ATP5B Is an Essential Factor for Hepatitis B Virus Entry
Abstract
1. Introduction
2. Results
2.1. Identification of ATP5B as a Factor Interacting with preS1
2.2. Myristoylation of PS1 Affects the Interaction
2.3. Colocalization of PS1 (a Large S Protein: LS) and ATP5B in the Cells
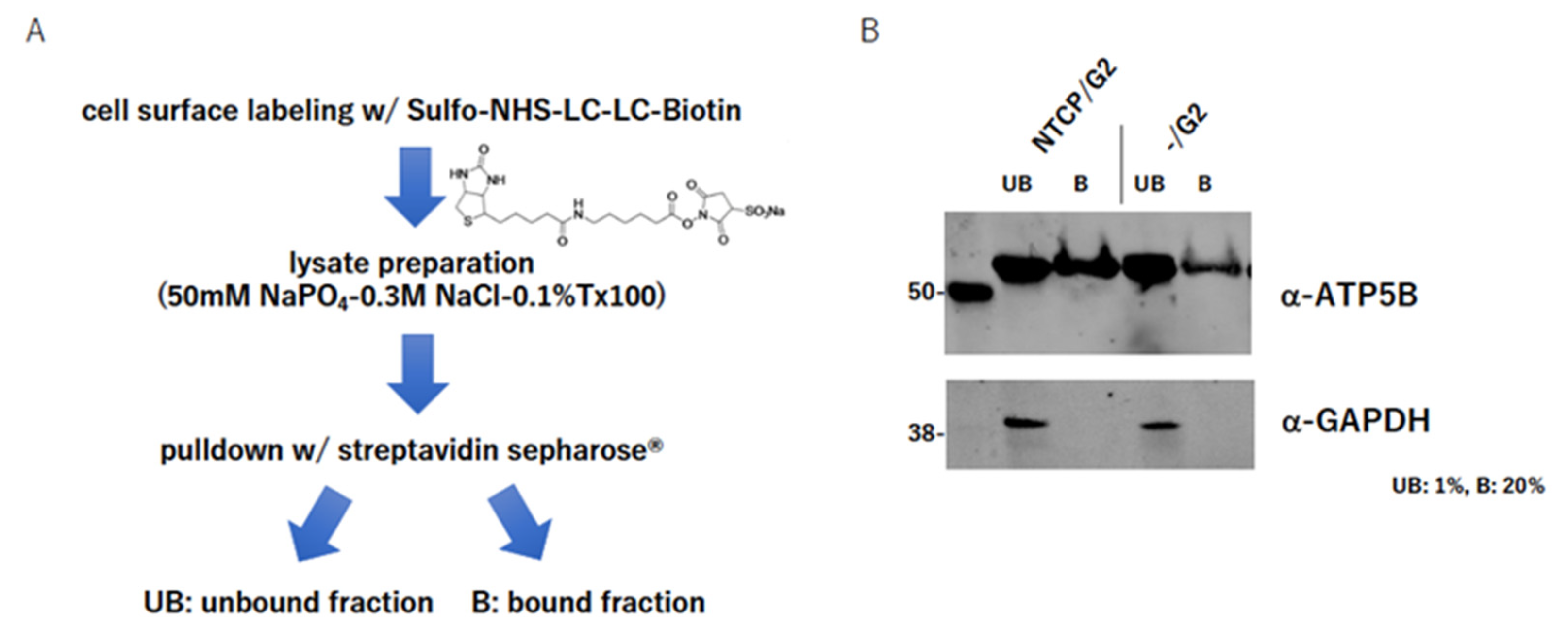
2.4. ATP5B Is Present on the Cell Surface
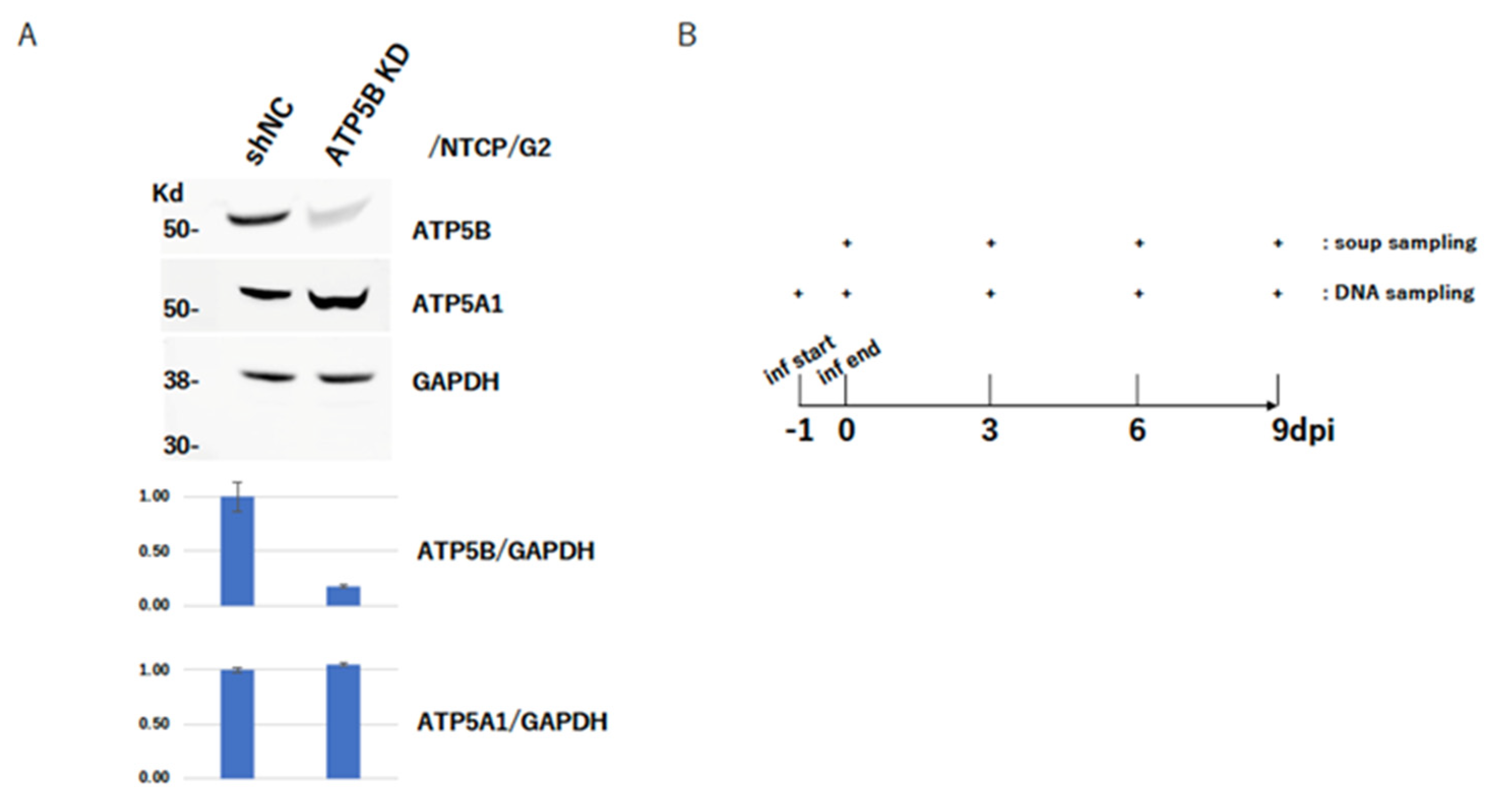
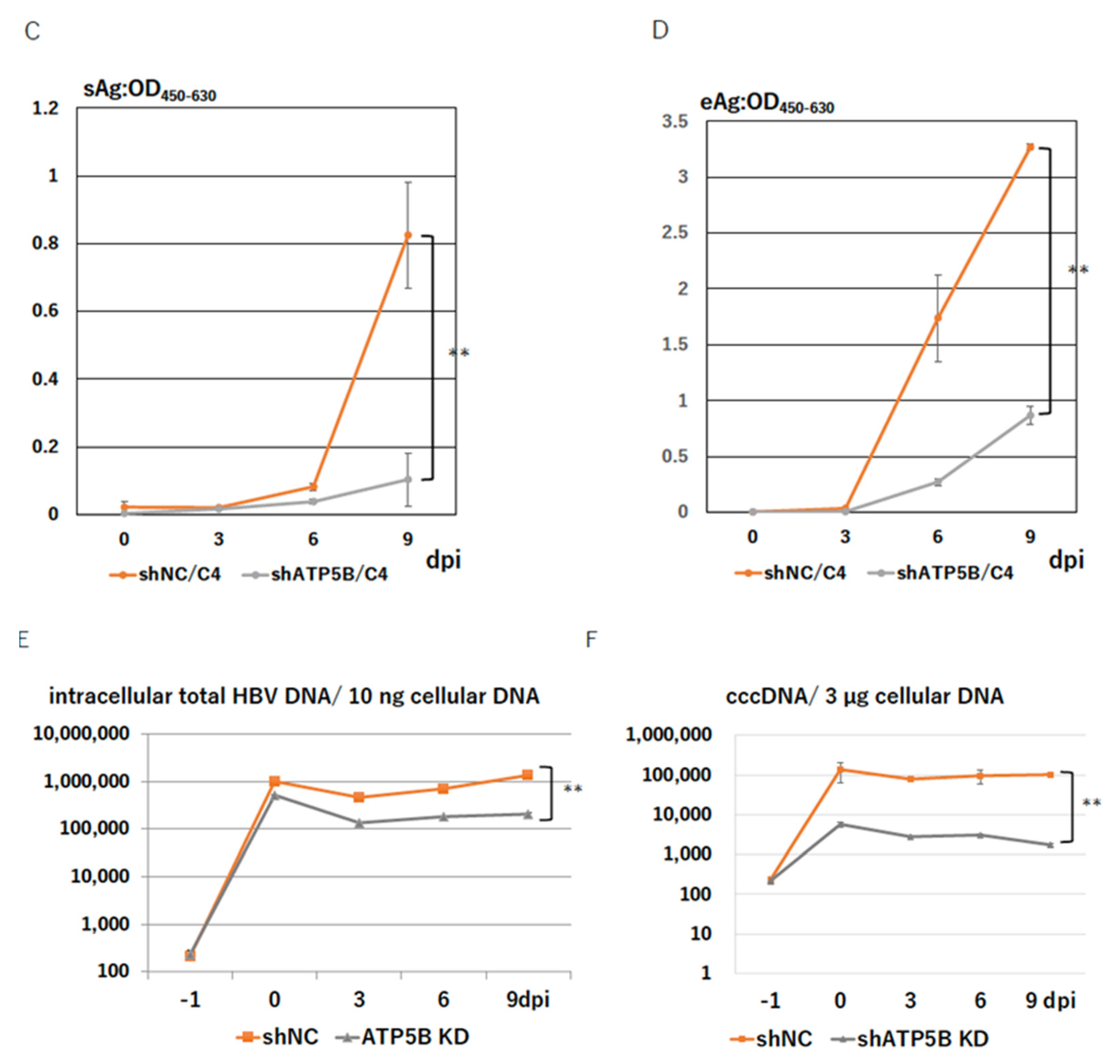
2.5. ATP5B Knockdown Loses HBV Infectivity
2.6. ATP5B Is Required for HBV Attachment/Entry
2.7. ATP5B Interacts with myrPS1:2-47 at the Front of the ATP-Binding Domain
2.8. An ATP5B Regional Peptide Blocks HBV Infection
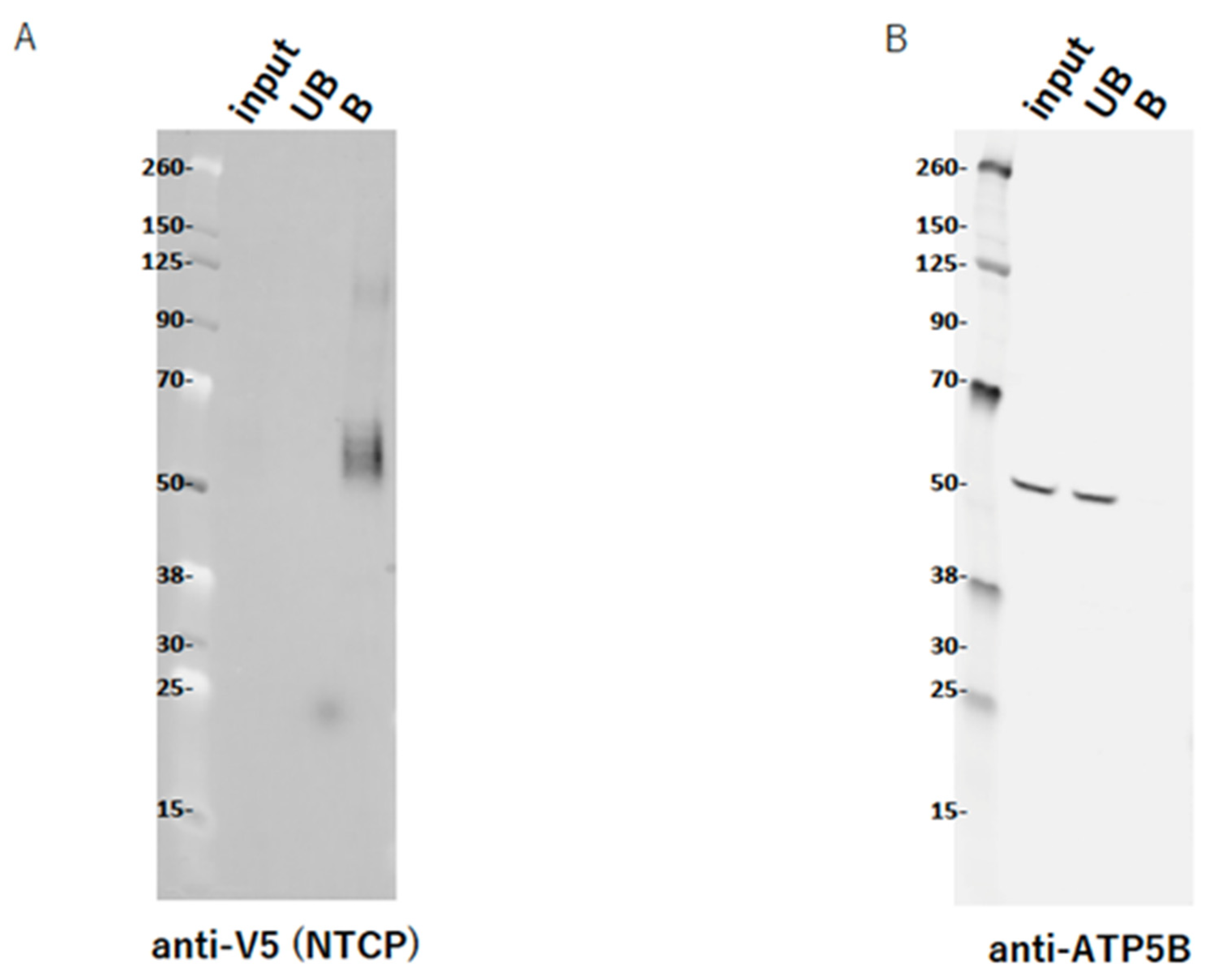
2.9. ATP5B Does Not Interact with NTCP
3. Discussion
4. Materials and Methods
4.1. preS1 Peptide Pulldown
4.2. Cells
4.3. Plasmids
4.4. Knockdown of ATP5B
4.5. Cell Surface Labeling
4.6. HBV Infection Experiment with NTCP/G2
4.7. HBV Genome Quantification
4.8. Transfection and Pull-Down of NTCP V5-His
4.9. Western Blot
4.10. Immunofluorescence Assay (IFA)
4.11. Primary Antibodies
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Seeger, C.; Zoulim, F.; Mason, W.S. Hepadnaviridae, 7th ed.; Wolters Kluwer: Philadelphia, PA, USA, 2022; Volume 2. [Google Scholar]
- Schweitzer, A.; Horn, J.; Mikolajczyk, R.T.; Krause, G.; Ott, J.J. Estimations of worldwide prevalence of chronic hepatitis B virus infection: A systematic review of data published between 1965 and 2013. Lancet 2015, 386, 1546–1555. [Google Scholar] [CrossRef]
- Blumberg, B.S.; Gerstley, B.J.; Hungerford, D.A.; London, W.T.; Sutnick, A.I. A serum antigen (Australia antigen) in Down’s syndrome, leukemia, and hepatitis. Ann. Intern. Med. 1967, 66, 924–931. [Google Scholar] [CrossRef] [PubMed]
- Blumberg, B.S.; Alter, H.J.; Visnich, S. A “New” Antigen in Leukemia Sera. JAMA 1965, 191, 541–546. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Zhong, G.; Xu, G.; He, W.; Jing, Z.; Gao, Z.; Huang, Y.; Qi, Y.; Peng, B.; Wang, H.; et al. Sodium taurocholate cotransporting polypeptide is a functional receptor for human hepatitis B and D virus. eLife 2012, 1, e00049. [Google Scholar] [CrossRef]
- Mi, S.; Li, Y.; Yan, J.; Gao, G.F. Na(+)/K (+)-ATPase beta1 subunit interacts with M2 proteins of influenza A and B viruses and affects the virus replication. Sci. China Life Sci. 2010, 53, 1098–1105. [Google Scholar] [CrossRef]
- Cui, X.; Sun, Z.R.; Ren, G.W.; Wang, G.L.; Qi, Y.; Ma, Y.P.; Ruan, Q. Interaction between human cytomegalovirus UL136 protein and ATP1B1 protein. Braz. J. Med. Biol. Res. 2011, 44, 1251–1255. [Google Scholar] [CrossRef][Green Version]
- Nishitsuji, H.; Sugiyama, R.; Abe, M.; Takaku, H. ATP1B3 Protein Modulates the Restriction of HIV-1 Production and Nuclear Factor kappa Light Chain Enhancer of Activated B Cells (NF-kappaB) Activation by BST-2. J. Biol. Chem. 2016, 291, 4754–4762. [Google Scholar] [CrossRef]
- Lu, Y.; Hou, H.; Wang, F.; Qiao, L.; Wang, X.; Yu, J.; Liu, W.; Sun, Z. ATP1B3: A virus-induced host factor against EV71 replication by up-regulating the production of type-I interferons. Virology 2016, 496, 28–34. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Li, Y.X.; Zhang, Y.; Li, X.; Tang, H. MiR-101 regulates HSV-1 replication by targeting ATP5B. Antivir. Res. 2011, 89, 219–226. [Google Scholar] [CrossRef]
- Ren, L.; Ding, S.; Song, Y.; Li, B.; Ramanathan, M.; Co, J.; Amieva, M.R.; Khavari, P.A.; Greenberg, H.B. Profiling of rotavirus 3’UTR-binding proteins reveals the ATP synthase subunit ATP5B as a host factor that supports late-stage virus replication. J. Biol. Chem. 2019, 294, 5993–6006. [Google Scholar] [CrossRef]
- Neupane, P.; Bhuju, S.; Thapa, N.; Bhattarai, H.K. ATP Synthase: Structure, Function and Inhibition. Biomol. Concepts 2019, 10, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Glebe, D.; Urban, S. Viral and cellular determinants involved in hepadnaviral entry. World J. Gastroenterol. 2007, 13, 22–38. [Google Scholar] [CrossRef] [PubMed]
- Martinez, L.O.; Jacquet, S.; Esteve, J.P.; Rolland, C.; Cabezon, E.; Champagne, E.; Pineau, T.; Georgeaud, V.; Walker, J.E.; Terce, F.; et al. Ectopic beta-chain of ATP synthase is an apolipoprotein A-I receptor in hepatic HDL endocytosis. Nature 2003, 421, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.H.; Kamanna, V.S.; Zhang, M.C.; Kashyap, M.L. Niacin inhibits surface expression of ATP synthase beta chain in HepG2 cells: Implications for raising HDL. J. Lipid. Res. 2008, 49, 1195–1201. [Google Scholar] [CrossRef]
- Moser, T.L.; Stack, M.S.; Asplin, I.; Enghild, J.J.; Hojrup, P.; Everitt, L.; Hubchak, S.; Schnaper, H.W.; Pizzo, S.V. Angiostatin binds ATP synthase on the surface of human endothelial cells. Proc. Natl. Acad. Sci. USA 1999, 96, 2811–2816. [Google Scholar] [CrossRef]
- Abou-Jaoude, G.; Molina, S.; Maurel, P.; Sureau, C. Myristoylation signal transfer from the large to the middle or the small HBV envelope protein leads to a loss of HDV particles infectivity. Virology 2007, 365, 204–209. [Google Scholar] [CrossRef]
- Bremer, C.M.; Sominskaya, I.; Skrastina, D.; Pumpens, P.; El Wahed, A.A.; Beutling, U.; Frank, R.; Fritz, H.J.; Hunsmann, G.; Gerlich, W.H.; et al. N-terminal myristoylation-dependent masking of neutralizing epitopes in the preS1 attachment site of hepatitis B virus. J. Hepatol. 2011, 55, 29–37. [Google Scholar] [CrossRef]
- Tsurimoto, T.; Fujiyama, A.; Matsubara, K. Stable expression and replication of hepatitis B virus genome in an integrated state in a human hepatoma cell line transfected with the cloned viral DNA. Proc. Natl. Acad. Sci. USA 1987, 84, 444–448. [Google Scholar] [CrossRef]
- Chi, S.L.; Pizzo, S.V. Cell surface F1Fo ATP synthase: A new paradigm? Ann. Med. 2006, 38, 429–438. [Google Scholar] [CrossRef]
- Schwiebert, E.M.; Zsembery, A. Extracellular ATP as a signaling molecule for epithelial cells. Biochim. Biophys. Acta 2003, 1615, 7–32. [Google Scholar] [CrossRef]
- Chi, S.L.; Pizzo, S.V. Angiostatin is directly cytotoxic to tumor cells at low extracellular pH: A mechanism dependent on cell surface-associated ATP synthase. Cancer Res. 2006, 66, 875–882. [Google Scholar] [CrossRef] [PubMed]
- Ishida, Y.; Yamasaki, C.; Yanagi, A.; Yoshizane, Y.; Fujikawa, K.; Watashi, K.; Abe, H.; Wakita, T.; Hayes, C.N.; Chayama, K.; et al. Novel robust in vitro hepatitis B virus infection model using fresh human hepatocytes isolated from humanized mice. Am. J. Pathol. 2015, 185, 1275–1285. [Google Scholar] [CrossRef]
- Verrier, E.R.; Colpitts, C.C.; Bach, C.; Heydmann, L.; Weiss, A.; Renaud, M.; Durand, S.C.; Habersetzer, F.; Durantel, D.; Abou-Jaoude, G.; et al. A targeted functional RNA interference screen uncovers glypican 5 as an entry factor for hepatitis B and D viruses. Hepatology 2016, 63, 35–48. [Google Scholar] [CrossRef]
- Schulze, A.; Gripon, P.; Urban, S. Hepatitis B virus infection initiates with a large surface protein-dependent binding to heparan sulfate proteoglycans. Hepatology 2007, 46, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Saso, W.; Sugiyama, R.; Ishii, K.; Ohki, M.; Nagamori, S.; Suzuki, R.; Aizaki, H.; Ryo, A.; Yun, J.H.; et al. Epidermal growth factor receptor is a host-entry cofactor triggering hepatitis B virus internalization. Proc. Natl. Acad. Sci. USA 2019, 116, 8487–8492. [Google Scholar] [CrossRef] [PubMed]
- Neurath, A.R.; Kent, S.B.; Strick, N.; Parker, K. Identification and chemical synthesis of a host cell receptor binding site on hepatitis B virus. Cell 1986, 46, 429–436. [Google Scholar] [CrossRef]
- Park, J.H.; Iwamoto, M.; Yun, J.H.; Uchikubo-Kamo, T.; Son, D.; Jin, Z.; Yoshida, H.; Ohki, M.; Ishimoto, N.; Mizutani, K.; et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 2022, 606, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Asami, J.; Kimura, K.T.; Fujita-Fujiharu, Y.; Ishida, H.; Zhang, Z.; Nomura, Y.; Liu, K.; Uemura, T.; Sato, Y.; Ono, M.; et al. Structure of the bile acid transporter and HBV receptor NTCP. Nature 2022, 606, 1021–1026. [Google Scholar] [CrossRef]
- Hong, S.; Pedersen, P.L. ATP synthase and the actions of inhibitors utilized to study its roles in human health, disease, and other scientific areas. Microbiol. Mol. Biol. Rev. 2008, 72, 590–641. [Google Scholar] [CrossRef]
- Kirschberg, M.; Heuser, S.; Marcuzzi, G.P.; Hufbauer, M.; Seeger, J.M.; Dukic, A.; Tomaic, V.; Majewski, S.; Wagner, S.; Wittekindt, C.; et al. ATP synthase modulation leads to an increase of spare respiratory capacity in HPV associated cancers. Sci. Rep. 2020, 10, 17339. [Google Scholar] [CrossRef]
- Ziegler, C.M.; Eisenhauer, P.; Kelly, J.A.; Dang, L.N.; Beganovic, V.; Bruce, E.A.; King, B.R.; Shirley, D.J.; Weir, M.E.; Ballif, B.A.; et al. A Proteomics Survey of Junin Virus Interactions with Human Proteins Reveals Host Factors Required for Arenavirus Replication. J. Virol. 2018, 92, e01565-17. [Google Scholar] [CrossRef] [PubMed]
- Meier, A.; Mehrle, S.; Weiss, T.S.; Mier, W.; Urban, S. Myristoylated PreS1-domain of the hepatitis B virus L-protein mediates specific binding to differentiated hepatocytes. Hepatology 2013, 58, 31–42. [Google Scholar] [CrossRef]
- Cheng, D.; Han, B.; Zhang, W.; Wu, W. Clinical effects of NTCP-inhibitor myrcludex B. J. Viral Hepat. 2021, 28, 852–858. [Google Scholar] [CrossRef] [PubMed]
- Donkers, J.M.; Appelman, M.D.; van de Graaf, S.F.J. Mechanistic insights into the inhibition of NTCP by myrcludex B. JHEP Rep. 2019, 1, 278–285. [Google Scholar] [CrossRef] [PubMed]
- Donkers, J.M.; Zehnder, B.; van Westen, G.J.P.; Kwakkenbos, M.J.; AP, I.J.; Oude Elferink, R.P.J.; Beuers, U.; Urban, S.; van de Graaf, S.F.J. Reduced hepatitis B and D viral entry using clinically applied drugs as novel inhibitors of the bile acid transporter NTCP. Sci. Rep. 2017, 7, 15307. [Google Scholar] [CrossRef]
- Muriungi, N.G.; Ueda, K. TIMM29 interacts with hepatitis B virus preS1 to modulate the HBV life cycle. Microbiol. Immunol. 2020, 64, 792–809. [Google Scholar] [CrossRef] [PubMed]
- Beil, W.; Birkholz, C.; Wagner, S.; Sewing, K.F. Bismuth subcitrate and omeprazole inhibit Helicobacter pyloriF1-ATPase. Pharmacology 1995, 50, 333–337. [Google Scholar] [CrossRef]
- De Clercq, E. 1984—Discovery of the First Anti-HIV Drug, Suramin. Viruses 2021, 13, 1646. [Google Scholar] [CrossRef]
- Tsiquaye, K.; Zuckerman, A. Suramin inhibits duck hepatitis B virus DNA polymerase activity. J. Hepatol. 1985, 1, 663–669. [Google Scholar] [CrossRef]
- Taylor, J.M.; Han, Z. Purinergic receptor functionality is necessary for infection of human hepatocytes by hepatitis delta virus and hepatitis B virus. PLoS ONE 2010, 5, e15784. [Google Scholar] [CrossRef]
- Martinez, L.O.; Najib, S.; Perret, B.; Cabou, C.; Lichtenstein, L. Ecto-F1-ATPase/P2Y pathways in metabolic and vascular functions of high density lipoproteins. Atherosclerosis 2015, 238, 89–100. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, M.; Watashi, K.; Tsukuda, S.; Aly, H.H.; Fukasawa, M.; Fujimoto, A.; Suzuki, R.; Aizaki, H.; Ito, T.; Koiwai, O.; et al. Evaluation and identification of hepatitis B virus entry inhibitors using HepG2 cells overexpressing a membrane transporter NTCP. Biochem. Biophys. Res. Commun. 2014, 443, 808–813. [Google Scholar] [CrossRef]
- Tsukuda, S.; Watashi, K.; Iwamoto, M.; Suzuki, R.; Aizaki, H.; Okada, M.; Sugiyama, M.; Kojima, S.; Tanaka, Y.; Mizokami, M.; et al. Dysregulation of retinoic acid receptor diminishes hepatocyte permissiveness to hepatitis B virus infection through modulation of sodium taurocholate cotransporting polypeptide (NTCP) expression. J. Biol. Chem. 2015, 290, 5673–5684. [Google Scholar] [CrossRef] [PubMed]
- Ladner, S.K.; Otto, M.J.; Barker, C.S.; Zaifert, K.; Wang, G.H.; Guo, J.T.; Seeger, C.; King, R.W. Inducible expression of human hepatitis B virus (HBV) in stably transfected hepatoblastoma cells: A novel system for screening potential inhibitors of HBV replication. Antimicrob. Agents Chemother. 1997, 41, 1715–1720. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ueda, K.; Suwanmanee, Y. ATP5B Is an Essential Factor for Hepatitis B Virus Entry. Int. J. Mol. Sci. 2022, 23, 9570. https://doi.org/10.3390/ijms23179570
Ueda K, Suwanmanee Y. ATP5B Is an Essential Factor for Hepatitis B Virus Entry. International Journal of Molecular Sciences. 2022; 23(17):9570. https://doi.org/10.3390/ijms23179570
Chicago/Turabian StyleUeda, Keiji, and Yadarat Suwanmanee. 2022. "ATP5B Is an Essential Factor for Hepatitis B Virus Entry" International Journal of Molecular Sciences 23, no. 17: 9570. https://doi.org/10.3390/ijms23179570
APA StyleUeda, K., & Suwanmanee, Y. (2022). ATP5B Is an Essential Factor for Hepatitis B Virus Entry. International Journal of Molecular Sciences, 23(17), 9570. https://doi.org/10.3390/ijms23179570





