Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction
Abstract
:1. Introduction
2. Results
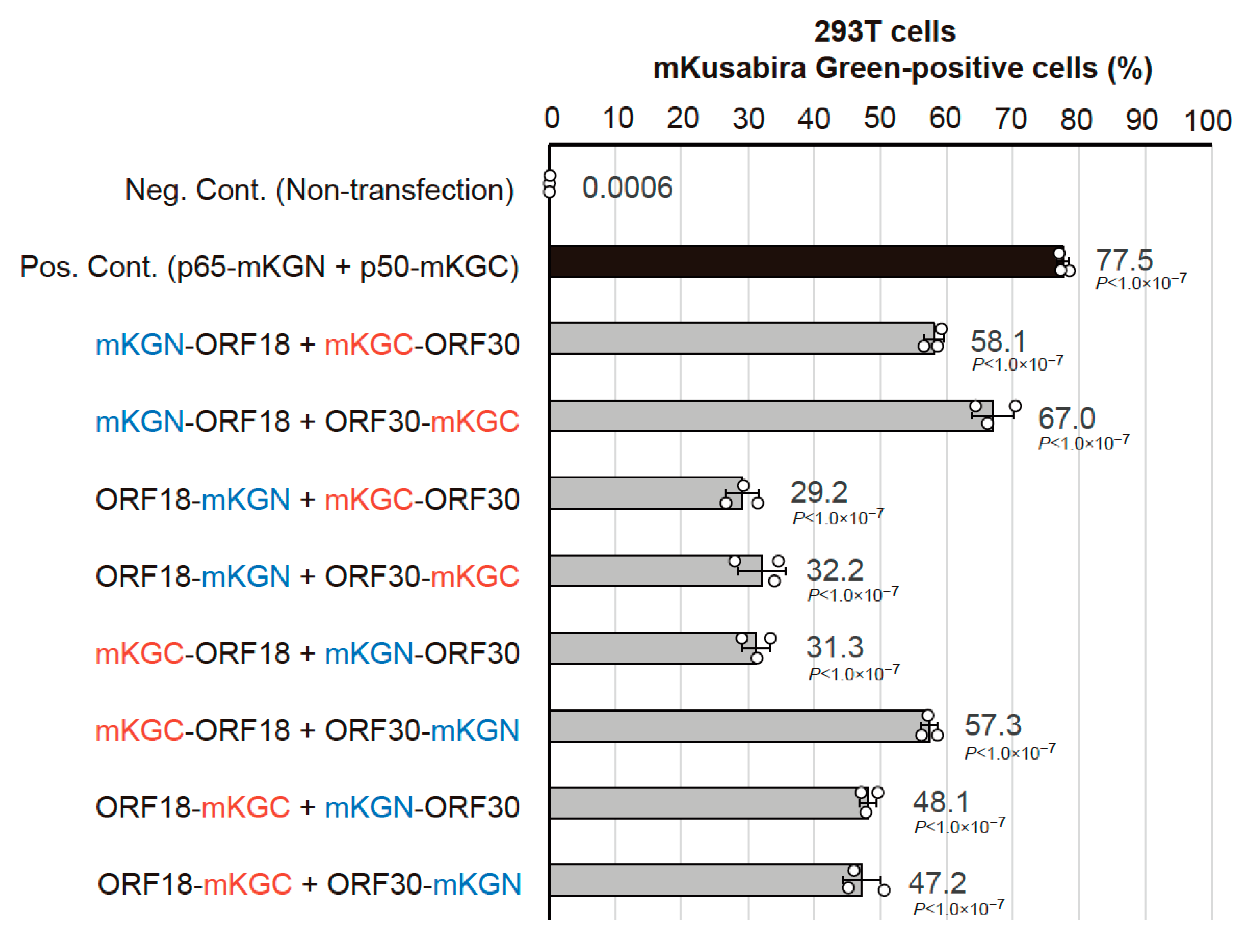
2.1. Optimization of the BiFC Assay to Assess vPIC Interactions
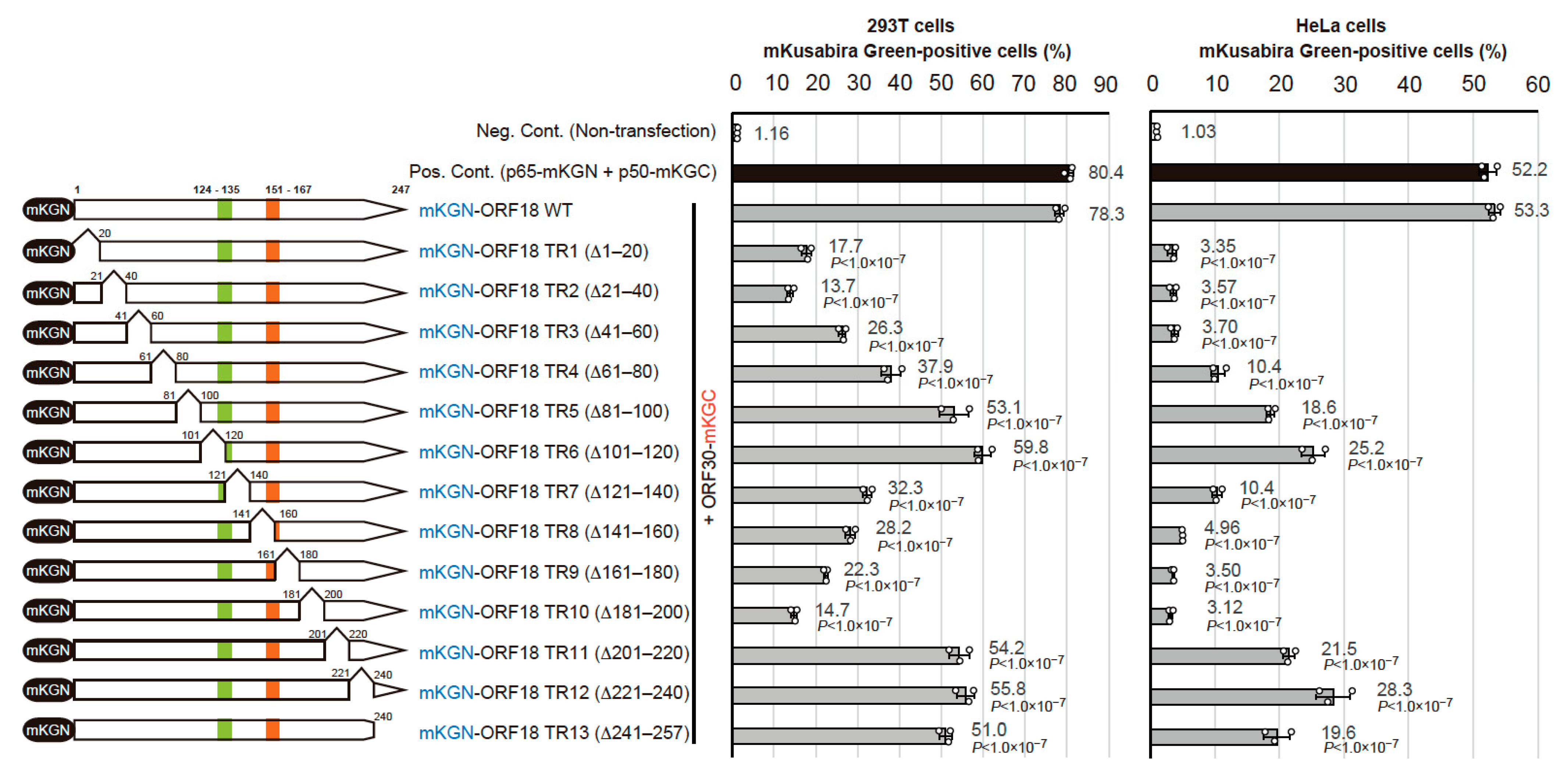
2.2. Identification of the Regions of ORF18 That Interact with ORF30
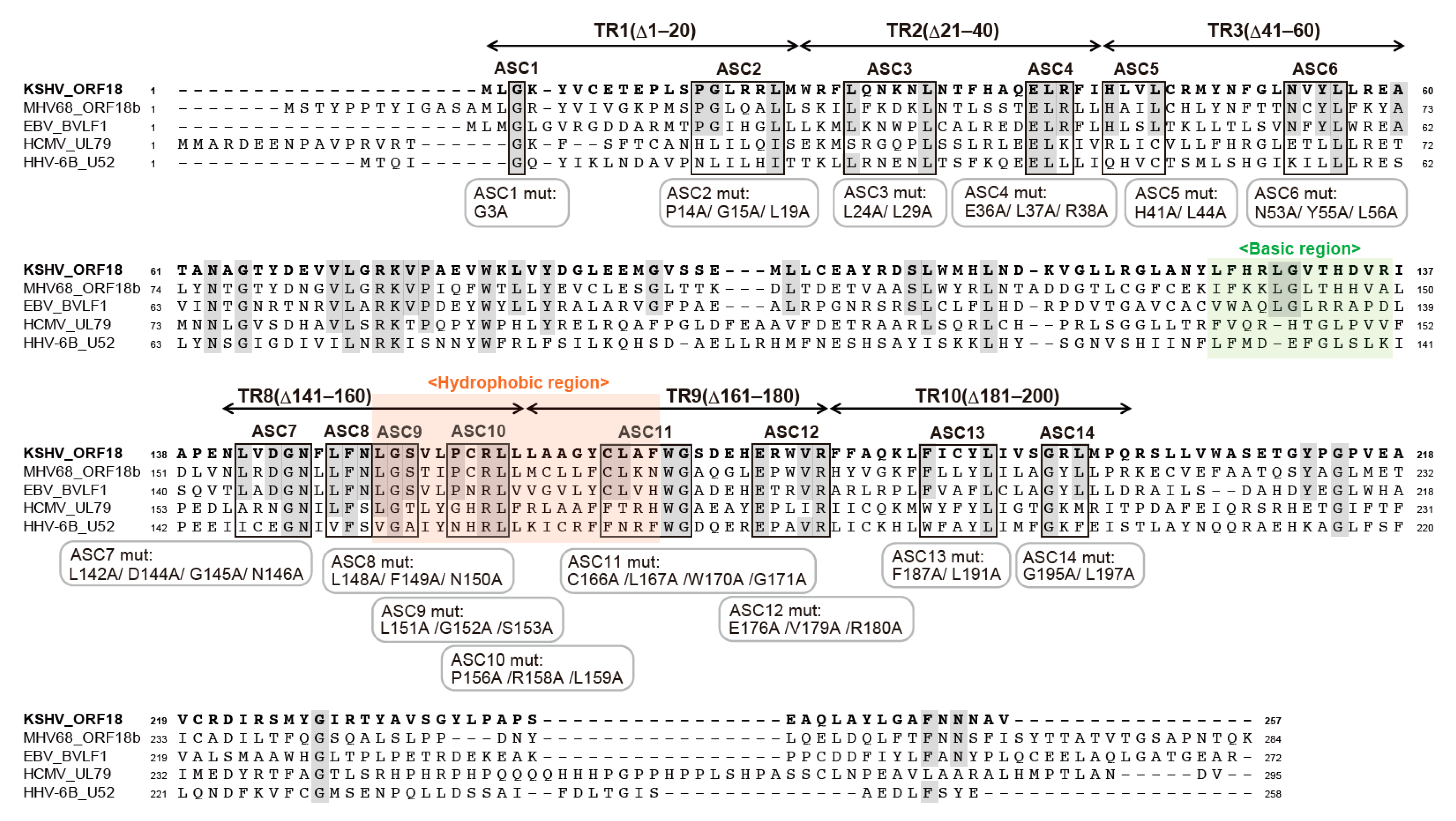
2.3. Alignment of KSHV ORF18 to Its Viral Homologs
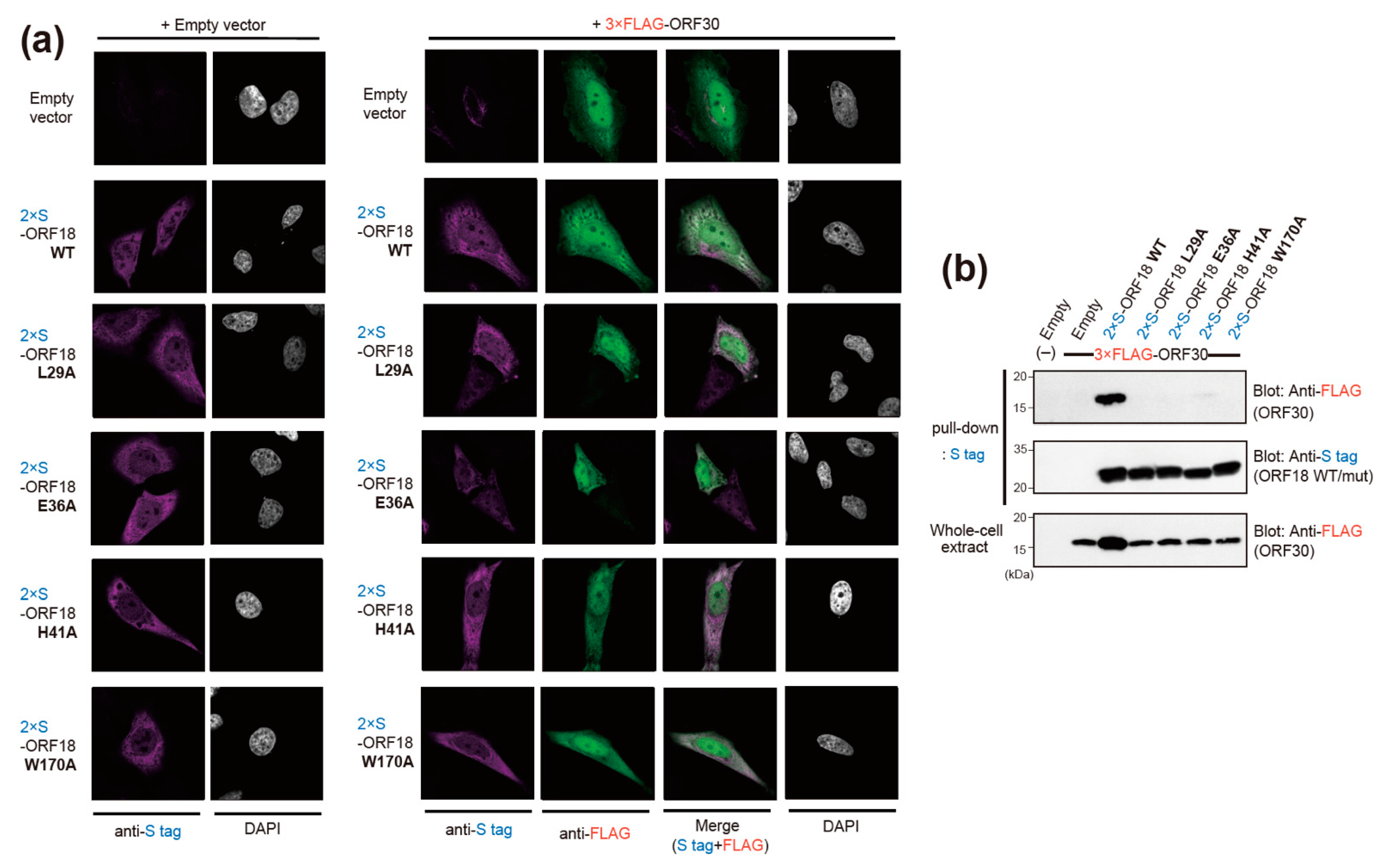
2.4. Narrowing the ORF18 Interaction Blocks to Single Amino-Acid Residues
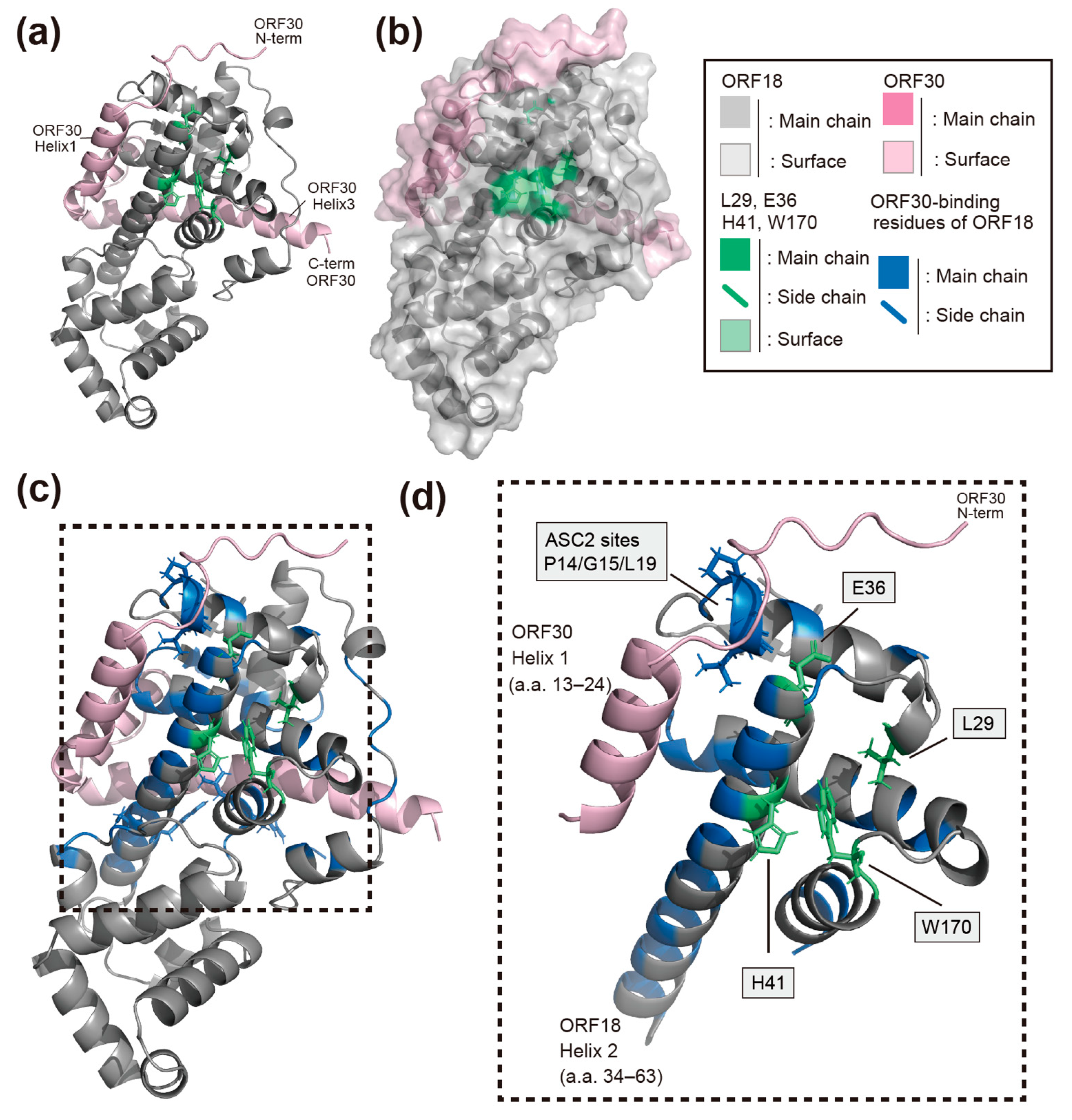
2.5. AI-Predicted Structure Model of ORF18 and the Identified Residues That Mediate Its Interaction with ORF30
3. Discussion
4. Materials and Methods
4.1. Cell Culture and Reagents
4.2. Plasmids
4.3. BiFC Interaction Assay
4.4. Western Blotting and Pull-Down Assay
4.5. Immunofluorescence Analysis (IFA)
4.6. Statistics
4.7. Protein Structure Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Chang, Y.; Cesarman, E.; Pessin, M.S.; Lee, F.; Culpepper, J.; Knowles, D.M.; Moore, P.S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef] [PubMed]
- Nador, R.G.; Cesarman, E.; Chadburn, A.; Dawson, D.B.; Ansari, M.Q.; Sald, J.; Knowles, D.M. Primary effusion lymphoma: A distinct clinicopathologic entity associated with the Kaposi’s sarcoma-associated herpes virus. Blood 1996, 88, 645–656. [Google Scholar] [CrossRef] [PubMed]
- Soulier, J.; Grollet, L.; Oksenhendler, E.; Cacoub, P.; Cazals-Hatem, D.; Babinet, P.; d’Agay, M.F.; Clauvel, J.P.; Raphael, M.; Degos, L.; et al. Kaposi’s sarcoma-associated herpesvirus-like DNA sequences in multicentric Castleman’s disease. Blood 1995, 86, 1276–1280. [Google Scholar] [CrossRef]
- Russo, J.J.; Bohenzky, R.A.; Chien, M.C.; Chen, J.; Yan, M.; Maddalena, D.; Parry, J.P.; Peruzzi, D.; Edelman, I.S.; Chang, Y.; et al. Nucleotide sequence of the Kaposi sarcoma-associated herpesvirus (HHV8). Proc. Natl. Acad. Sci. USA 1996, 93, 14862–14867. [Google Scholar] [CrossRef]
- Sudomova, M.; Berchova-Bimova, K.; Mazurakova, A.; Samec, D.; Kubatka, P.; Hassan, S.T.S. Flavonoids target human herpesviruses that infect the nervous system: Mechanisms of action and therapeutic insights. Viruses 2022, 14, 592. [Google Scholar] [CrossRef]
- Tso, F.Y.; Sawyer, A.; Kwon, E.H.; Mudenda, V.; Langford, D.; Zhou, Y.; West, J.; Wood, C. Kaposi’s sarcoma-associated herpesvirus infection of neurons in HIV-Positive patients. J. Infect. Dis. 2017, 215, 1898–1907. [Google Scholar] [CrossRef]
- Baldini, F.; Baiocchini, A.; Schininà, V.; Agrati, C.; Giancola, M.L.; Alba, L.; Grisetti, S.; Del Nonno, F.; Capobianchi, M.R.; Antinori, A. Brain localization of Kaposi’s sarcoma in a patient treated by combination antiretroviral therapy. BMC Infect. Dis. 2013, 13, 600. [Google Scholar] [CrossRef]
- Jha, H.C.; Mehta, D.; Lu, J.; El-Naccache, D.; Shukla, S.K.; Kovacsics, C.; Kolson, D.; Robertson, E.S. Gammaherpesvirus Infection of Human Neuronal Cells. mBio 2015, 6, e01844–e01915. [Google Scholar] [CrossRef]
- Krug, L.T.; Pellet, P.E. The Family Herpesviridae: A Brief Introduction; Damania, B.A., Cohen, J.I., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2022; Volume 2. [Google Scholar]
- Gruffat, H.; Marchione, R.; Manet, E. Herpesvirus late gene expression: A viral-specific pre-initiation complex is key. Front. Microbiol. 2016, 7, 869. [Google Scholar] [CrossRef]
- Davis, Z.H.; Hesser, C.; Park, J.; Glaunsinger, B.A. Interaction between ORF24 and ORF34 in the Kaposi’s sarcoma-associated herpesvirus late gene transcription factor complex is essential for viral late gene expression. J. Virol. 2015, 90, 599–604. [Google Scholar] [CrossRef] [Green Version]
- Nishimura, M.; Watanabe, T.; Yagi, S.; Yamanaka, T.; Fujimuro, M. Kaposi’s sarcoma-associated herpesvirus ORF34 is essential for late gene expression and virus production. Sci. Rep. 2017, 7, 329. [Google Scholar] [CrossRef] [PubMed]
- Brulois, K.; Wong, L.Y.; Lee, H.R.; Sivadas, P.; Ensser, A.; Feng, P.; Gao, S.J.; Toth, Z.; Jung, J.U. Association of Kaposi’s Sarcoma-associated herpesvirus ORF31 with ORF34 and ORF24 is critical for late gene expression. J. Virol. 2015, 89, 6148–6154. [Google Scholar] [CrossRef]
- Castañeda, A.F.; Glaunsinger, B.A. The Interaction between ORF18 and ORF30 Is Required for Late Gene Expression in Kaposi’s Sarcoma-Associated Herpesvirus. J. Virol. 2019, 93, e01488–e01518. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, T.; Nishimura, M.; Izumi, T.; Kuriyama, K.; Iwaisako, Y.; Hosokawa, K.; Takaori-Kondo, A.; Fujimuro, M. Kaposi’s Sarcoma-associated herpesvirus orf66 is essential for late gene expression and virus production via interaction with ORF34. J. Virol. 2020, 94, e01300–e01319. [Google Scholar] [CrossRef] [PubMed]
- Didychuk, A.L.; Castaneda, A.F.; Kushnir, L.O.; Huang, C.J.; Glaunsinger, B.A. Conserved CxnC Motifs in Kaposi’s Sarcoma-associated herpesvirus ORF66 are required for viral late gene expression and are essential for its interaction with ORF34. J. Virol. 2020, 94, e01299–e01319. [Google Scholar] [CrossRef]
- Kodama, Y.; Hu, C.D. Bimolecular fluorescence complementation (BiFC): A 5-year update and future perspectives. Biotechniques 2012, 53, 285–298. [Google Scholar] [CrossRef]
- David, A.; Islam, S.; Tankhilevich, E.; Sternberg, M.J.E. The AlphaFold database of protein structures: A biologist’s guide. J. Mol. Biol. 2022, 434, 167336. [Google Scholar] [CrossRef]
- Fersht, A.R. AlphaFold—A personal perspective on the impact of machine learning. J. Mol. Biol. 2021, 433, 167088. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Zidek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Arumugaswami, V.; Wu, T.T.; Martinez-Guzman, D.; Jia, Q.; Deng, H.; Reyes, N.; Sun, R. ORF18 is a transfactor that is essential for late gene transcription of a gammaherpesvirus. J. Virol. 2006, 80, 9730–9740. [Google Scholar] [CrossRef] [Green Version]
- Gong, D.; Wu, N.C.; Xie, Y.; Feng, J.; Tong, L.; Brulois, K.F.; Luan, H.; Du, Y.; Jung, J.U.; Wang, C.Y.; et al. Kaposi’s sarcoma-associated herpesvirus ORF18 and ORF30 are essential for late gene expression during lytic replication. J. Virol. 2014, 88, 11369–11382. [Google Scholar] [CrossRef] [PubMed]
- Wu, T.-T.; Park, T.; Kim, H.; Tran, T.; Tong, L.; Martinez-Guzman, D.; Reyes, N.; Deng, H.; Sun, R. ORF30 and ORF34 are essential for expression of late genes in Murine Gammaherpesvirus 68. J. Virol. 2009, 83, 2265–2273. [Google Scholar] [CrossRef] [PubMed]
- Perng, Y.C.; Campbell, J.A.; Lenschow, D.J.; Yu, D. Human cytomegalovirus pUL79 is an elongation factor of RNA polymerase II for viral gene transcription. PLoS Pathog. 2014, 10, e1004350. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Han, T.; Tang, S.; Xu, W.; Bao, Q.; Sun, Y.; Xuan, B.; Qian, Z. Murine Cytomegalovirus Protein pM91 Interacts with pM79 and Is Critical for Viral Late Gene Expression. J. Virol. 2018, 92, e00675–e00718. [Google Scholar] [CrossRef]
- Brulois, K.F.; Chang, H.; Lee, A.S.; Ensser, A.; Wong, L.Y.; Toth, Z.; Lee, S.H.; Lee, H.R.; Myoung, J.; Ganem, D.; et al. Construction and manipulation of a new Kaposi’s sarcoma-associated herpesvirus bacterial artificial chromosome clone. J. Virol. 2012, 86, 9708–9720. [Google Scholar] [CrossRef] [PubMed]
- Nabhan, J.F.; Hu, R.; Oh, R.S.; Cohen, S.N.; Lu, Q. Formation and release of arrestin domain-containing protein 1-mediated microvesicles (ARMMs) at plasma membrane by recruitment of TSG101 protein. Proc. Natl. Acad. Sci. USA 2012, 109, 4146–4151. [Google Scholar] [CrossRef]
- Yoshida, T.; Ebina, H.; Koyanagi, Y. N-linked glycan-dependent interaction of CD63 with CXCR4 at the Golgi apparatus induces downregulation of CXCR4. Microbiol. Immunol. 2009, 53, 629–635. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maeda, Y.; Watanabe, T.; Izumi, T.; Kuriyama, K.; Ohno, S.; Fujimuro, M. Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction. Int. J. Mol. Sci. 2022, 23, 9647. https://doi.org/10.3390/ijms23179647
Maeda Y, Watanabe T, Izumi T, Kuriyama K, Ohno S, Fujimuro M. Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction. International Journal of Molecular Sciences. 2022; 23(17):9647. https://doi.org/10.3390/ijms23179647
Chicago/Turabian StyleMaeda, Yoshiko, Tadashi Watanabe, Taisuke Izumi, Kazushi Kuriyama, Shinji Ohno, and Masahiro Fujimuro. 2022. "Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction" International Journal of Molecular Sciences 23, no. 17: 9647. https://doi.org/10.3390/ijms23179647
APA StyleMaeda, Y., Watanabe, T., Izumi, T., Kuriyama, K., Ohno, S., & Fujimuro, M. (2022). Biomolecular Fluorescence Complementation Profiling and Artificial Intelligence Structure Prediction of the Kaposi’s Sarcoma-Associated Herpesvirus ORF18 and ORF30 Interaction. International Journal of Molecular Sciences, 23(17), 9647. https://doi.org/10.3390/ijms23179647





