Impact of Growth Conditions on Pseudomonas fluorescens Morphology Characterized by Atomic Force Microscopy
Abstract
1. Introduction
2. Results and Discussion
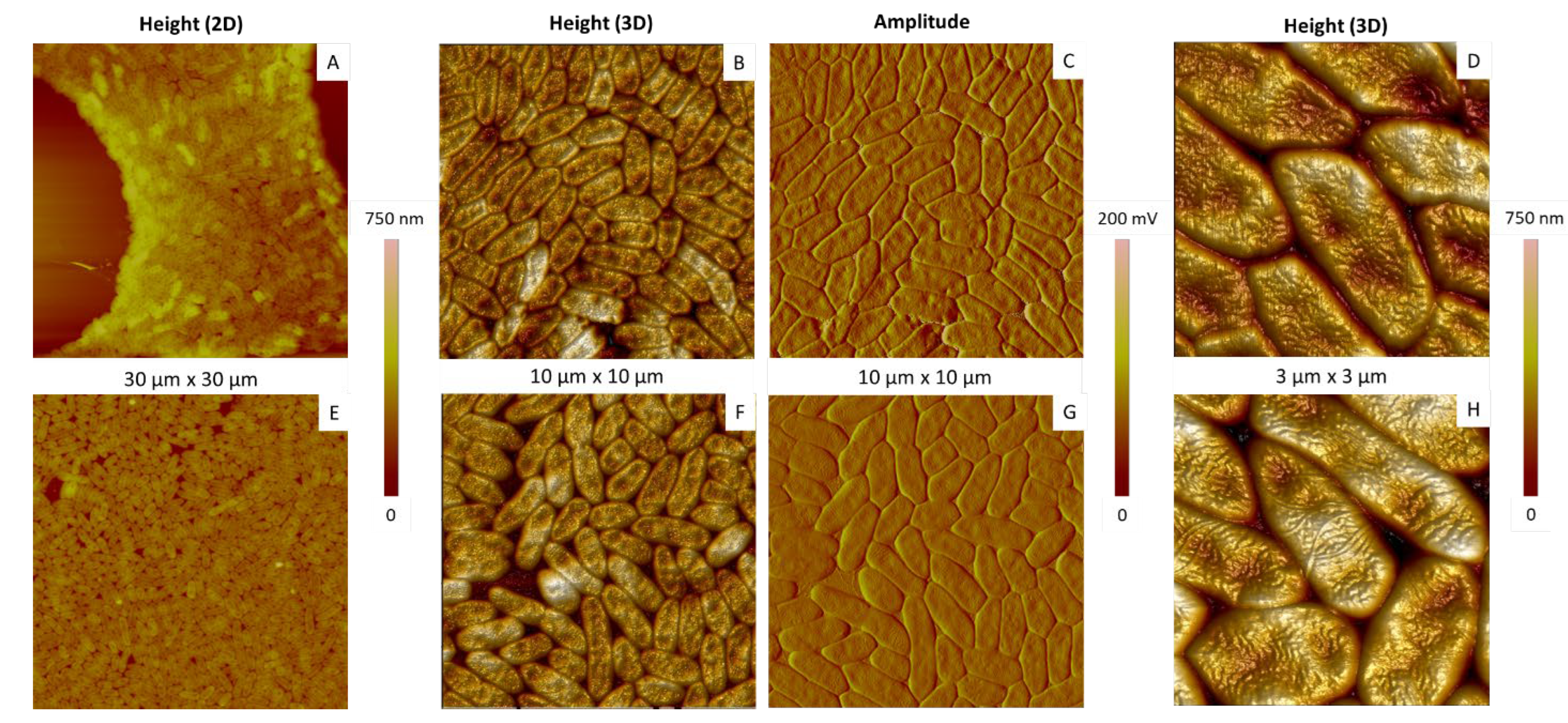
2.1. Determination of the Optimal Bacterial Concentration in Suspensions for AFM
2.2. Characterization by AFM of P. fluorescens Grown under Optimal Conditions
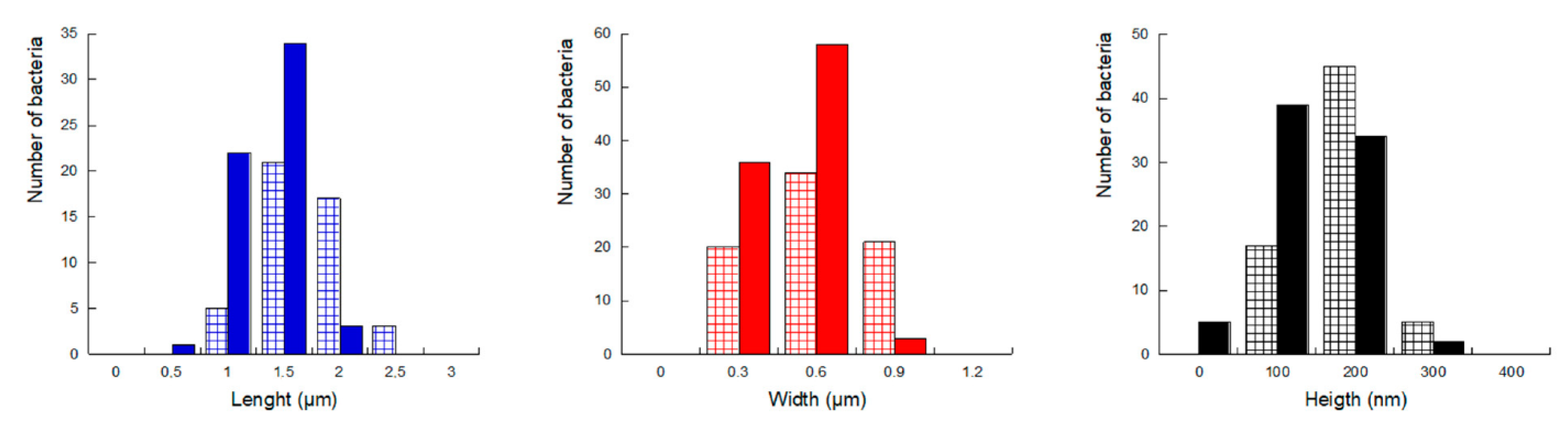
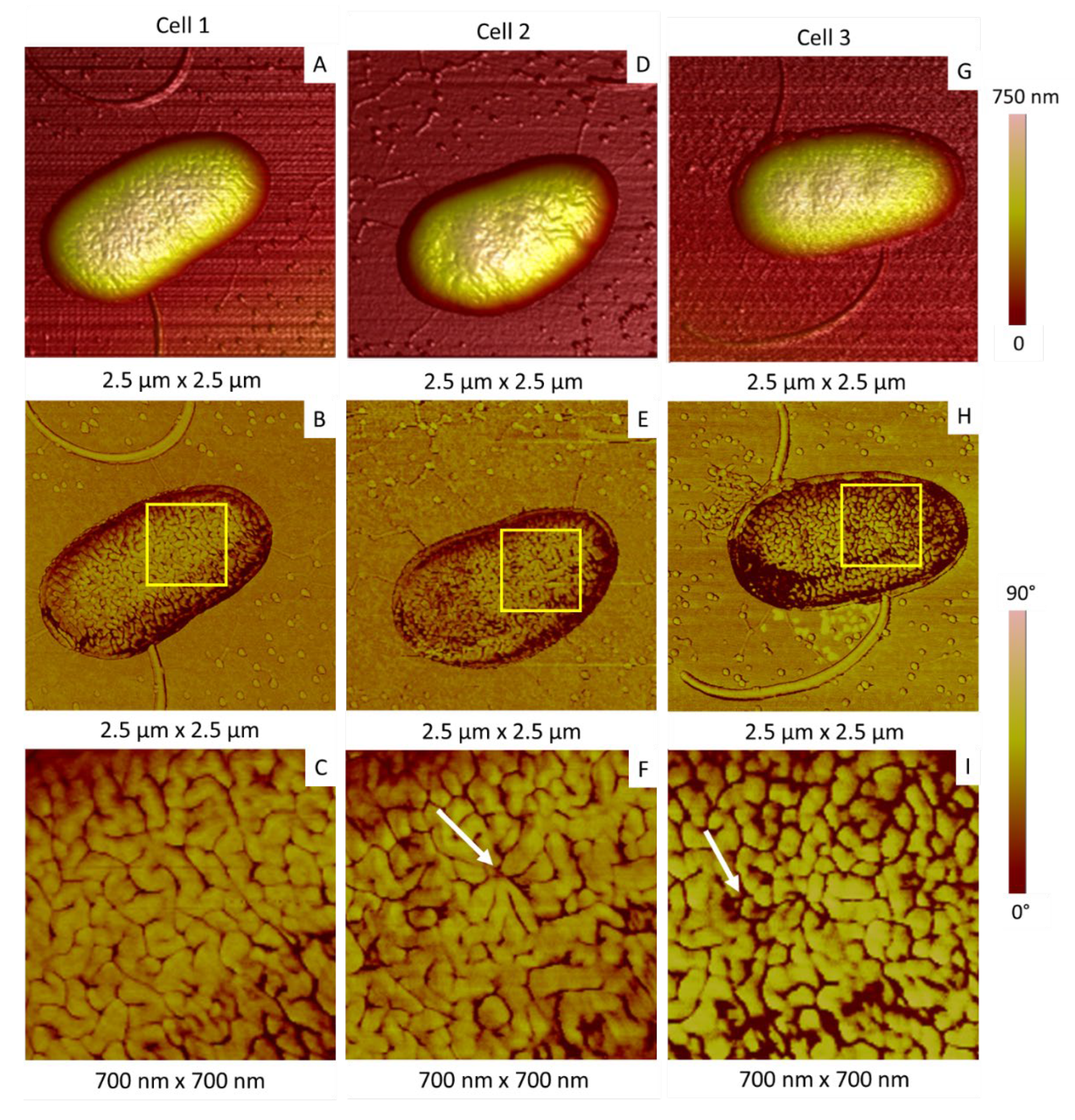
2.2.1. Morphology and Dimensions
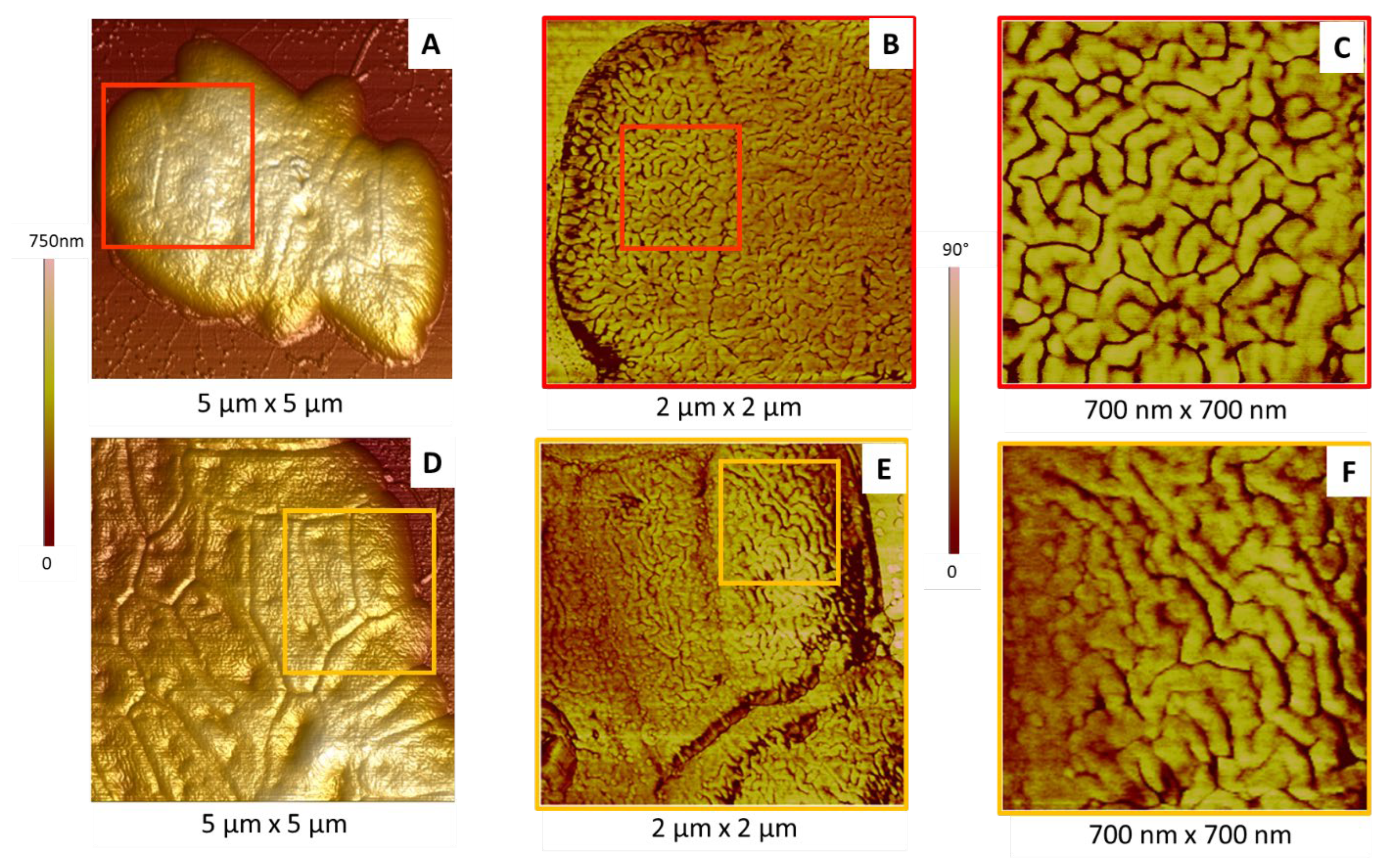
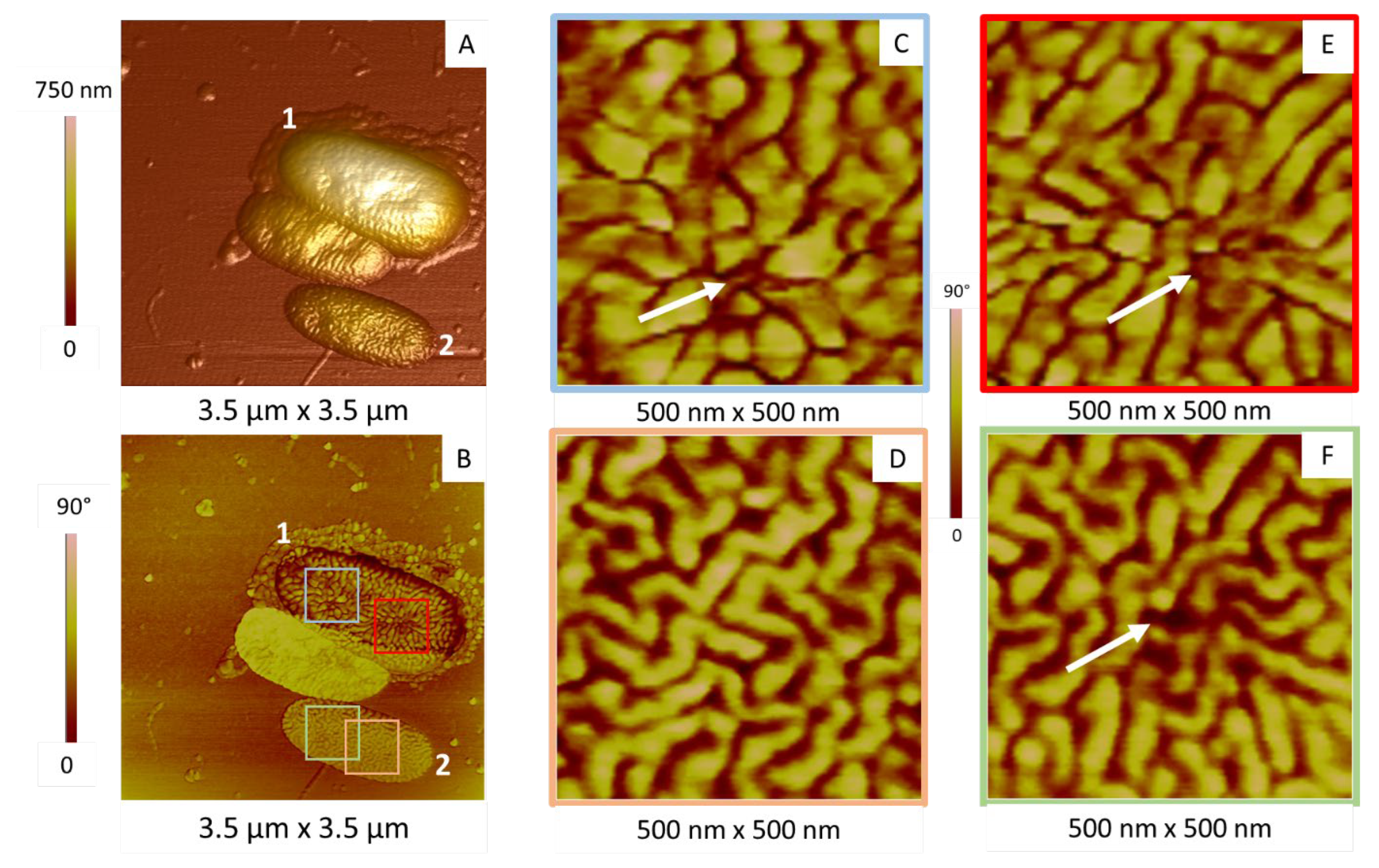
2.2.2. Membrane Organization
2.3. Impact of Less Favourable Culture Conditions on P. fluorescens Morphology
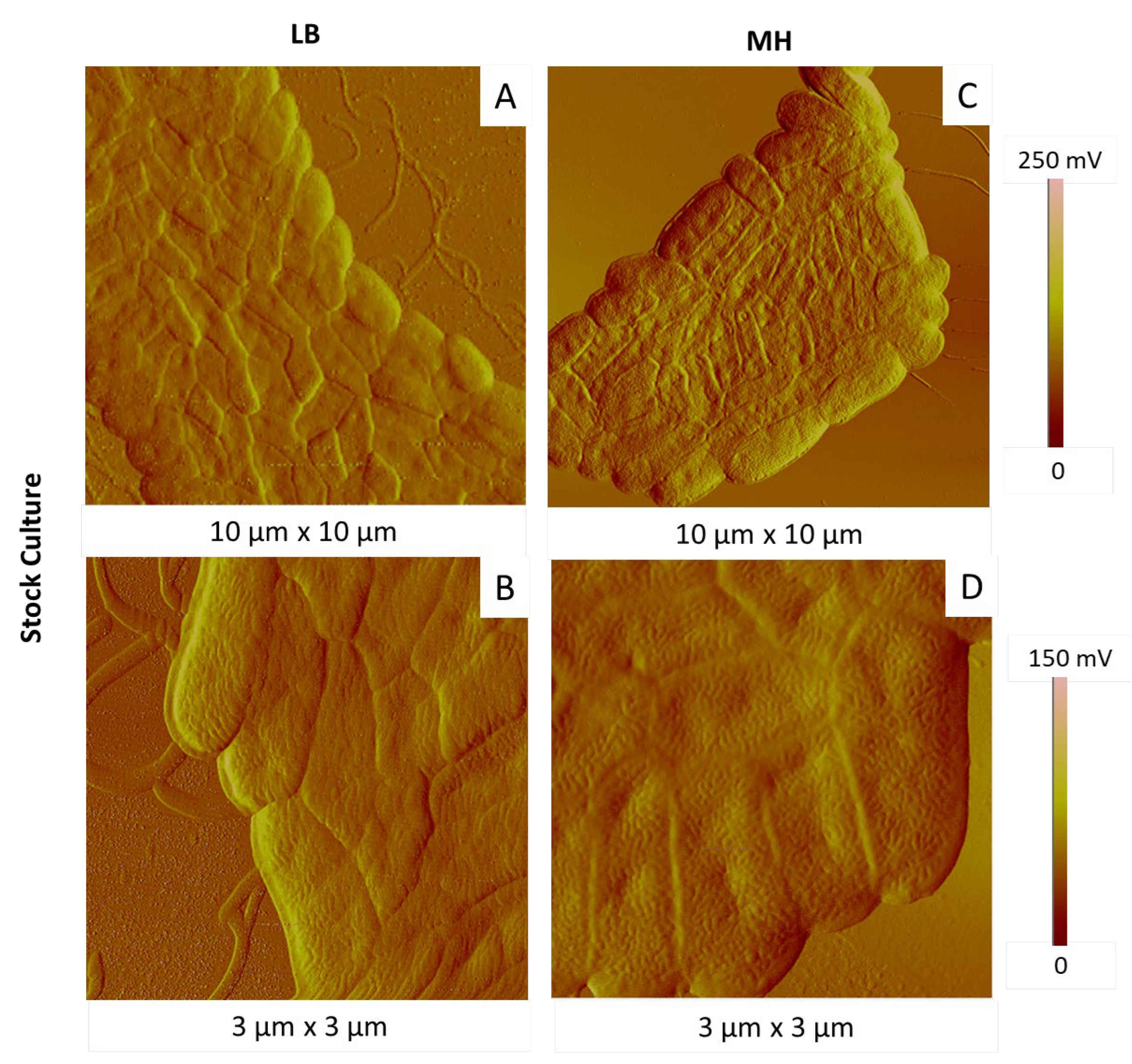
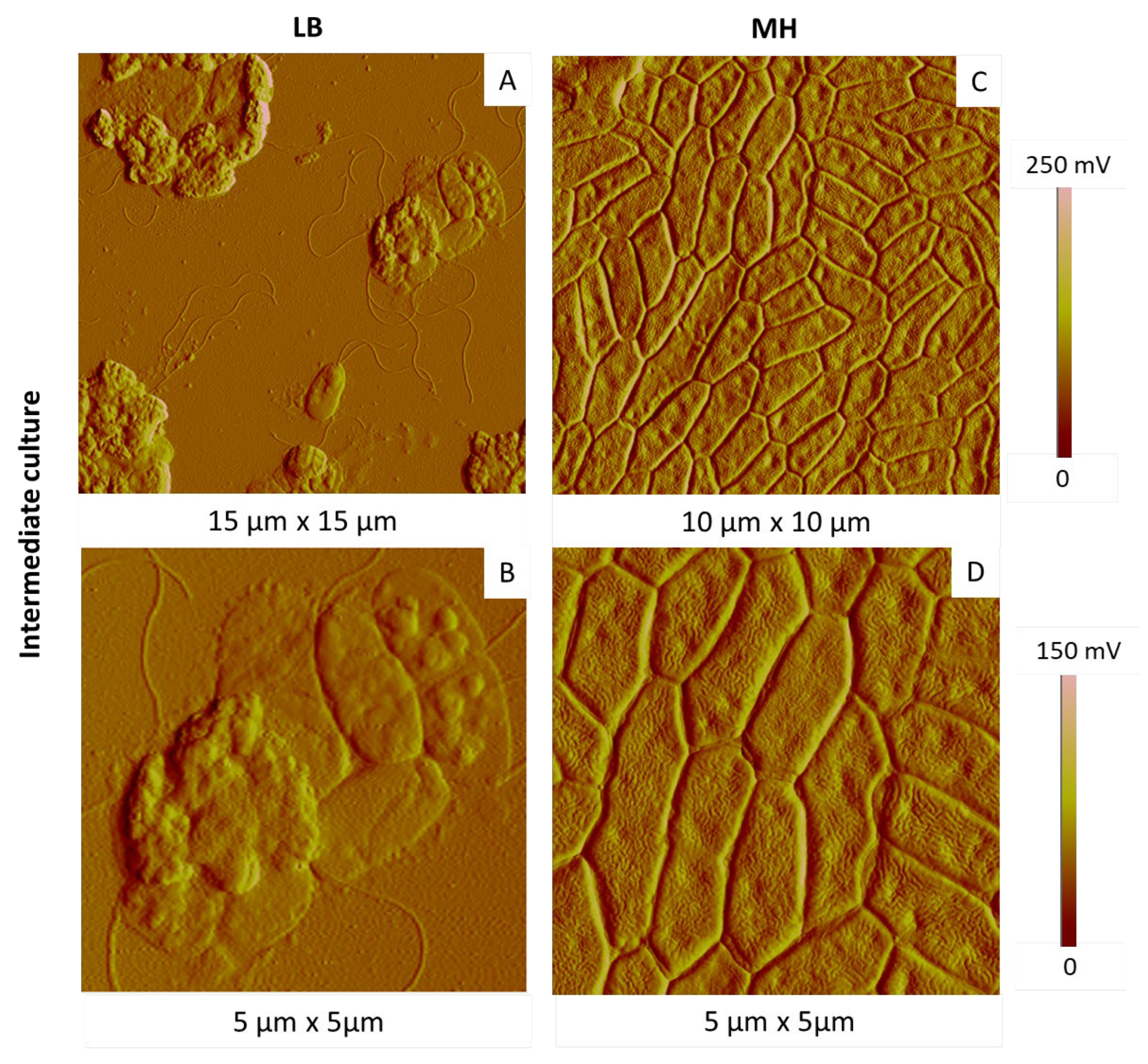
2.3.1. Influence of the Culture Medium
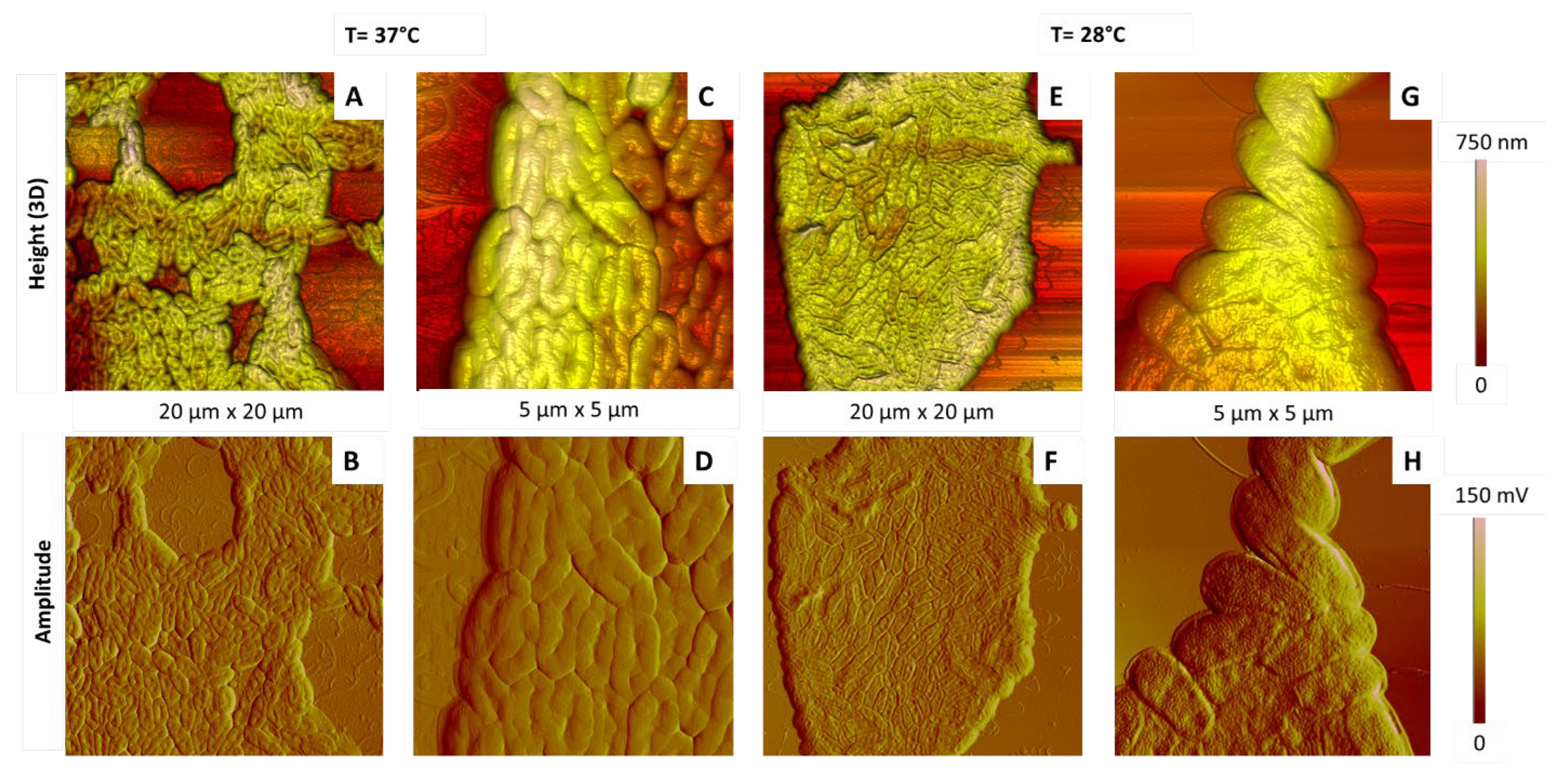
2.3.2. Influence of the Culture Temperature
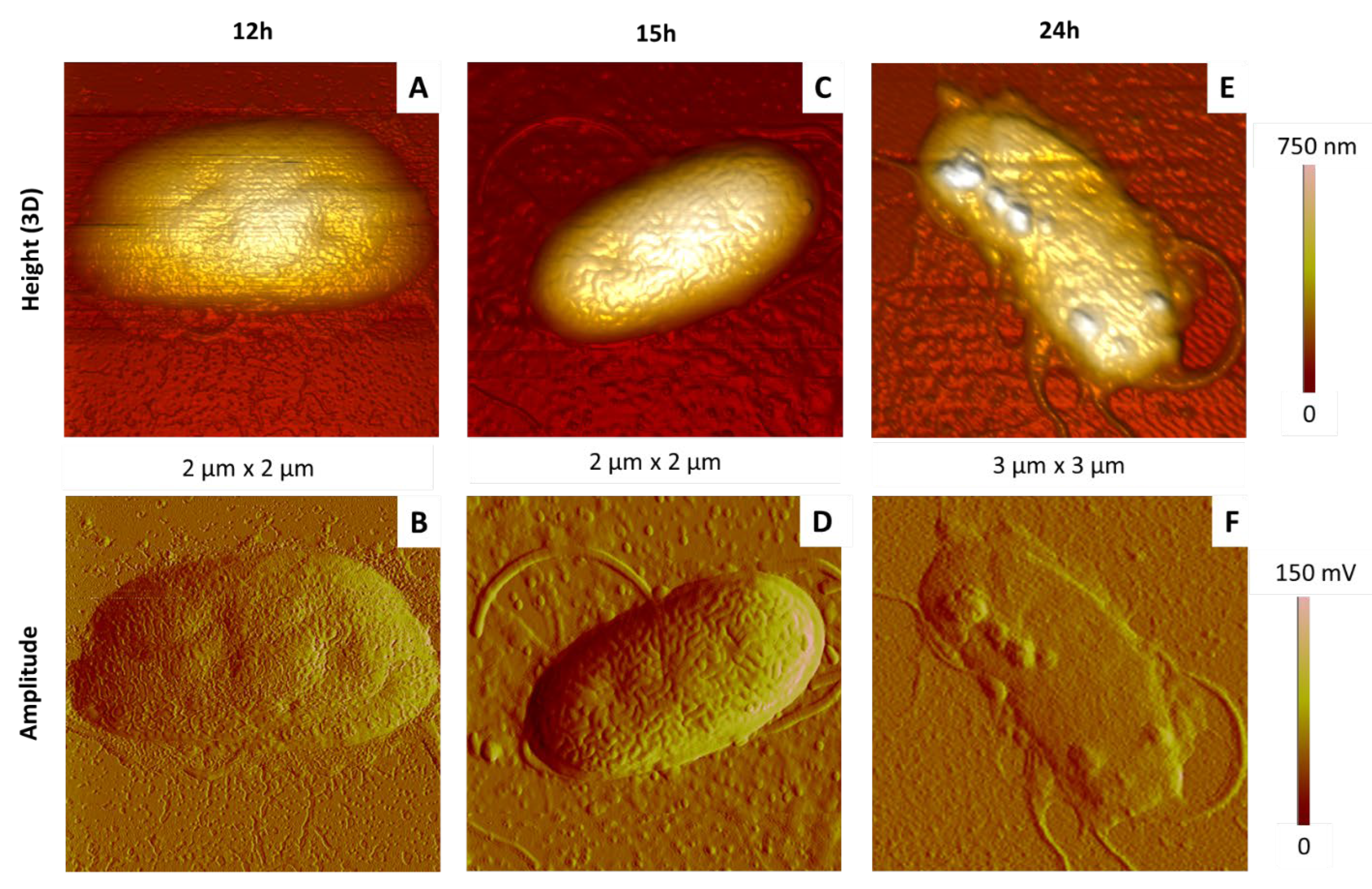
2.3.3. Influence of the Growth Incubation Time
3. Materials and Methods
3.1. Bacterial Strain, Reagents and Growth Conditions
3.2. Preparation of Samples for Imaging Experiments
3.3. Optical Microscopy (OM)
3.4. Atomic Force Microscopy
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Lemanceau, P. Effets bénéfiques de rhizobactéries sur les plantes: Exemple des Pseudomonas spp. fluorescents. Agronomie 1992, 12, 413–437. [Google Scholar] [CrossRef]
- Meyer, J.-M. Pyoverdines: Pigments, siderophores and potential taxonomic markers of Fluorescent Pseudomonas Species. Arch. Microbiol. 2000, 174, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Baranski, R.; Klocke, E.; Nothnagel, T. enhancing resistance of transgenic carrot to fungal pathogens by the expression of Pseudomonas Fluorescens Microbial Factor 3 (MF3) Gene. Physiol. Mol. Plant Pathol. 2007, 71, 88–95. [Google Scholar] [CrossRef]
- Wu, B.; He, T.; Wang, Z.; Qiao, S.; Wang, Y.; Xu, F.; Xu, H. Insight into the mechanisms of plant growth promoting strain SNB6 on Enhancing the Phytoextraction in Cadmium Contaminated Soil. J. Hazard. Mater. 2020, 385, 121587. [Google Scholar] [CrossRef]
- Bolwerk, A.; Lagopodi, A.L.; Wijfjes, A.H.M.; Lamers, G.E.M.; Chin-A-Woeng, T.F.C.; Lugtenberg, B.J.J.; Bloemberg, G.V. Interactions in the Tomato Rhizosphere of Two Pseudomonas Biocontrol Strains with the Phytopathogenic Fungus Fusarium oxysporum f. Sp. Radicis-Lycopersici. MPMI 2003, 16, 983–993. [Google Scholar] [CrossRef]
- Watson, T.T.; Forge, T.A.; Nelson, L.M. Pseudomonads Contribute to Regulation of Pratylenchus Penetrans (Nematoda) Populations on Apple. Can. J. Microbiol. 2018, 64, 775–785. [Google Scholar] [CrossRef]
- Garbeva, P.; van Veen, J.A.; van Elsas, J.D. Assessment of the Diversity, and Antagonism towards Rhizoctonia solani AG3, of Pseudomonas Species in Soil from Different Agricultural Regimes. FEMS Microbiol. Ecol. 2004, 47, 51–64. [Google Scholar] [CrossRef]
- Höfte, M.; Bakker, P.A.H.M. Competition for Iron and Induced Systemic Resistance by Siderophores of Plant Growth Promoting Rhizobacteria. In Microbial Siderophores; Varma, A., Chincholkar, S.B., Eds.; Soil Biology; Springer: Berlin/Heidelberg, Germany, 2007; pp. 121–133. ISBN 978-3-540-71160-5. [Google Scholar]
- Meyer, J.M.; Abdallah, M.A.Y. 1978 The Fluorescent Pigment of Pseudomonas fluorescens: Biosynthesis, Purification and Physicochemical Properties. Microbiology 1978, 107, 319–328. [Google Scholar] [CrossRef]
- Meyer, J.M.; Hornsperger, J.M.Y. 1978 Role of PyoverdinePf, the Iron-Binding Fluorescent Pigment of Pseudomonas fluorescens, in Iron Transport. Microbiology 1978, 107, 329–331. [Google Scholar] [CrossRef]
- Latour, X.; Lemanceau, P. Carbon and energy metabolism of oxidase-positive saprophytic fluorescent Pseudomonas spp. Agronomie 1997, 9–10, 427–443. [Google Scholar] [CrossRef]
- Coates, T.; Bax, R.; Coates, A. Nasal Decolonization of Staphylococcus aureus with Mupirocin: Strengths, Weaknesses and Future Prospects. J. Antimicrob. Chemother. 2009, 64, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Aponte, A.; Castillo, O.; Cabrera, G.; Pernia, M.; Hernandez, Y. Rhizobacteria Pseudomonas fluorescens and Azospirillum Sp. Association Enhances Growth of Lactuca Sativa L. under Tropical Conditions. J. Cent. Eur. Agric. 2017, 18, 424–440. [Google Scholar] [CrossRef]
- Gamalero, E.; Trotta, A.; Massa, N.; Copetta, A.; Martinotti, M.G.; Berta, G. Impact of Two Fluorescent Pseudomonads and an Arbuscular Mycorrhizal Fungus on Tomato Plant Growth, Root Architecture and P Acquisition. Mycorrhiza 2004, 14, 185–192. [Google Scholar] [CrossRef] [PubMed]
- Picot, L.; Mezghani-Abdelmoula, S.; Chevalier, S.; Merieau, A.; Lesouhaitier, O.; Guerillon, J.; Cazin, L.; Orange, N.; Feuilloley, M.G.J. Regulation of the Cytotoxic Effects of Pseudomonas fluorescens by Growth Temperature. Res. Microbiol. 2004, 155, 39–46. [Google Scholar] [CrossRef]
- Scales, B.S.; Dickson, R.P.; LiPuma, J.J.; Huffnagle, G.B. Microbiology, Genomics, and Clinical Significance of the Pseudomonas fluorescens Species Complex, an Unappreciated Colonizer of Humans. Clin. Microbiol. Rev. 2014, 27, 927–948. [Google Scholar] [CrossRef]
- Raja, C.E.; Anbazhagan, K.; Selvam, G.S. Isolation and Characterization of A Metal-Resistant Pseudomonas aeruginosa Strain. World J. Microbiol. Biotechnol. 2006, 22, 577–585. [Google Scholar] [CrossRef]
- Edward Raja, C.; Sasikumar, S.; Selvam, G.S. Adaptive and Cross Resistance to Cadmium (II) and Zinc (II) by Pseudomonas aeruginosa BC15. Biologia 2008, 63, 461. [Google Scholar] [CrossRef]
- Raja, C.E.; Selvam, G.S. Plasmid Profile and Curing Analysis of Pseudomonas aeruginosa as Metal Resistant. Int. J. Environ. Sci. Technol. 2009, 6, 259–266. [Google Scholar] [CrossRef]
- Chellaiah, E.R. Cadmium (Heavy Metals) Bioremediation by Pseudomonas aeruginosa: A Minireview. Appl. Water Sci. 2018, 8, 154. [Google Scholar] [CrossRef]
- Pollock, H.M.; Minshew, B.H.; Kenny, M.A.; Schoenknecht, F.D. Effect of Different Lots of Mueller-Hinton Agar on the Interpretation of the Gentamicin Susceptibility of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 1978, 14, 360–367. [Google Scholar] [CrossRef]
- Fujita, S.; Tonohata, A.; Matsuoka, T.; Okado, N.; Hashimoto, T. Identification of Pseudomonas aeruginosa by Using a Disk of Phenanthroline and 9-Chloro-9-[4-(Diethylamino)Phenyl]-9,10-Dihydro-10-Phenylacridine Hydrochloride and by Cell Agglutination Testing with Monoclonal Antibodies. J. Clin. Microbiol. 1992, 30, 2728–2729. [Google Scholar] [CrossRef] [PubMed]
- Munna, M.S.; Zeba, Z.; Noor, R. Influence of Temperature on the Growth of Pseudomonas putida. Stamford J. Microbiol. 2015, 5, 9–12. [Google Scholar] [CrossRef]
- Picot, L.; Chevalier, S.; Mezghani-Abdelmoula, S.; Merieau, A.; Lesouhaitier, O.; Leroux, P.; Cazin, L.; Orange, N.; Feuilloley, M.G.J. Cytotoxic Effects of the Lipopolysaccharide from Pseudomonas fluorescens on Neurons and Glial Cells. Microb. Pathog. 2003, 35, 95–106. [Google Scholar] [CrossRef]
- Auerbach, I.D.; Sorensen, C.; Hansma, H.G.; Holden, P.A. Physical Morphology and Surface Properties of Unsaturated Pseudomonas putida Biofilms. J. Bacteriol. 2000, 182, 3809–3815. [Google Scholar] [CrossRef] [PubMed]
- Spiers, A.J.; Bohannon, J.; Gehrig, S.M.; Rainey, P.B. Biofilm Formation at the Air–Liquid Interface by the Pseudomonas Fluorescens SBW25 Wrinkly Spreader Requires an Acetylated Form of Cellulose. Mol. Microbiol. 2003, 50, 15–27. [Google Scholar] [CrossRef]
- Capdevila, S.; Martínez-Granero, F.M.; Sánchez-Contreras, M.; Rivilla, R.; Martín, M. Analysis of Pseudomonas fluorescens F113 Genes Implicated in Flagellar Filament Synthesis and Their Role in Competitive Root Colonization. Microbiology 2004, 150, 3889–3897. [Google Scholar] [CrossRef]
- Mitik-Dineva, N.; Wang, J.; Truong, V.K.; Stoddart, P.; Malherbe, F.; Crawford, R.J.; Ivanova, E.P. Escherichia coli, Pseudomonas aeruginosa, and Staphylococcus Aureus Attachment Patterns on Glass Surfaces with Nanoscale Roughness. Curr. Microbiol. 2009, 58, 268–273. [Google Scholar] [CrossRef]
- Yuan, S.J.; Pehkonen, S.O. AFM Study of Microbial Colonization and Its Deleterious Effect on 304 Stainless Steel by Pseudomonas NCIMB 2021 and Desulfovibrio desulfuricans in Simulated Seawater. Corros. Sci. 2009, 51, 1372–1385. [Google Scholar] [CrossRef]
- Kamaeva, A.A.; Vasilchenko, A.S.; Deryabin, D.G. Atomic Force Microscopy Reveals a Morphological Differentiation of Chromobacterium violaceum Cells Associated with Biofilm Development and Directed by N-Hexanoyl-L-Homoserine Lactone. PLoS ONE 2014, 9, e103741. [Google Scholar] [CrossRef]
- Cai, Y.; King, R.B.; Law, W.; McInerney, D.M. Which Comes First? Modeling the Relationships among Future Goals, Metacognitive Strategies and Academic Achievement Using Multilevel Cross-Lagged SEM. Learn. Individ. Differ. 2019, 74, 101750. [Google Scholar] [CrossRef]
- Wang, Q.; Yang, Q.; Wu, W. Progress on Structured Biosensors for Monitoring Aflatoxin B1 From Biofilms: A Review. Front. Microbiol. 2020, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.Y.; Llewellyn, E.; Chen, J.; Olinares, P.D.B.; Brewer, J.; Chait, B.T.; Campbell, E.A.; Darst, S.A. Structural Basis for Transcription Complex Disruption by the Mfd Translocase. eLife 2021, 10, e62117. [Google Scholar] [CrossRef] [PubMed]
- Simon, A.; Cohen-Bouhacina, T.; Porté, M.C.; Aimé, J.P.; Amédée, J.; Bareille, R.; Baquey, C. Characterization of Dynamic Cellular Adhesion of Osteoblasts Using Atomic Force Microscopy. Cytom. Part A 2003, 54A, 36–47. [Google Scholar] [CrossRef]
- Dufrêne, Y.F. Application of Atomic Force Microscopy to Microbial Surfaces: From Reconstituted Cell Surface Layers to Living Cells. Micron 2001, 32, 153–165. [Google Scholar] [CrossRef]
- Baniasadi, M.; Xu, Z.; Gandee, L.; Du, Y.; Lu, H.; Zimmern, P.; Minary-Jolandan, M. Nano indentation Of Pseudomonas aeruginosa bacterial Biofilm Using Atomic Force Microscopy. Mater. Res. Express 2014, 1, 045411. [Google Scholar] [CrossRef]
- Gammoudi, I.; Mathelie-guinlet, M.; Morote, F.; Beven, L.; Moynet, D.; Grauby-heywang, C.; Cohen-bouhacina, T. Morphological and Nanostructural Surface Changes in Escherichia Coli over Time, Monitored by Atomic Force Microscopy. Colloids Surf. B Biointerfaces 2016, 141, 355–364. [Google Scholar] [CrossRef]
- Fernandez, M.; Godino, A.; Príncipe, A.; Morales, G.M.; Fischer, S. Effect of a Pseudomonas fluorescens Tailocin against Phytopathogenic Xanthomonas Observed by Atomic Force Microscopy. J. Biotechnol. 2017, 256, 13–20. [Google Scholar] [CrossRef]
- Mathelié-Guinlet, M.; Béven, L.; Moroté, F.; Moynet, D.; Grauby-Heywang, C.; Gammoudi, I.; Delville, M.-H.; Cohen-Bouhacina, T. Probing the Threshold of Membrane Damage and Cytotoxicity Effects Induced by Silica Nanoparticles in Escherichia coli Bacteria. Adv. Colloid Interface Sci. 2017, 245, 81–91. [Google Scholar] [CrossRef] [PubMed]
- Díaz, C.; Schilardi, P.L.; Salvarezza, R.C.; Fernández Lorenzo de Mele, M. Have Flagella a Preferred Orientation during Early Stages of Biofilm Formation?: AFM Study Using Patterned Substrates. Colloids Surf. B Biointerfaces 2011, 82, 536–542. [Google Scholar] [CrossRef]
- Allen, A.; Habimana, O.; Casey, E. The Effects of Extrinsic Factors on the Structural and Mechanical Properties of Pseudomonas fluorescens Biofilms: A Combined Study of Nutrient Concentrations and Shear Conditions. Colloids Surf. B Biointerfaces 2018, 165, 127–134. [Google Scholar] [CrossRef]
- Lindsay, D.; Brözel, V.S.; Mostert, J.F.; von Holy, A. Differential Efficacy of a Chlorine Dioxide-Containing Sanitizer against Single Species and Binary Biofilms of a Dairy-Associated Bacillus Cereus and a Pseudomonas fluorescens Isolate. J. Appl. Microbiol. 2002, 92, 352–361. [Google Scholar] [CrossRef] [PubMed]
- Nony, L.; Boisgard, R.; Aimé, J.-P. DNA Properties Investigated by Dynamic Force Microscopy. Biomacromolecules 2001, 2, 827–835. [Google Scholar] [CrossRef][Green Version]
- Mathelié-Guinlet, M.; Cohen-Bouhacina, T.; Gammoudi, I.; Martin, A.; Béven, L.; Delville, M.-H.; Grauby-Heywang, C. Silica Nanoparticles-Assisted Electrochemical Biosensor for the Rapid, Sensitive and Specific Detection of Escherichia coli. Sens. Actuators B Chem. 2019, 292, 314–320. [Google Scholar] [CrossRef]
- Wilkinson, S.G. Composition and Structure of Lipopolysaccharides from Pseudomonas aeruginosa. Rev. Infect. Dis. 1983, 5, S941–S949. [Google Scholar] [CrossRef] [PubMed]
- Dowhan, W. Role of Phospholipids in Escherichia coli Cell Function. In Advances in Cellular and Molecular Biology of Membranes and Organelles; Tartakoff, A.M., Dalbey, R.E., Eds.; Protein Export and Membrane Biogenesis; JAI Press: Greenwich, CT, USA, 1995; Volume 4, pp. 189–217. [Google Scholar]
- Gerald B, P. Pseudomonas aeruginosa Lipopolysaccharide: A Major Virulence Factor, Initiator of Inflammation and Target for Effective Immunity. Int. J. Med. Microbiol. 2007, 297, 277–295. [Google Scholar] [CrossRef]
- Ruhal, R.; Antti, H.; Rzhepishevska, O.; Boulanger, N.; Barbero, D.R.; Wai, S.N.; Uhlin, B.E.; Ramstedt, M. A Multivariate Approach to Correlate Bacterial Surface Properties to Biofilm Formation by Lipopolysaccharide Mutants of Pseudomonas aeruginosa. Colloids Surf. B Biointerfaces 2015, 127, 182–191. [Google Scholar] [CrossRef]
- Ellis, T.N.; Leiman, S.A.; Kuehn, M.J. Naturally Produced Outer Membrane Vesicles from Pseudomonas aeruginosa Elicit a Potent Innate Immune Response via Combined Sensing of Both Lipopolysaccharide and Protein Components. Infect. Immun. 2010, 78, 3822–3831. [Google Scholar] [CrossRef]
- Cooke, A.C.; Nello, A.V.; Ernst, R.K.; Schertzer, J.W. Analysis of Pseudomonas aeruginosa Biofilm Membrane Vesicles Supports Multiple Mechanisms of Biogenesis. PLoS ONE 2019, 14, e0212275. [Google Scholar] [CrossRef]
- Armstrong, D.A.; Lee, M.K.; Hazlett, H.F.; Dessaint, J.A.; Mellinger, D.L.; Aridgides, D.S.; Hendricks, G.M.; Abdalla, M.A.K.; Christensen, B.C.; Ashare, A. Extracellular Vesicles from Pseudomonas aeruginosa Suppress MHC-Related Molecules in Human Lung Macrophages. ImmunoHorizons 2020, 4, 508–519. [Google Scholar] [CrossRef]
- Abraham, T.; Schooling, S.R.; Beveridge, T.J.; Katsaras, J. Monolayer Film Behavior of Lipopolysaccharide from Pseudomonas aeruginosa at the Air-Water Interface. Biomacromolecules 2008, 9, 2799–2804. [Google Scholar] [CrossRef]
- Meler, M.J.; Subasinghe, R.M.; Beaudette, L.A. Draft Genome Sequence of the Industrially Significant Bacterium Pseudomonas fluorescens ATCC 13525. Microbiol. Resour. Announc. 2018, 7, e01368-18. [Google Scholar] [CrossRef]
- Haddad, S.; Elliot, M.; Savard, T.; Deschênes, L.; Smith, T.; Ells, T. Variations in Biofilms Harbouring Listeria Monocytogenes in Dual and Triplex Cultures with Pseudomonas fluorescens and Lactobacillus plantarum Produced under a Model System of Simulated Meat Processing Conditions, and Their Resistance to Benzalkonium Chloride. Food Control 2021, 123, 107720. [Google Scholar] [CrossRef]
- Islam, M.A.; Hassen, W.M.; Tayabali, A.F.; Dubowski, J.J. Short Ligand, Cysteine-Modified Warnericin RK Antimicrobial Peptides Favor Highly Sensitive Detection of Legionella pneumophila. ACS Omega 2021, 6, 1299–1308. [Google Scholar] [CrossRef]
- Ben-David, A.; Davidson, C.E. Estimation Method for Serial Dilution Experiments. J. Microbiol. Methods 2014, 107, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Goldman, E.; Green, L.H. Practical Handbook of Microbiology; CRC Press: Boca Taton, FL, USA, 2015; ISBN 978-1-4665-8740-3. [Google Scholar]
- Nanjo, H.; Nony, L.; Yoneya, M.; Aimé, J.P. Simulation of Section Curve by Phase Constant Dynamic Mode Atomic Force Microscopy in Non-Contact Situation. Appl. Surf. Sci. 2003, 210, 49–53. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kahli, H.; Béven, L.; Grauby-Heywang, C.; Debez, N.; Gammoudi, I.; Moroté, F.; Sbartai, H.; Cohen-Bouhacina, T. Impact of Growth Conditions on Pseudomonas fluorescens Morphology Characterized by Atomic Force Microscopy. Int. J. Mol. Sci. 2022, 23, 9579. https://doi.org/10.3390/ijms23179579
Kahli H, Béven L, Grauby-Heywang C, Debez N, Gammoudi I, Moroté F, Sbartai H, Cohen-Bouhacina T. Impact of Growth Conditions on Pseudomonas fluorescens Morphology Characterized by Atomic Force Microscopy. International Journal of Molecular Sciences. 2022; 23(17):9579. https://doi.org/10.3390/ijms23179579
Chicago/Turabian StyleKahli, Houssem, Laure Béven, Christine Grauby-Heywang, Nesrine Debez, Ibtissem Gammoudi, Fabien Moroté, Hana Sbartai, and Touria Cohen-Bouhacina. 2022. "Impact of Growth Conditions on Pseudomonas fluorescens Morphology Characterized by Atomic Force Microscopy" International Journal of Molecular Sciences 23, no. 17: 9579. https://doi.org/10.3390/ijms23179579
APA StyleKahli, H., Béven, L., Grauby-Heywang, C., Debez, N., Gammoudi, I., Moroté, F., Sbartai, H., & Cohen-Bouhacina, T. (2022). Impact of Growth Conditions on Pseudomonas fluorescens Morphology Characterized by Atomic Force Microscopy. International Journal of Molecular Sciences, 23(17), 9579. https://doi.org/10.3390/ijms23179579





