concentration on Plant Development
2.1.1. Plant Growth
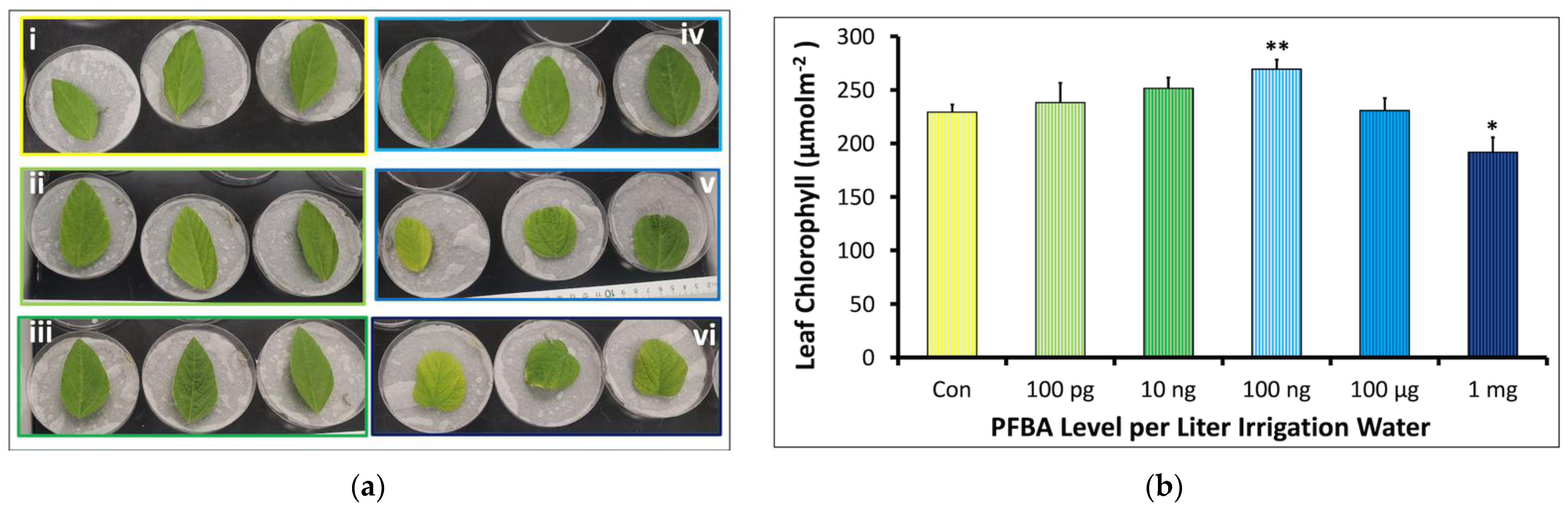
2.1.2. Leaf Morphology and Chlorophyll Content
2.1.3. Effect of PFBA on Flavonoid Content
2.2. Effect of PFBA on Stress Molecules, Antioxidant Stress Enzymes, and Metabolites
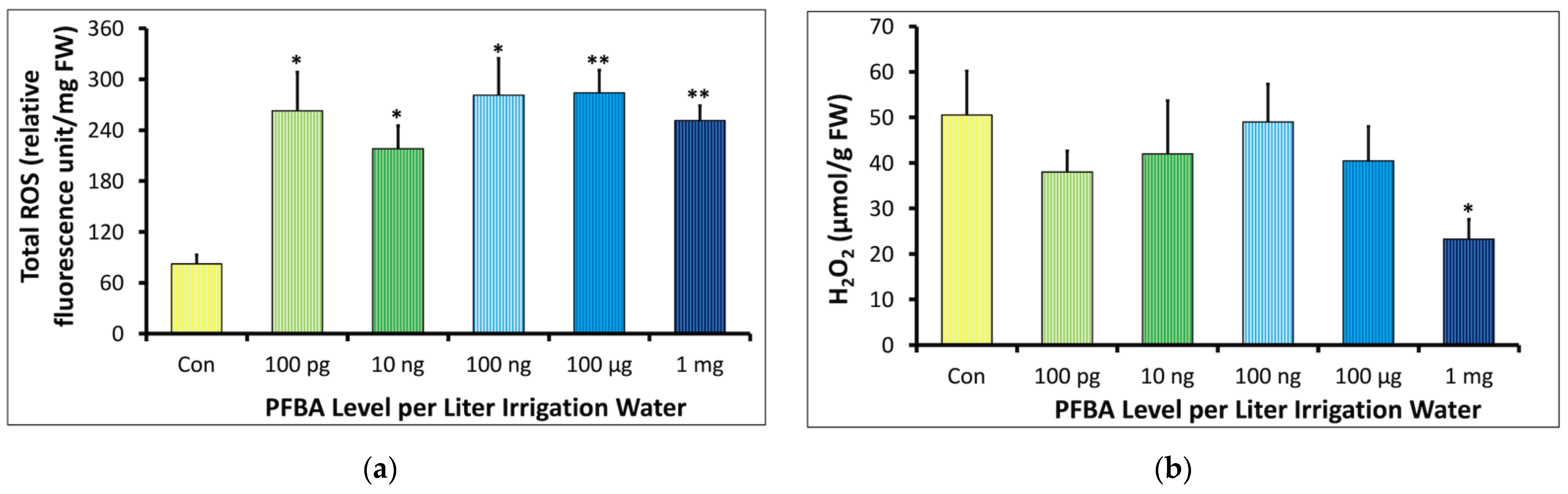
2.2.1. Total Reactive Oxygen Species (ROS) and Hydrogen Peroxide (H2O2)
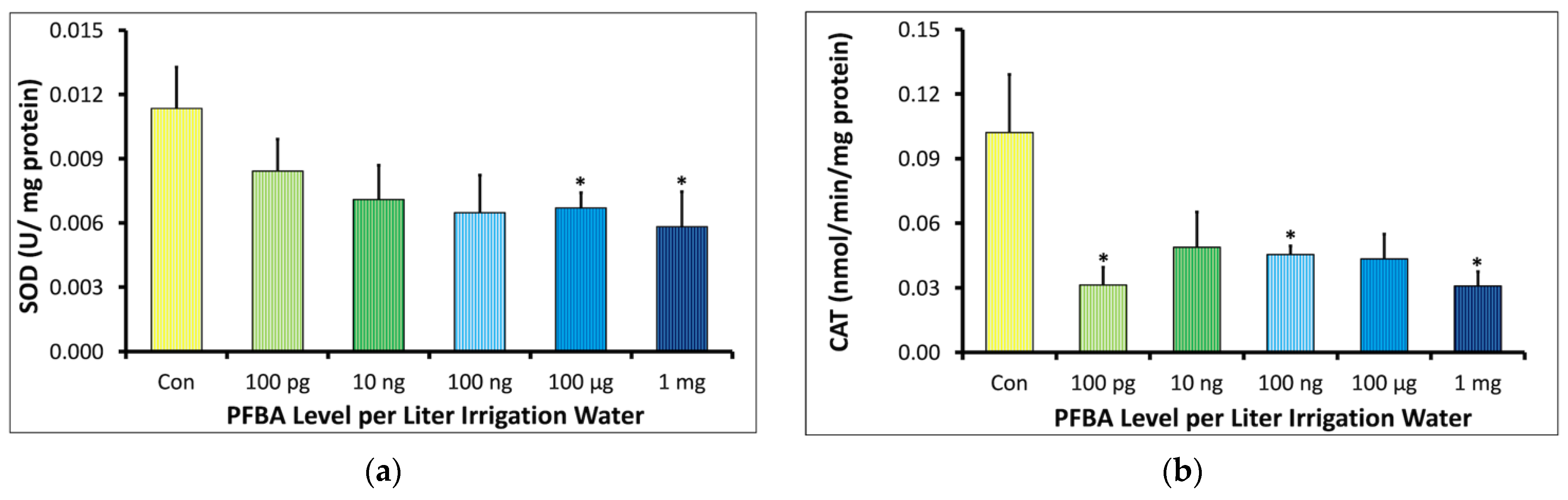
2.2.2. Superoxide Dismutase (SOD) and Catalase (CAT)
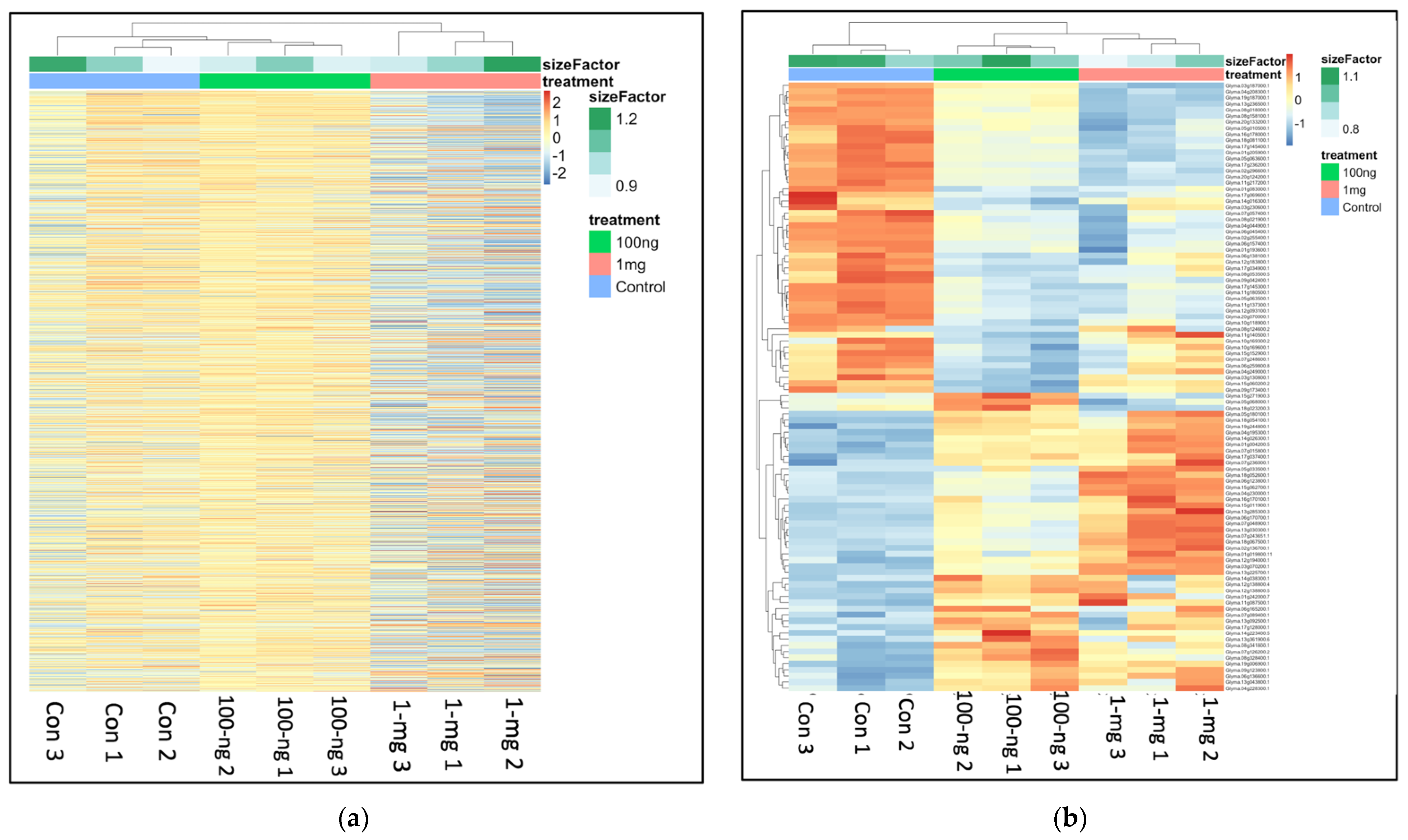
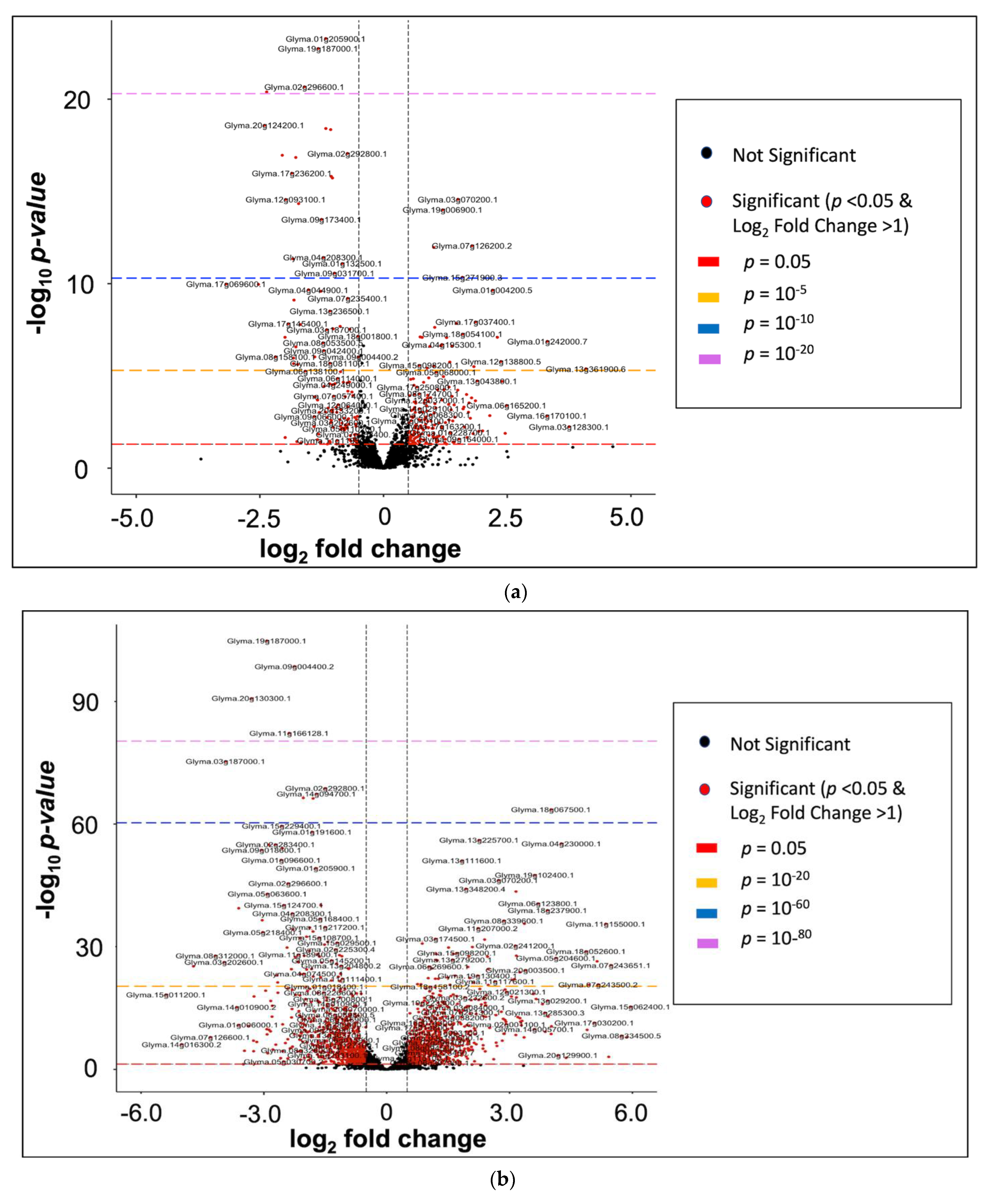
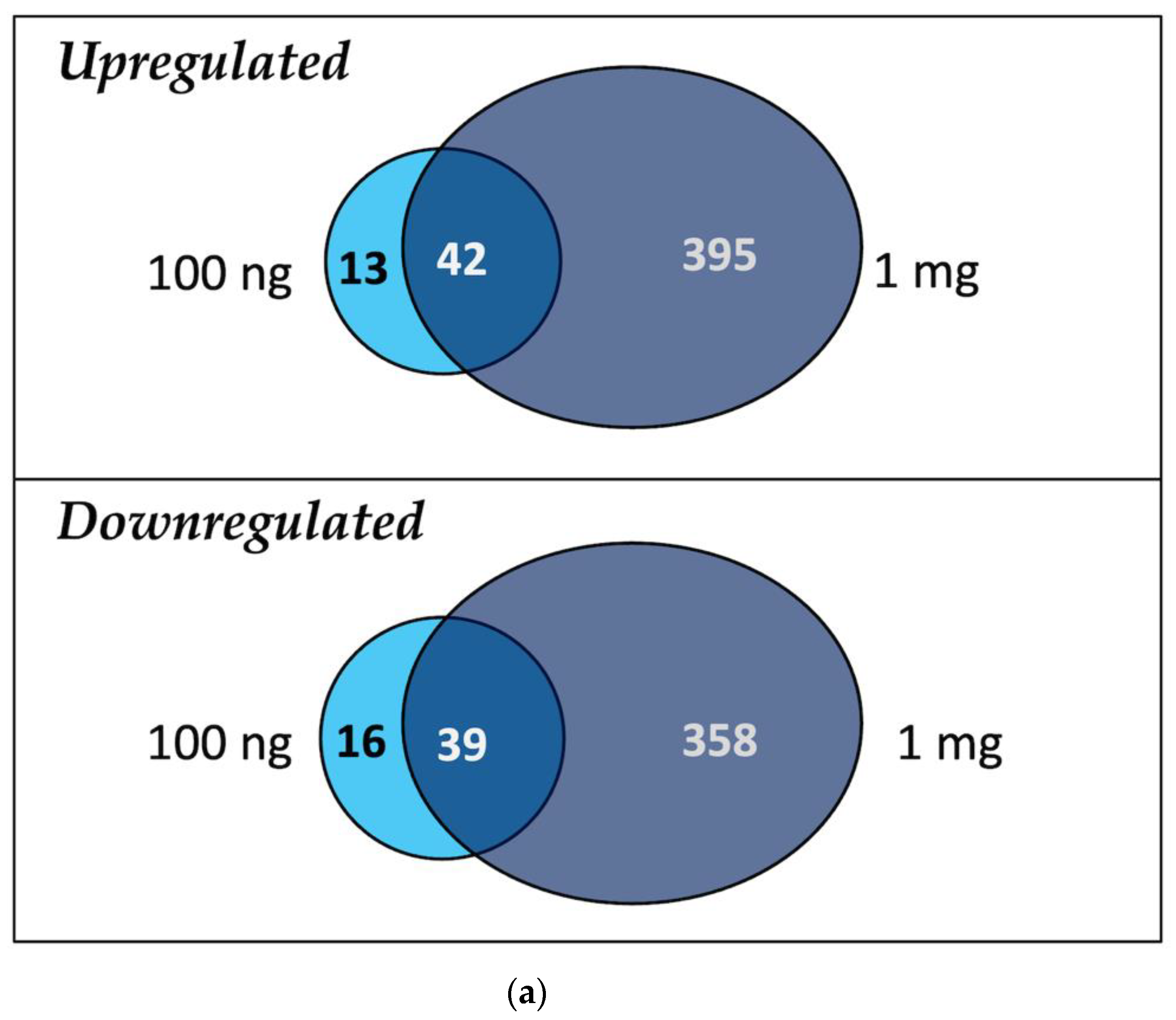
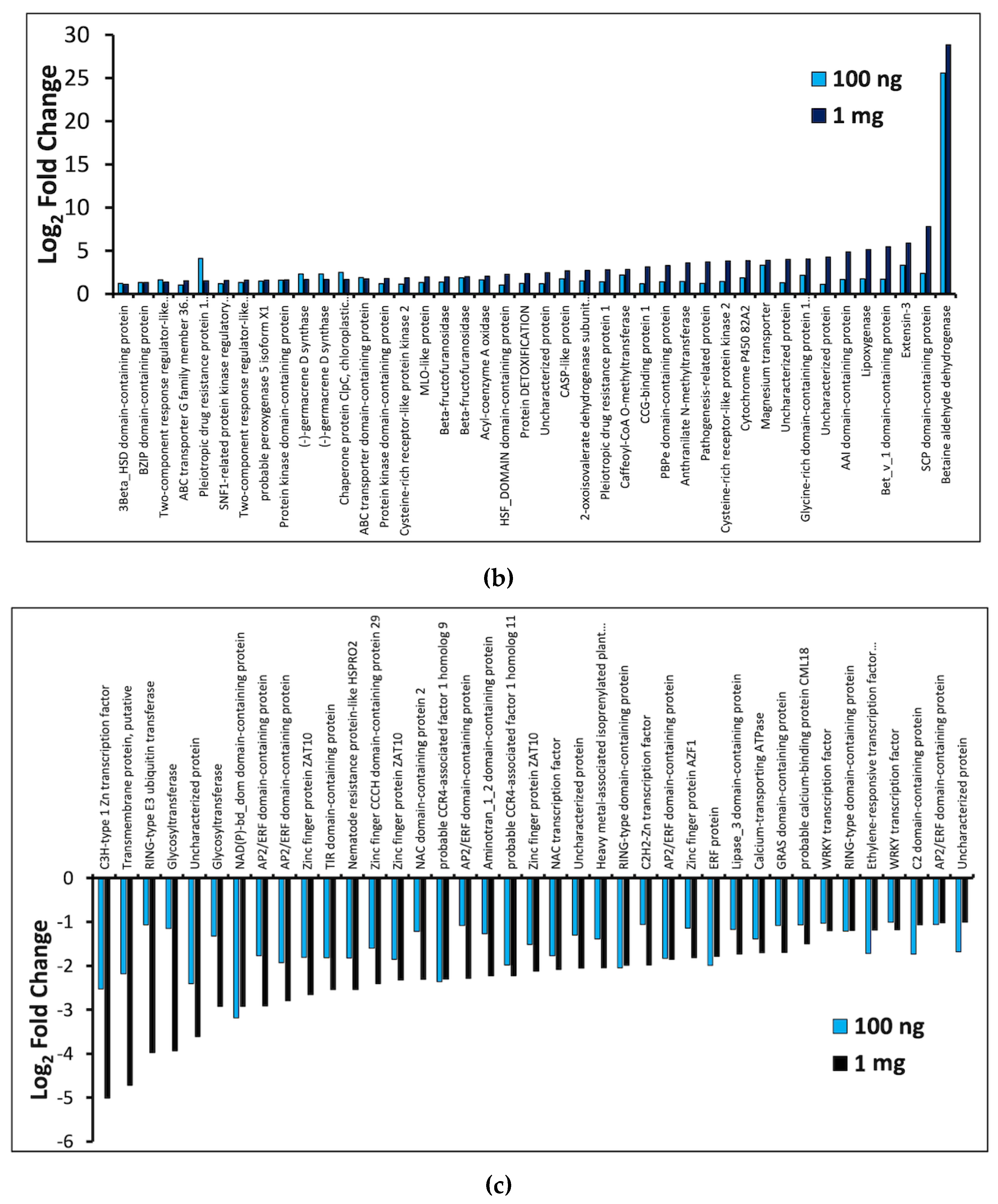
2.3. General Transcriptome and Differential Gene Expression Profile
3. Discussion
3.1. PFBA and Plant Stress
3.2. PFBA-Induced Toxicity and Stress Response Genes
3.3. PFBA and Hormesis
4. Materials and Methods
4.1. Chemicals, Standards, and Reagents
4.2. Pot Experiments
4.3. Physiological Endpoints Determination
4.4. Flavonoid Content Determination
4.5. Stress Enzymes and Metabolites Quantification
4.5.1. Total Reactive Oxygen Species
4.5.2. Superoxide Dismutase Assay
4.5.3. Hydrogen Peroxide Content
4.5.4. Catalase Assay
4.6. RNA Isolation and Sequencing
4.7. Transcriptome Analysis
4.8. Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Costello, M.C.S.; Lee, L.S. Sources, Fate, and Plant Uptake in Agricultural Systems of Per-and Polyfluoroalkyl Substances. Curr. Pollut. Rep. 2020, 1–21. [Google Scholar] [CrossRef]
- Dalahmeh, S.; Tirgani, S.; Komakech, A.J.; Niwagaba, C.B.; Ahrens, L. Per- and polyfluoroalkyl substances (PFASs) in water, soil and plants in wetlands and agricultural areas in Kampala, Uganda. Sci. Total Environ. 2018, 631–632, 660–667. [Google Scholar] [CrossRef]
- Ghisi, R.; Vamerali, T.; Manzetti, S. Accumulation of perfluorinated alkyl substances (PFAS) in agricultural plants: A review. Environ. Res. 2019, 169, 326–341. [Google Scholar] [CrossRef]
- Gobelius, L.; Lewis, J.; Ahrens, L. Plant uptake of per-and polyfluoroalkyl substances at a contaminated fire training facility to evaluate the phytoremediation potential of various plant species. Environ. Sci. Technol. 2017, 51, 12602–12610. [Google Scholar] [CrossRef]
- Vedagiri, U.K.; Anderson, R.H.; Loso, H.M.; Schwach, C.M. Ambient levels of PFOS and PFOA in multiple environmental media. Remediation 2018, 28, 9–51. [Google Scholar] [CrossRef]
- Yamazaki, E.; Taniyasu, S.; Noborio, K.; Eun, H.; Thaker, P.; Kumar, N.J.I.; Wang, X.; Yamashita, N. Accumulation of perfluoroalkyl substances in lysimeter-grown rice in Japan using tap water and simulated contaminated water. Chemosphere 2019, 231, 502–509. [Google Scholar] [CrossRef]
- Carneiro, J.M.T.; Chacón-Madrid, K.; Galazzi, R.M.; Campos, B.K.; Arruda, S.C.C.; Azevedo, R.A.; Arruda, M.A.Z. Evaluation of silicon influence on the mitigation of cadmium-stress in the development of Arabidopsis thaliana through total metal content, proteomic and enzymatic approaches. J. Trace Elem. Med. Biol. 2017, 44, 50–58. [Google Scholar] [CrossRef]
- Felizeter, S.; McLachlan, M.S.; Voogt, P.M. Root uptake and translocation of perfluorinated alkyl acids by three hydroponically grown crops. J. Agric. Food Chem. 2014, 62, 3334–3342. [Google Scholar] [CrossRef]
- García-Valcárcel, A.I.; Molero, E.; Escorial, M.C.; Chueca, M.C.; Tadeo, J.L. Uptake of perfluorinated compounds by plants grown in nutrient solution. Sci. Total Environ. 2014, 472, 20–26. [Google Scholar] [CrossRef]
- Mudumbi, J.B.N.; Ntwampe, S.K.O.; Mekuto, L.; Itoba-Tombo, E.F.; Matsha, T.E. Are aquaporins (AQPs) the gateway that conduits nutrients, persistent organic pollutants and perfluoroalkyl substances (PFASs) into plants. Springer Sci. Rev. 2017, 5, 31–48. [Google Scholar] [CrossRef]
- McLachlan, M.S.; Felizeter, S.; Klein, M.; Kotthoff, M.; Voogt, P.M. Fate of a perfluoroalkyl acid mixture in an agricultural soil studied in lysimeters. Chemosphere 2019, 223, 180–187. [Google Scholar] [CrossRef] [PubMed]
- Gredelj, A.; Polesel, F.; Trapp, S. Model-based analysis of the uptake of perfluoroalkyl acids (PFAAs) from soil into plants. Chemosphere 2020, 244, 125534. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.Q.; Wang, M.; He, Q.; Niu, X.; Liang, Y. Distribution of perfluoroalkyl substances (PFASs) in aquatic plant-based systems: From soil adsorption and plant uptake to effects on microbial community. Environ. Pollut. 2020, 257, 113575. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Ye, C.; Liu, Y.; Zhao, F. Accumulation and phytotoxicity of perfluorooctanoic acid in the model plant species Arabidopsis thaliana. Environ. Pollut. 2015, 206, 560–566. [Google Scholar] [CrossRef]
- Zhao, S.; Liang, T.; Zhou, T.; Li, D.; Wang, B.; Zhan, J.; Liu, L. Biotransformation and responses of antioxidant enzymes in hydroponically cultured soybean and pumpkin exposed to perfluorooctane sulfonamide (FOSA). Ecotoxicol. Environ. Saf. 2018, 161, 669–675. [Google Scholar] [CrossRef]
- Qu, B.; Zhao, H.; Zhou, J. Toxic effects of perfluorooctane sulfonate (PFOS) on wheat (Triticum aestivum L.) plant. Chemosphere 2010, 79, 555–560. [Google Scholar] [CrossRef]
- Zhao, S.; Fan, Z.; Sun, L.; Zhou, T.; Xing, Y.; Liu, L. Interaction effects on uptake and toxicity of perfluoroalkyl substances and cadmium in wheat (Triticum aestivum L.) and rapeseed (Brassica campestris L.) from co-contaminated soil. Ecotoxicol. Environ. Saf. 2017, 137, 194–201. [Google Scholar] [CrossRef]
- Zhou, L.; Xia, M.; Wang, L.; Mao, H. Toxic effect of perfluorooctanoic acid (PFOA) on germination and seedling growth of wheat (Triticum aestivum L.). Chemosphere 2016, 159, 420–425. [Google Scholar] [CrossRef]
- Li, P.; Sun, J.; Xie, X.; Li, Z.; Huang, B.; Zhang, G.; Li, J.; Xiao, Z. Stress response and tolerance to perfluorooctane sulfonate (PFOS) in lettuce (Lactuca sativa). J. Hazard. Mater. 2021, 404, 124213. [Google Scholar] [CrossRef]
- Li, P.; Oyang, X.; Xie, X.; Li, Z.; Yang, H.; Xi, J.; Guo, Y.; Tian, X.; Liu, B.; Li, J. Phytotoxicity induced by perfluorooctanoic acid and perfluorooctane sulfonate via metabolomics. J. Hazard. Mater. 2020, 389, 121852. [Google Scholar] [CrossRef]
- Fan, L.; Tang, J.; Zhang, D.; Ma, M.; Wang, Y.; Han, Y. Investigations on the phytotoxicity of perfluorooctanoic acid in Arabidopsis thaliana. Environ. Sci. Pollut. Res. 2020, 27, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Sun, J.; Li, P. Exposure routes, bioaccumulation and toxic effects of per-and polyfluoroalkyl substances (PFASs) on plants: A critical review. Environ. Int. 2022, 158, 106891. [Google Scholar] [CrossRef] [PubMed]
- Omagamre, E.W.; Ojo, F.; Zebelo, S.A.; Pitula, J.S. Influence of Perfluorobutanoic Acid (PFBA) on the Developmental Cycle and Damage Potential of the Beet Armyworm Spodoptera exigua (Hübner) (Insecta: Lepidoptera: Noctuidae). Arch. Environ. Contam. Toxicol. 2020, 79, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, J.; Zhang, L.; Shi, J. Toxicity assessment of perfluorinated carboxylic acids (PFCAs) towards the rotifer Brachionus calyciflorus. Sci. Total Environ. 2014, 491–492, 266–270. [Google Scholar] [CrossRef]
- Kim, W.S.; Haj-Ahmod, Y. Evaluation of Plant RNA Integrity Number (RIN) Generated Using an Agilent BioAnalyzer 2100; Application Note 80 Plant/Fungi RNA Sample Preparation; Norgen Biotek Corp.: Thorold, ON, Canada, 2014. [Google Scholar]
- Calabrese, E.J. Biphasic dose responses in biology, toxicology and medicine: Accounting for their generalizability and quantitative features. Environ. Pollut. 2013, 182, 452–460. [Google Scholar] [CrossRef]
- Cedergreen, N.; Ritz, C.; Streibig, J.C. Improved empirical models describing hormesis. Environ. Toxicol. Chem. 2005, 24, 3166–3172. [Google Scholar] [CrossRef] [PubMed]
- Hong, G.; Zhang, W.; Li, H.; Shen, X.; Guo, Z. Separate enrichment analysis of pathways for up-and downregulated genes. J. R. Soc. Interface 2014, 11, 20130950. [Google Scholar] [CrossRef] [PubMed]
- Ge, S.X.; Jung, D.; Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 2020, 36, 2628–2629. [Google Scholar] [CrossRef]
- Li, P.; Xiao, Z.; Xie, X.; Li, Z.; Yang, H.; Ma, X.; Sun, J.; Li, J. Perfluorooctanoic acid (PFOA) changes nutritional compositions in lettuce (Lactuca sativa) leaves by activating oxidative stress. Environ. Pollut. 2021, 285, 117246. [Google Scholar] [CrossRef]
- Zhang, W.; Zhang, D.; Zagorevski, D.V.; Liang, Y. Exposure of Juncus effusus to seven perfluoroalkyl acids: Uptake, accumulation and phytotoxicity. Chemosphere 2019, 233, 300–308. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, L.; Dong, F.; Gao, J.; Galbraith, D.W.; Song, C.-P. Hydrogen peroxide is involved in abscisic acid-induced stomatal closure in Vicia faba. Plant Physiol. 2001, 126, 1438–1448. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guan, Y.; Zhang, G.; Zhang, Z.; Tan, F.; Quan, X.; Chen, J. Uptake of perfluorooctane sulfonate (PFOS) by wheat (Triticum aestivum L.) plant. Chemosphere 2013, 91, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Qu, B.; Guan, Y.; Jiang, J.; Chen, X. Influence of salinity and temperature on uptake of perfluorinated carboxylic acids (PFCAs) by hydroponically grown wheat (Triticum aestivum L.). Springerplus 2016, 5, 541. [Google Scholar] [CrossRef]
- Lan, Z.; Zhou, M.; Yao, Y.; Sun, H. Plant uptake and translocation of perfluoroalkyl acids in a wheat–soil system. Environ. Sci. Pollut. Res. 2018, 25, 30907–30916. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Qin, L.; Chong, E.L.C.; Choong, T.-W.; Lee, S.K. Plant growth and photosynthetic characteristics of Mesembryanthemum crystallinum grown aeroponically under different blue-and red-LEDs. Front. Plant Sci. 2017, 8, 361. [Google Scholar] [CrossRef]
- Kirschbaum, M.U.F. Does enhanced photosynthesis enhance growth? Lessons learned from CO2 enrichment studies. Plant Physiol. 2011, 155, 117–124. [Google Scholar] [CrossRef] [PubMed]
- Ferreyra, M.L.F.; Serra, P.; Casati, P. Recent advances on the roles of flavonoids as plant protective molecules after UV and high light exposure. Physiol. Plant. 2021, 173, 736–749. [Google Scholar] [CrossRef] [PubMed]
- Dao, T.T.H.; Linthorst, H.J.M.; Verpoorte, R. Chalcone synthase and its functions in plant resistance. Phytochem. Rev. 2011, 10, 397–412. [Google Scholar] [CrossRef]
- Golestan Hashemi, F.S.; Ismail, M.R.; Rafii, M.Y.; Aslani, F.; Miah, G.; Muharam, F.M. Critical multifunctional role of the betaine aldehyde dehydrogenase gene in plants. Biotechnol. Biotechnol. Equip. 2018, 32, 815–829. [Google Scholar] [CrossRef]
- Yu, H.; Zhou, X.; Wang, Y.; Zhou, S.; Fu, F.; Li, W. A betaine aldehyde dehydrogenase gene from Ammopiptanthus nanus enhances tolerance of Arabidopsis to high salt and drought stresses. Plant Growth Regul. 2017, 83, 265–276. [Google Scholar] [CrossRef]
- Fouad, A.; Hegazy, A.E.; Azab, E.; Khojah, E.; Kapiel, T. Boosting of Antioxidants and Alkaloids in Catharanthus roseus Suspension Cultures Using Silver Nanoparticles with Expression of CrMPK3 and STR Genes. Plants 2021, 10, 2202. [Google Scholar] [CrossRef] [PubMed]
- Matsuura, H.N.; Rau, M.R.; Fett-Neto, A.G. Oxidative stress and production of bioactive monoterpene indole alkaloids: Biotechnological implications. Biotechnol. Lett. 2014, 36, 191–200. [Google Scholar] [CrossRef] [PubMed]
- Saleem, S.; Mushtaq, N.U.; Shah, W.H.; Rasool, A.; Hakeem, K.R.; Rehman, R. Beneficial Role of Phytochemicals in Oxidative Stress Mitigation in Plants. In Antioxidant Defense in Plants; Springer: Singapore, 2022; pp. 435–451. [Google Scholar]
- Vera-Reyes, I.; Huerta-Heredia, A.A.; Ponce-Noyola, T.; Cerda-Garcia-Rojas, C.M.; Trejo-Tapia, G.; Ramos-Valdivia, A.C. Monoterpenoid indole alkaloids and phenols are required antioxidants in glutathione depleted Uncaria tomentosa root cultures. Front. Environ. Sci. 2015, 3, 27. [Google Scholar] [CrossRef]
- Vanderauwera, S.; Zimmermann, P.; Rombauts, S.; Vandenabeele, S.; Langebartels, C.; Gruissem, W.; Inzé, D.; Van Breusegem, F. Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 2005, 139, 806–821. [Google Scholar] [CrossRef]
- Wu, A.; Allu, A.D.; Garapati, P.; Siddiqui, H.; Dortay, H.; Zanor, M.-I.; Asensi-Fabado, M.A.; Munne-Bosch, S.; Antonio, C.; Tohge, T. JUNGBRUNNEN1, a reactive oxygen species–responsive NAC transcription factor, regulates longevity in Arabidopsis. Plant Cell 2012, 24, 482–506. [Google Scholar] [CrossRef]
- Gadjev, I.; Vanderauwera, S.; Gechev, T.S.; Laloi, C.; Minkov, I.N.; Shulaev, V.; Apel, K.; Inzé, D.; Mittler, R.; Van Breusegem, F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006, 141, 436–445. [Google Scholar] [CrossRef]
- Li, R.; Tang, T.; Qiao, W.; Huang, J. Toxic effect of perfluorooctane sulfonate on plants in vertical-flow constructed wetlands. J. Environ. Sci. 2020, 92, 176–186. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, X.; Zheng, X.; Yang, M.; Gao, X.; Huang, J.; Zhang, L.; Fan, Z. Toxic effects and mechanisms of PFOA and its substitute GenX on the photosynthesis of Chlorella pyrenoidosa. Sci. Total Environ. 2021, 765, 144431. [Google Scholar] [CrossRef]
- Dodd, A.N.; Salathia, N.; Hall, A.; Kévei, E.; Tóth, R.; Nagy, F.; Hibberd, J.M.; Millar, A.J.; Webb, A.A.R. Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science 2005, 309, 630–633. [Google Scholar] [CrossRef]
- Graf, A.; Schlereth, A.; Stitt, M.; Smith, A.M. Circadian control of carbohydrate availability for growth in Arabidopsis plants at night. Proc. Acad. Nat. Sci. USA 2010, 107, 9458–9463. [Google Scholar] [CrossRef] [Green Version]
- Graf, A.; Smith, A.M. Starch and the clock: The dark side of plant productivity. Trends Plant Sci. 2011, 16, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Linde, A.M.; Eklund, D.M.; Kubota, A.; Perderson, E.R.A.; Holm, K.; Gyllenstrand, N.; Nishihama, R.; Cronberg, N.; Muranaka, T.; Oyama, T. Early evolution of the land plant circadian clock. New Phytol. 2017, 216, 576–590. [Google Scholar] [CrossRef]
- Srivastava, D.; Shamim, M.; Kumar, M.; Mishra, A.; Maurya, R.; Sharma, D.; Pandey, P.; Singh, K.N. Role of circadian rhythm in plant system: An update from development to stress response. Environ Exp. Bot. 2019, 162, 256–271. [Google Scholar] [CrossRef]
- Covington, M.F.; Maloof, J.N.; Straume, M.; Kay, S.A.; Harmer, S.L. Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol. 2008, 9, R130. [Google Scholar] [CrossRef]
- Grundy, J.; Stoker, C.; Carré, I.A. Circadian regulation of abiotic stress tolerance in plants. Front. Plant Sci. 2015, 6, 648. [Google Scholar] [CrossRef] [PubMed]
- Habte, E.; Müller, L.M.; Shtaya, M.; Davis, S.J.; Von Korf, M. Osmotic stress at the barley root affects expression of circadian clock genes in the shoot. Plant 2014, 37, 1321–1337. [Google Scholar] [CrossRef] [PubMed]
- Marcolino-Gomes, J.; Rodrigues, F.A.; Fuganti-Pagliarini, R.B.C.; Nakayama, T.J.; Celaya, B.; Molinari, H.B.C.; Nevesde Oliveira, M.C.; Harmon, F.G.; Nepomuceno, A. Diurnal oscillations of soybean circadian clock and drought responsive genes. PLoS ONE 2014, 9, e86402. [Google Scholar] [CrossRef]
- Li, M.; Cao, L.; Mwimba, M.; Zhou, Y.; Li, L.; Zhou, M.; Schnable, P.S.; O’Rourke, J.A.; Dong, X.; Wang, W. Comprehensive mapping of abiotic stress inputs into the soybean circadian clock. Pro. Acad. Nat. Sci. USA 2019, 116, 23840–23849. [Google Scholar] [CrossRef]
- Chaves, I.; Pokorny, R.; Byrdin, M.; Hoang, N.; Ritz, T.; Brettel, K.; Essen, L.; van der Horst, G.T.J.; Batschauer, A.; Ahmad, M. The cryptochromes: Blue light photoreceptors in plants and animals. Ann. Rev. Plant Biol. 2011, 62, 335–364. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Liu, B.; Zhao, C.; Pepper, M.; Lin, C. The action mechanisms of plant cryptochromes. Trends Plant Sci. 2011, 16, 684–691. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Liu, H.; Klejnot, J.; Lin, C. The cryptochrome blue-light receptors. Arab. Book 2010, 8, e0135. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Su, T.; He, W.; Ren, H.; Liu, S.; Chen, Y.; Gao, L.; Hu, X.; Lu, H.; Cao, S.; et al. Photooligomerization determines photosensitivity and photoreactivity of plant cryptochromes. Mol. Plant. 2020, 13, 398–413. [Google Scholar] [CrossRef] [PubMed]
- Jiao, L.; Wang, L.; Qiu, Z.; Wang, Q.; Zhou, Q.; Huang, X. Effects of bisphenol A on chlorophyll synthesis in soybean seedlings. Environ. Sci. Pollut. Res. 2015, 22, 5877–5886. [Google Scholar] [CrossRef] [PubMed]
- Ghasemi, K.; Ghasemi, Y.; Ebrahimzadeh, M.A. Antioxidant activity, phenol and flavonoid contents of 13 citrus species peels and tissues. Pak. J. Pharm. Sci. 2009, 22, 277–281. [Google Scholar]
- Jambunathan, N. Determination and detection of reactive oxygen species (ROS), lipid peroxidation, and electrolyte leakage in plants. In Plant Stress Tolerance; Springer: Berlin/Heidelberg, Germany, 2010; pp. 291–297. [Google Scholar]
- Lara-Nunez, A.; Romero-Romero, T.; Ventura, J.L.; Blancas, V.; Anaya, A.L.; Cruz-Ortega, R. Allelochemical stress causes inhibition of growth and oxidative damage in Lycopersicon esculentum Mill. Plant Cell Environ. 2006, 29, 2009–2016. [Google Scholar] [CrossRef]
- Aebi, H. Catalase in vitro. In Methods in Enzymology 105; Elsevier: Amsterdam, The Netherlands, 1984; pp. 121–126. [Google Scholar]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Team, R.S. RStudio: Integrated Development Environment for R; RStudio, PBC: Boston, MA, USA, 2020; Volume 2021. [Google Scholar]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Grant, D.; Nelson, R.T.; Cannon, S.B.; Shoemaker, R.C. SoyBase, the USDA-ARS soybean genetics and genomics database. Nucleic Acids Res. 2010, 38, D843–D846. [Google Scholar] [CrossRef]
- Ritz, C.; Strebig, J.C.; Ritz, M.C. Package ‘drc’; Creative Commons: Mountain View, CA, USA, 2016. [Google Scholar]
- Brain, P.; Cousens, R. An equation to describe dose responses where there is stimulation of growth at low doses. Weed Res. 1989, 29, 93–96. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Sato, Y.; Ishiguro-Watanabe, M.; Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 2021, 49, D545–D551. [Google Scholar] [CrossRef]
- Filichkin, S.A.; Cumbie, J.S.; Dharmawardhana, P.; Pankaj, J.P.; Chang, J.H.; Palusa, S.G.; Reddy, A.S.N.; Megraw, M.; Mockler, T.C. Environmental stresses modulate abundance and timing of alternatively spliced circadian transcripts in Arabidopsis. Mol. Plant. 2015, 8, 207–227. [Google Scholar] [CrossRef] [Green Version]
- James, A.B.; Syed, N.H.; Bordage, S.; Marshall, J.; Nimmo, G.A.; Jenkins, G.I.; Herzyk, P.; Brown, J.W.; Nimmo, H.G. Alternative Splicing Mediates Responses of the Arabidopsis Circadian Clock to Temperature Changes. Plant Cell 2012, 24, 961–981. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.J.; Park, M.J.; Kim, S.G.; Baldwin, I.T.; Chung-Mo, P. Alternative splicing and nonsense-mediated decay of circadian clock genes under environmental stress conditions in Arabidopsis. BMC Plant Biol. 2014, 14, 136. [Google Scholar] [CrossRef] [PubMed]
- Agathokleous, E.; Kitao, M.; Calabrese, E.J. Biphasic effect of abscisic acid on plants: An hormetic viewpoint. Botany 2018, 96, 637–642. [Google Scholar] [CrossRef]
- Cedergreen, N.; Streibig, J.C.; Kudsk, P.; Mathiassen, S.K.; Duke, S.O. The occurrence of hormesis in plants and algae. Dose-Response 2007, 5, 150–162. [Google Scholar] [CrossRef] [PubMed]
- Gressel, J.; Dodds, J. Commentary: Hormesis can be used in enhancing plant productivity and health; but not as previously envisaged. Plant Sci. 2013, 213, 123–127. [Google Scholar] [CrossRef]
- Huseynova, I.M. Photosynthetic characteristics and enzymatic antioxidant capacity of leaves from wheat cultivars exposed to drought. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Song, N.H.; Le Yin, X.; Chen, G.F.; Yang, H. Biological responses of wheat (Triticum aestivum) plants to the herbicide chlorotoluron in soils. Chemosphere 2007, 68, 1779–1787. [Google Scholar] [CrossRef]
- Calabrese, E.J.; Blain, R.B. The hormesis database: The occurrence of hormetic dose responses in the toxicological literature. Regul. Toxicol. Pharmacol. 2011, 61, 73–81. [Google Scholar] [CrossRef]
- Belz, R.G.; Piepho, H. Statistical modeling of the hormetic dose zone and the toxic potency completes the quantitative description of hormetic dose responses. Environ. Toxicol. and Chem. 2015, 34, 1169–1177. [Google Scholar] [CrossRef]
- Belz, R.G.; Piepho, H.P. Interspecies variability of plant hormesis by the antiauxin PCIB in a laboratory bioassay. J. Plant Growth Regul. 2014, 33, 499–512. [Google Scholar] [CrossRef]
- Belz, R.G.; Piepho, H.P. Variability of hormetic dose responses of the antiauxin PCIB on L actuca sativa in a plant bioassay. Weed Res. 2013, 53, 418–428. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Omagamre, E.W.; Mansourian, Y.; Liles, D.; Tolosa, T.; Zebelo, S.A.; Pitula, J.S. Perfluorobutanoic Acid (PFBA) Induces a Non-Enzymatic Oxidative Stress Response in Soybean (Glycine max L. Merr.). Int. J. Mol. Sci. 2022, 23, 9934. https://doi.org/10.3390/ijms23179934
Omagamre EW, Mansourian Y, Liles D, Tolosa T, Zebelo SA, Pitula JS. Perfluorobutanoic Acid (PFBA) Induces a Non-Enzymatic Oxidative Stress Response in Soybean (Glycine max L. Merr.). International Journal of Molecular Sciences. 2022; 23(17):9934. https://doi.org/10.3390/ijms23179934
Chicago/Turabian StyleOmagamre, Eguono W., Yeganeh Mansourian, Diamond Liles, Tigist Tolosa, Simon A. Zebelo, and Joseph S. Pitula. 2022. "Perfluorobutanoic Acid (PFBA) Induces a Non-Enzymatic Oxidative Stress Response in Soybean (Glycine max L. Merr.)" International Journal of Molecular Sciences 23, no. 17: 9934. https://doi.org/10.3390/ijms23179934
APA StyleOmagamre, E. W., Mansourian, Y., Liles, D., Tolosa, T., Zebelo, S. A., & Pitula, J. S. (2022). Perfluorobutanoic Acid (PFBA) Induces a Non-Enzymatic Oxidative Stress Response in Soybean (Glycine max L. Merr.). International Journal of Molecular Sciences, 23(17), 9934. https://doi.org/10.3390/ijms23179934





