Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species
Abstract
:1. Introduction
2. Results
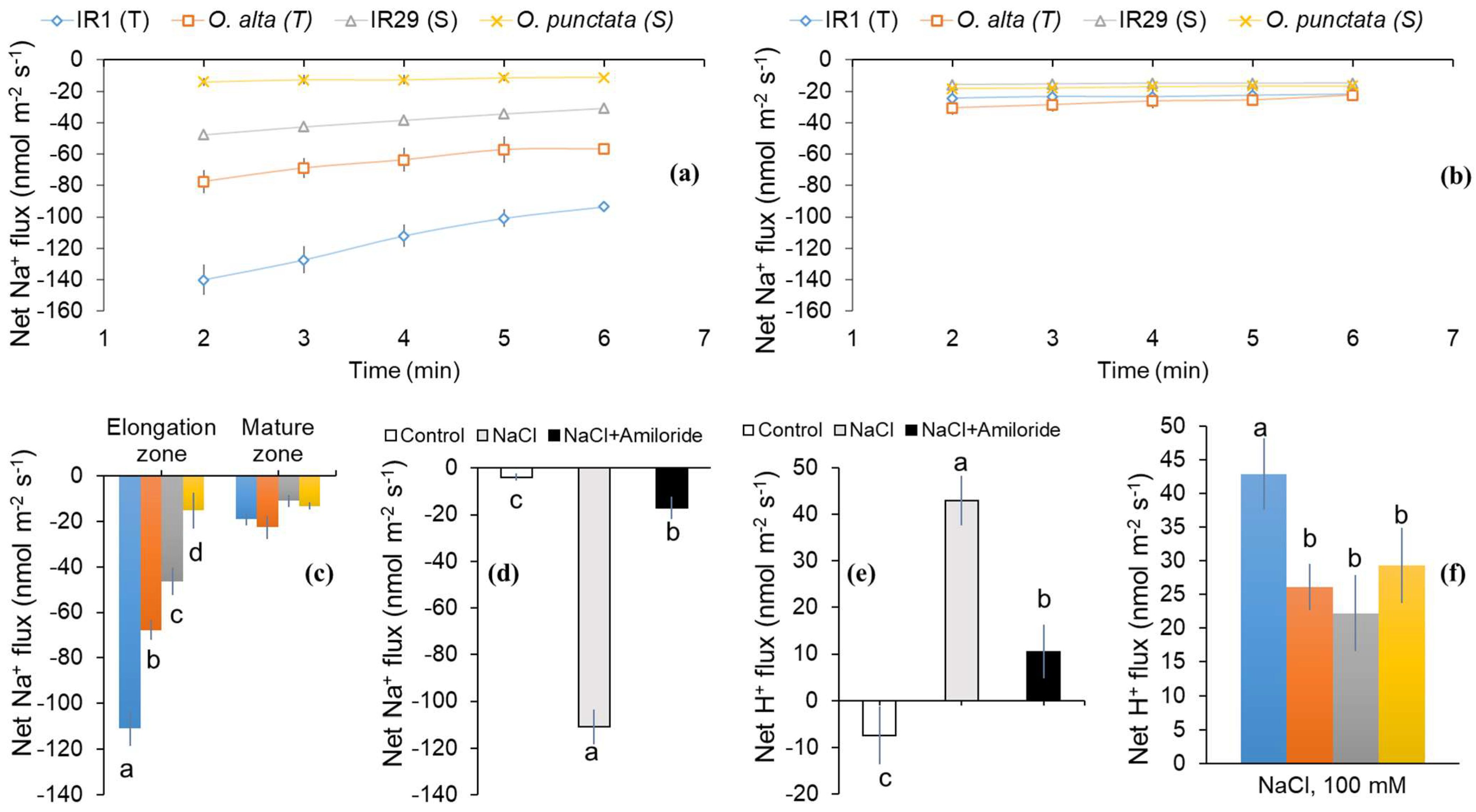
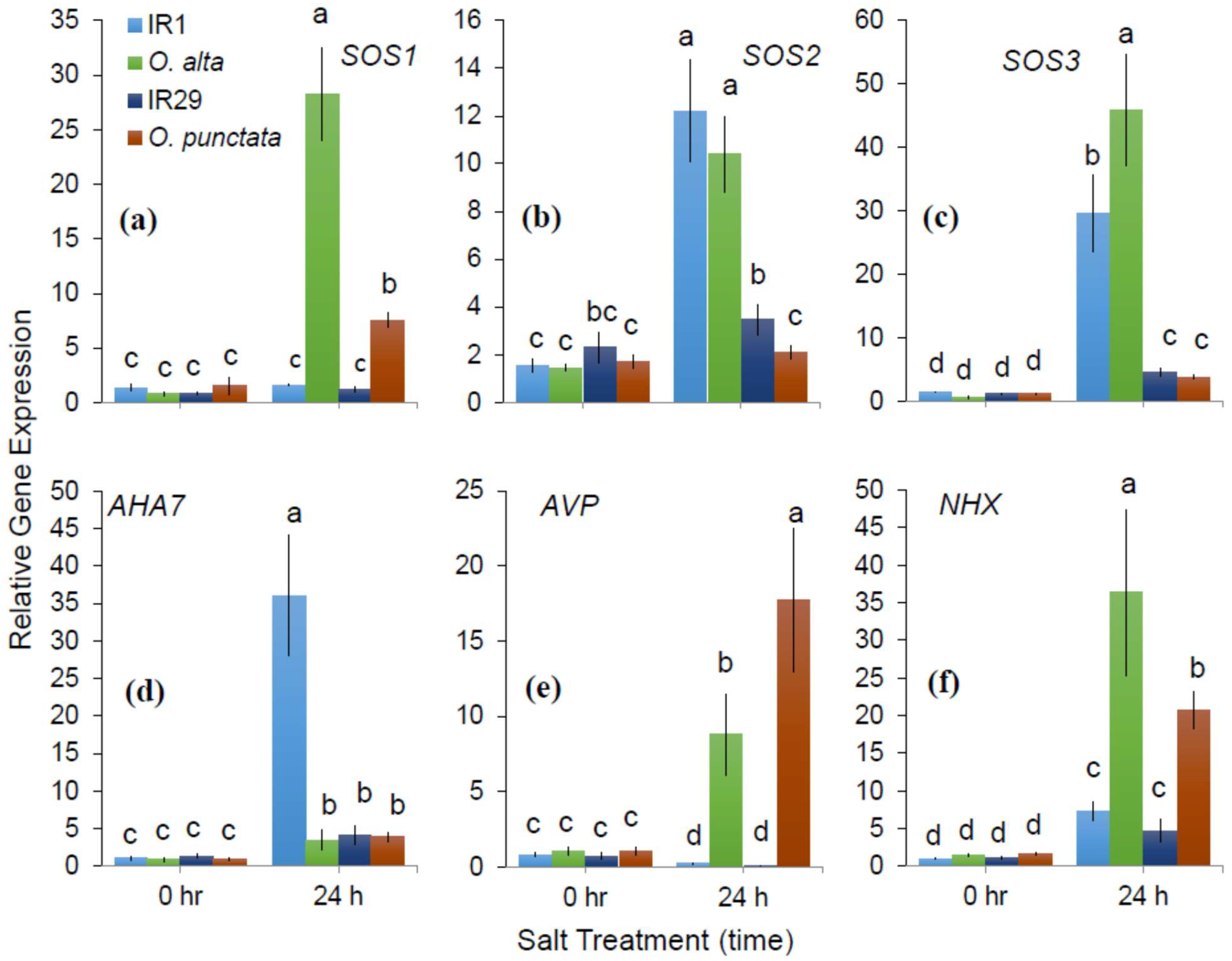
2.1. Differences in SOS1-Mediated Na+ Exclusion between the Cultivated and Wild Rice Species
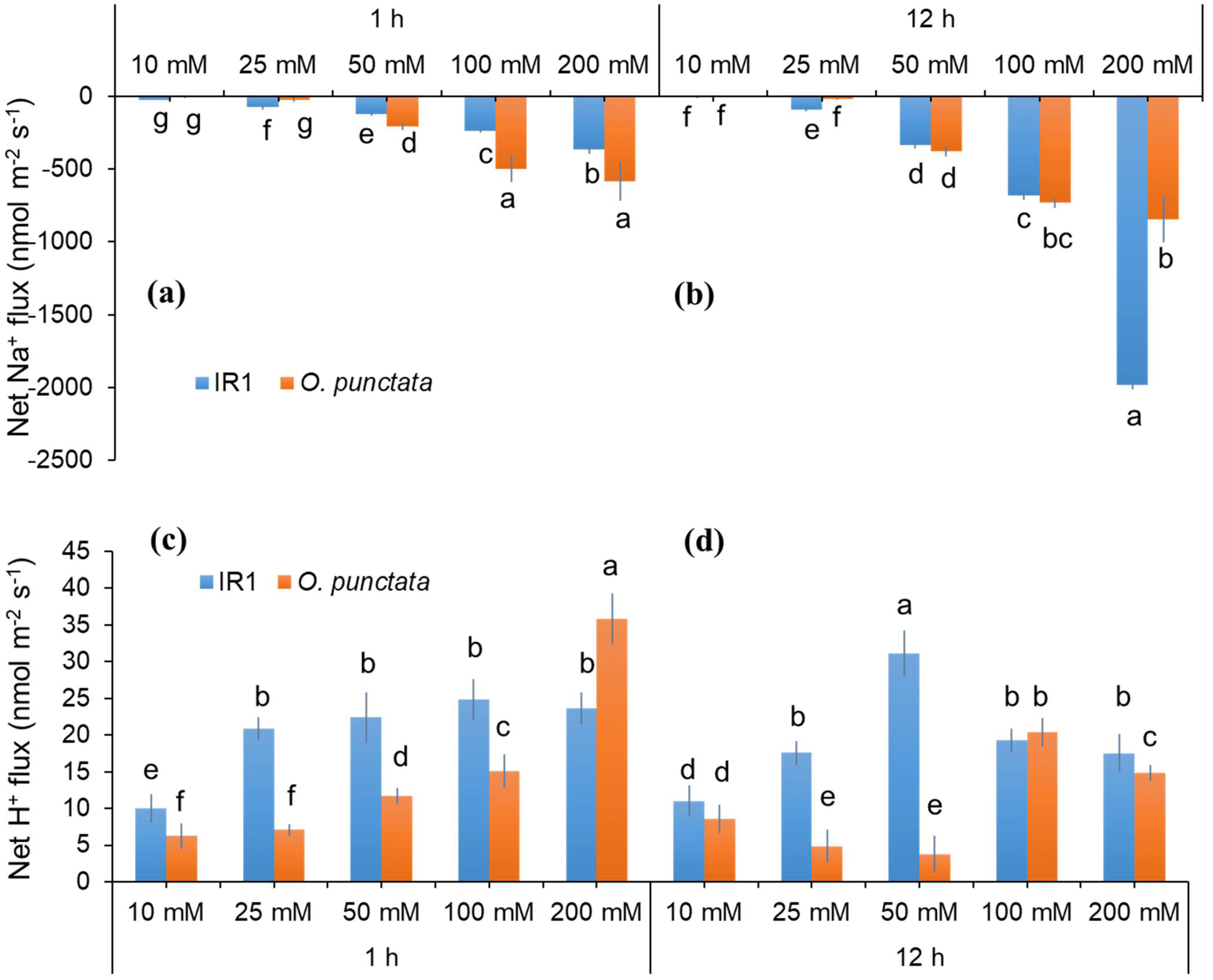
2.2. Dose- and Time-Dependency of Cellular Na+ Exclusion in Contrasting Rice Genotypes
3. Discussion
3.1. Dose- and Time-Dependent Na+ Efflux of Contrasting Pairs of Rice Species
3.2. The Role of SOS Activity in Conferring Salinity Stress Tolerance in Rice Species
4. Materials and Methods
4.1. Plant Material and Growth Conditions
4.2. Non-Invasive ion Flux Measurements (MIFE)
4.3. Net Na+ and H+ Flux Measurements Using “Recovery Protocols”
4.4. Quantitative Real Time PCR
4.5. Dose- and Time-Dependence of Na+ Exclusion
4.6. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J.; Drechsel, P.; Noble, A.D. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Shabala, S.; Pottosin, I. Regulation of potassium transport in plants under hostile conditions: Implications for abiotic and biotic stress tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, B.; Rehman, A.; Tanveer, M.; Wang, L.; Park, S.K.; Ali, A. Salt stress in brassica: Effects, tolerance mechanisms, and management. J. Plant Growth Regul. 2022, 41, 781–795. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Blumwald, E. Developing salt-tolerant crop plants: Challenges and opportunities. Trends Plant Sci. 2005, 10, 615–620. [Google Scholar] [CrossRef] [PubMed]
- Shahbaz, M.; Ashraf, M. Improving salinity tolerance in cereals. Crit. Rev. Plant Sci. 2013, 32, 237–249. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Rahnama, A.; James, R.A.; Poustini, K.; Munns, R. Stomatal conductance as a screen for osmotic stress tolerance in durum wheat growing in saline soil. Funct. Plant Biol. 2010, 37, 255–263. [Google Scholar] [CrossRef]
- Shahzad, B.; Fahad, S.; Tanveer, M.; Saud, S.; Khan, I.A. Plant responses and tolerance to salt stress. In Approaches for Enhancing Abiotic Stress Tolerance in Plants; CRC Press: Boca Raton, FL, USA, 2019; pp. 61–78. [Google Scholar]
- Zhao, C.; Zhang, H.; Song, C.; Zhu, J.-K.; Shabala, S. Mechanisms of plant responses and adaptation to soil salinity. Innovation 2020, 1, 100017. [Google Scholar] [CrossRef]
- Cuin, T.A.; Bose, J.; Stefano, G.; Jha, D.; Tester, M.; Mancuso, S.; Shabala, S. Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: In planta quantification methods. Plant Cell Environ. 2011, 34, 947–961. [Google Scholar] [CrossRef]
- Bose, J.; Shabala, L.; Pottosin, I.; Zeng, F.; Velarde-Buendía, A.-M.; Massart, A.; Poschenrieder, C.; Hariadi, Y.; Shabala, S. Kinetics of xylem loading, membrane potential maintenance, and sensitivity of K+-permeable channels to reactive oxygen species: Physiological traits that differentiate salinity tolerance between pea and barley. Plant Cell Environ. 2014, 37, 589–600. [Google Scholar] [CrossRef]
- Munns, R.; Passioura, J.B.; Colmer, T.D.; Byrt, C.S. Osmotic adjustment and energy limitations to plant growth in saline soil. New Phytol. 2020, 225, 1091–1096. [Google Scholar] [CrossRef] [PubMed]
- Prusty, M.R.; Kim, S.-R.; Vinarao, R.; Entila, F.; Egdane, J.; Diaz, M.G.Q.; Jena, K.K. Newly identified wild rice accessions conferring high salt tolerance might use a tissue tolerance mechanism in leaf. Front. Plant Sci. 2018, 9, 417. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Shabala, S.; Shabala, L.; Zhou, M.; Meinke, H.; Venkataraman, G.; Chen, Z.; Zeng, F.; Zhao, Q. Tissue-specific regulation of Na+ and K+ transporters explains genotypic differences in salinity stress tolerance in rice. Front. Plant Sci. 2019, 10, 1361. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Mondal, S.; Ray, S.; Samal, P.; Pradhan, B.; Chattopadhyay, K.; Kar, M.K.; Swain, P.; Sarkar, R.K. Tissue tolerance coupled with ionic discrimination can potentially minimize the energy cost of salinity tolerance in rice. Front. Plant Sci. 2020, 11, 265. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z. The importance of Cl− exclusion and vacuolar Cl− sequestration: Revisiting the role of Cl− transport in plant salt tolerance. Front. Plant Sci. 2019, 10, 1418. [Google Scholar] [CrossRef]
- Martínez-Atienza, J.; Jiang, X.; Garciadeblas, B.; Mendoza, I.; Zhu, J.-K.; Pardo, J.M.; Quintero, F.J. Conservation of the Salt Overly Sensitive Pathway in rice. Plant Physiol. 2006, 143, 1001–1012. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.-T.; Venkataraman, G.; Milham, P.; Zhou, M.; Shabala, L.; Holford, P.; Shabala, S.; Chen, Z.-H. Sodium sequestration confers salinity tolerance in an ancestral wild rice. Physiol. Plant. 2021, 172, 1594–1608. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.-T.; Zhou, M.; Venkataraman, G.; Shabala, L.; Holford, P.; Shabala, S.; Chen, Z.-H. Evolutionary Significance of NHX Family and NHX1 in Salinity Stress Adaptation in the Genus Oryza. Int. J. Mol. Sci. 2022, 23, 2092. [Google Scholar] [CrossRef]
- Blumwald, E.; Aharon, G.S.; Apse, M.P. Sodium transport in plant cells. Biochim. Et Biophys. Acta-Biomembr. 2000, 1465, 140–151. [Google Scholar] [CrossRef]
- Apse, M.P.; Blumwald, E. Na+ transport in plants. FEBS Lett. 2007, 581, 2247–2254. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Lee, B.-h.; Wu, S.-J.; Zhu, J.-K. Overexpression of a plasma membrane Na+/H+ antiporter gene improves salt tolerance in Arabidopsis thaliana. Nat. Biotechnol. 2003, 21, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Nublat, A.; Desplans, J.; Casse, F.; Berthomieu, P. Sas1, an Arabidopsis mutant overaccumulating sodium in the shoot, shows deficiency in the control of the root radial transport of sodium. Plant Cell 2001, 13, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Ranathunge, K.; Franke, R.; Prakash, H.S.; Schreiber, L.; Mathew, M.K. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 2009, 230, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Faiyue, B.; Vijayalakshmi, C.; Nawaz, S.; Nagato, Y.; Taketa, S.; Ichii, M.; Al-Azzawi, M.J.; Flowers, T.J. Studies on sodium bypass flow in lateral rootless mutants lrt1 and lrt2, and crown rootless mutant crl1 of rice (Oryza sativa L.). Plant Cell Environ. 2010, 33, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Faiyue, B.; Al-Azzawi, M.J.; Flowers, T.J. The role of lateral roots in bypass flow in rice (Oryza sativa L.). Plant Cell Environ. 2010, 33, 702–716. [Google Scholar] [CrossRef]
- El Mahi, H.; Pérez-Hormaeche, J.; De Luca, A.; Villalta, I.; Espartero, J.; Gámez-Arjona, F.; Fernández, J.L.; Bundó, M.; Mendoza, I.; Mieulet, D. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 2019, 180, 1046–1065. [Google Scholar] [CrossRef]
- Reddy, I.N.B.L.; Kim, B.-K.; Yoon, I.-S.; Kim, K.-H.; Kwon, T.-R. Salt tolerance in rice: Focus on mechanisms and approaches. Rice Sci. 2017, 24, 123–144. [Google Scholar] [CrossRef]
- Wang, Z.; Hong, Y.; Li, Y.; Shi, H.; Yao, J.; Liu, X.; Wang, F.; Huang, S.; Zhu, G.; Zhu, J.-K. Natural variations in SlSOS1 contribute to the loss of salt tolerance during tomato domestication. Plant Biotechnol. J. 2021, 19, 20–22. [Google Scholar] [CrossRef]
- Solis, C.A.; Yong, M.T.; Vinarao, R.; Jena, K.; Holford, P.; Shabala, L.; Zhou, M.; Shabala, S.; Chen, Z.-H. Back to the wild: On a quest for donors toward salinity tolerant rice. Front. Plant Sci. 2020, 11, 323. [Google Scholar] [CrossRef]
- Shabala, S.; Shabala, S.; Cuin, T.A.; Pang, J.; Percey, W.; Chen, Z.; Conn, S.; Eing, C.; Wegner, L.H. Xylem ionic relations and salinity tolerance in barley. Plant J. 2010, 61, 839–853. [Google Scholar] [CrossRef]
- Zhu, M.; Zhou, M.; Shabala, L.; Shabala, S. Physiological and molecular mechanisms mediating xylem Na+ loading in barley in the context of salinity stress tolerance. Plant Cell Environ. 2017, 40, 1009–1020. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, B.; Yun, P.; Rasouli, F.; Shabala, L.; Zhou, M.; Venkataraman, G.; Chen, Z.-H.; Shabala, S. Root K+ Homeostasis and Signalling as a Determinant of Salinity Stress Tolerance in Cultivated and Wild Rice Species. Environ. Exp. Bot. 2022, 21, 104944. [Google Scholar] [CrossRef]
- Shi, H.; Ishitani, M.; Kim, C.; Zhu, J.-K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 2000, 97, 6896–6901. [Google Scholar] [CrossRef] [PubMed]
- Shi, H.; Quintero, F.J.; Pardo, J.M.; Zhu, J.-K. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 2002, 14, 465–477. [Google Scholar] [CrossRef]
- Wu, H.; Zhang, X.; Giraldo, J.P.; Shabala, S. It is not all about sodium: Revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil 2018, 431, 1–17. [Google Scholar] [CrossRef]
- Putney, L.K.; Denker, S.P.; Barber, D.L. The changing face of the Na+/H+ exchanger, NHE1: Structure, regulation, and cellular actions. Annu. Rev. Pharmacol. Toxicol. 2002, 42, 527–552. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Zhou, M.; Su, N.; Wu, Q.; Ul-Haq, T.; Zhu, J.; Mancuso, S.; Azzarello, E.; Shabala, S. Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J. 2019, 100, 55–67. [Google Scholar] [CrossRef]
- Shabala, S.; Cuin, T.A. Potassium transport and plant salt tolerance. Physiol. Plant. 2008, 133, 651–669. [Google Scholar] [CrossRef]
- Wu, H.; Shabala, L.; Liu, X.; Azzarello, E.; Zhou, M.; Pandolfi, C.; Chen, Z.-H.; Bose, J.; Mancuso, S.; Shabala, S. Linking salinity stress tolerance with tissue-specific Na+ sequestration in wheat roots. Front. Plant Sci. 2015, 6, 71. [Google Scholar] [CrossRef]
- Møller, I.M.; Jensen, P.E.; Hansson, A. Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 2007, 58, 459–481. [Google Scholar] [CrossRef] [Green Version]
- Shabala, S.; Munns, R. Salinity stress: Physiological constraints and adaptive mechanisms. Plant Stress Physiol. 2012, 1, 59–93. [Google Scholar]
- Tyerman, S.D.; Munns, R.; Fricke, W.; Arsova, B.; Barkla, B.J.; Bose, J.; Bramley, H.; Byrt, C.; Chen, Z.; Colmer, T.D.; et al. Energy costs of salinity tolerance in crop plants. New Phytol. 2019, 221, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Shahzad, B.; Yun, P.; Shabala, L.; Zhou, M.; Sellamuthu, G.; Venkataraman, G.; Chen, Z.-H.; Shabala, S. Unravelling the physiological basis of salinity stress tolerance in cultivated and wild rice species. Funct. Plant Biol. 2022, 49, 351–364. [Google Scholar] [CrossRef] [PubMed]
- Bose, J.; Rodrigo-Moreno, A.; Lai, D.; Xie, Y.; Shen, W.; Shabala, S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann. Bot. 2014, 115, 481–494. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, K.; Bose, J.; Shabala, L.; Shabala, S. Difference in root K+ retention ability and reduced sensitivity of K+-permeable channels to reactive oxygen species confer differential salt tolerance in three Brassica species. J. Exp. Bot. 2016, 67, 4611–4625. [Google Scholar] [CrossRef]
- Vera-Estrella, R.; Barkla, B.J.; García-Ramírez, L.; Pantoja, O. Salt stress in Thellungiella halophila activates Na+ transport mechanisms required for salinity tolerance. Plant Physiol. 2005, 139, 1507–1517. [Google Scholar] [CrossRef]
- Shabala, S.; Mackay, A. Chapter 5—Ion transport in halophytes. In Advances in Botanical Research; Turkan, I., Ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 57, pp. 151–199. [Google Scholar]
- Niu, M.; Huang, Y.; Sun, S.; Sun, J.; Cao, H.; Shabala, S.; Bie, Z. Root respiratory burst oxidase homologue-dependent H2O2 production confers salt tolerance on a grafted cucumber by controlling Na+ exclusion and stomatal closure. J. Exp. Bot. 2017, 69, 3465–3476. [Google Scholar] [CrossRef]
- Chakraborty, K.; Basak, N.; Bhaduri, D.; Ray, S.; Vijayan, J.; Chattopadhyay, K.; Sarkar, R.K. Ionic basis of salt tolerance in plants: Nutrient homeostasis and oxidative stress tolerance. In Plant Nutrients and Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Oku, H., Nahar, K., Hawrylak-Nowak, B., Eds.; Springer: Singapore, 2018; pp. 325–362. [Google Scholar] [CrossRef]
- Huang, Y.; Cao, H.; Yang, L.; Chen, C.; Shabala, L.; Xiong, M.; Niu, M.; Liu, J.; Zheng, Z.; Zhou, L.; et al. Tissue-specific respiratory burst oxidase homolog-dependent H2O2 signaling to the plasma membrane H+-ATPase confers potassium uptake and salinity tolerance in Cucurbitaceae. J. Exp. Bot. 2019, 70, 5879–5893. [Google Scholar] [CrossRef]
- Ueno, K.; Kinoshita, T.; Inoue, S.-I.; Emi, T.; Shimazaki, K.-I. Biochemical characterization of plasma membrane H+-ATPase activation in guard cell protoplasts of Arabidopsis thaliana in response to blue light. Plant Cell Physiol. 2005, 46, 955–963. [Google Scholar] [CrossRef]
- Haruta, M.; Sussman, M.R. The effect of a genetically reduced plasma membrane protonmotive force on vegetative growth of Arabidopsis. Plant Physiol. 2012, 158, 1158–1171. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Singh, A.; Mithra, S.V.A.; Krishnamurthy, S.L.; Parida, S.K.; Jain, S.; Tiwari, K.K.; Kumar, P.; Rao, A.R.; Sharma, S.K.; et al. Genome-wide association mapping of salinity tolerance in rice (Oryza sativa). DNA Res. 2015, 22, 133–145. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Zhang, D.; Song, T.; Xu, F.; Lin, S.; Xu, W.; Li, Q.; Zhu, Y.; Liang, J.; Zhang, J. Arabidopsis plasma membrane H+-ATPase genes AHA2 and AHA7 have distinct and overlapping roles in the modulation of root tip H+ efflux in response to low-phosphorus stress. J. Exp. Bot. 2017, 68, 1731–1741. [Google Scholar] [CrossRef] [PubMed]
- Sze, H.; Li, X.; Palmgren, M.G. Energization of plant cell membranes by H+-pumping ATPases: Regulation and biosynthesis. Plant Cell 1999, 11, 677–689. [Google Scholar] [CrossRef] [PubMed]
- Maeshima, M. Vacuolar H+-pyrophosphatase. Biochim. Et Biophys. Acta-Biomembr. 2000, 1465, 37–51. [Google Scholar] [CrossRef]
- Ratajczak, R. Structure, function and regulation of the plant vacuolar H+-translocating ATPase. Biochim. Et Biophys. Acta-Biomembr. 2000, 1465, 17–36. [Google Scholar] [CrossRef]
- Ma, B.; Qian, D.; Nan, Q.; Tan, C.; An, L.; Xiang, Y. Arabidopsis vacuolar H+-ATPase (V-ATPase) B subunits are involved in actin cytoskeleton remodeling via binding to, bundling, and stabilizing F-actin. J. Biol. Chem. 2012, 287, 19008–19017. [Google Scholar] [CrossRef]
- Haruta, M.; Gray, W.M.; Sussman, M.R. Regulation of the plasma membrane proton pump (H+-ATPase) by phosphorylation. Curr. Opin. Plant Biol. 2015, 28, 68–75. [Google Scholar] [CrossRef]
- Gaxiola, R.A.; Regmi, K.; Paez-Valencia, J.; Pizzio, G.; Zhang, S. Plant H+-PPases: Reversible enzymes with contrasting functions dependent on membrane environment. Mol. Plant 2016, 9, 317–319. [Google Scholar] [CrossRef]
- Shabala, S.N.; Newman, I.A.; Morris, J. Oscillations in H+ and Ca2+ ion fluxes around the elongation region of corn roots and effects of external pH. Plant Physiol. 1997, 113, 111–118. [Google Scholar] [CrossRef]
- Shabala, L.; Ross, T.; McMeekin, T.; Shabala, S. Non-invasive microelectrode ion flux measurements to study adaptive responses of microorganisms to the environment. FEMS Microbiol. Rev. 2006, 30, 472–486. [Google Scholar] [CrossRef] [Green Version]
- Shabala, L.; Cuin, T.A.; Newman, I.A.; Shabala, S. Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 2005, 222, 1041–1050. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Demidchik, V.; Shabala, L.; Cuin, T.A.; Smith, S.J.; Miller, A.J.; Davies, J.M.; Newman, I.A. Extracellular Ca2+ ameliorates NaCl-Induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+-permeable channels. Plant Physiol. 2006, 141, 1653–1665. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Shabala, L.; Newman, I. Studying membrane transport processes by non-invasive microelectrodes: Basic principles and methods. In Plant Electrophysiology: Methods and Cell Electrophysiology; Volkov, A.G., Ed.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 167–186. [Google Scholar] [CrossRef]
- Takehisa, H.; Sato, Y.; Igarashi, M.; Abiko, T.; Antonio, B.A.; Kamatsuki, K.; Minami, H.; Namiki, N.; Inukai, Y.; Nakazono, M.; et al. Genome-wide transcriptome dissection of the rice root system: Implications for developmental and physiological functions. Plant J. 2012, 69, 126–140. [Google Scholar] [CrossRef]
- Yong, M.-T.; Solis, C.A.; Amatoury, S.; Sellamuthu, G.; Rajakani, R.; Mak, M.; Venkataraman, G.; Shabala, L.; Zhou, M.; Ghannoum, O. Proto Kranz-like leaf traits and cellular ionic regulation are associated with salinity tolerance in a halophytic wild rice. Stress Biol. 2022, 2, 8. [Google Scholar] [CrossRef]
| Primer Name | Sequence |
|---|---|
| NHX1_F | CTGTCGTTCTTTTTAGCACTATGG |
| NHX1_R | GGTGACAGGATGGCCTGA |
| OsV-PPase_ F | ATGGCTCTCTTCGGAAGGGTTG |
| OsV-PPase_ R | GTCACCGACATTGTCAGCAATCAC |
| OsSOS1_F | AGATCGCGCTTACTCTTGCTGTC |
| OsSOS1_R | AGACCTCCAGTGCATCTTGTGC |
| OsSOS2_ F | ACTTAGCACTTTGGCCCAGAAAG |
| OsSOS2_ R | ACCACATGACCAAACATCTGCTG |
| OsSOS3_ F | GAACATGTCACTTCCCTATTTGC |
| OsSOS3_ R | GTCATGGGCTTCTGAATGCATT |
| OsAHA_ F | ACAGAACCTGGCTTGAGTGTG |
| OsAHA_ R | GGGCAAGCAGCATAAACCCAAA |
| G6PDH_F | AAGCCAGCATCCTATGATCAGATT |
| G6PDH_R | CGTAACCCAGAATACCCTTGAGTTT |
| ELF-a-F | CAGCAACTTGACTATGGATTGGTGGA |
| ELF-a-R | CATCCAGCACAAACATCTTAATGTGGTC |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shahzad, B.; Shabala, L.; Zhou, M.; Venkataraman, G.; Solis, C.A.; Page, D.; Chen, Z.-H.; Shabala, S. Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species. Int. J. Mol. Sci. 2022, 23, 9900. https://doi.org/10.3390/ijms23179900
Shahzad B, Shabala L, Zhou M, Venkataraman G, Solis CA, Page D, Chen Z-H, Shabala S. Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species. International Journal of Molecular Sciences. 2022; 23(17):9900. https://doi.org/10.3390/ijms23179900
Chicago/Turabian StyleShahzad, Babar, Lana Shabala, Meixue Zhou, Gayatri Venkataraman, Celymar Angela Solis, David Page, Zhong-Hua Chen, and Sergey Shabala. 2022. "Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species" International Journal of Molecular Sciences 23, no. 17: 9900. https://doi.org/10.3390/ijms23179900
APA StyleShahzad, B., Shabala, L., Zhou, M., Venkataraman, G., Solis, C. A., Page, D., Chen, Z.-H., & Shabala, S. (2022). Comparing Essentiality of SOS1-Mediated Na+ Exclusion in Salinity Tolerance between Cultivated and Wild Rice Species. International Journal of Molecular Sciences, 23(17), 9900. https://doi.org/10.3390/ijms23179900










