Abstract
Apical Lesions of Endodontic Origin (ALEO) are initiated by polymicrobial endodontic canal infection. Porphyromonas gingivalis (Pg) and Porphyromonas endodontalis (Pe) lipopolysaccharides (LPS) can induce a pro-inflammatory macrophage response through their recognition by TLR2 and TLR4. However, polarization responses induced by Pg and/or Pe LPS in macrophages are not fully understood. We aimed to characterize the polarization profiles of macrophages differentiated from THP-1 cells following Pg and/or Pe LPS stimulation from reference strain and clinical isolates. A modified LPS purification protocol was implemented and the electrophoretic LPS profiles were characterized. THP-1 human monocytes differentiated to macrophages were stimulated with Pg and Pe LPS. Polarization profiles were characterized through cell surface markers and secreted cytokines levels after 24 h of stimulation. TLR2 and TLR4 cell surfaces and transcriptional levels were determined after 24 or 2 h of LPS stimulation, respectively. LPS from Pg induced a predominant M1 profile in macrophages evidenced by changes in the expression of the surface marker CD64 and pro-inflammatory cytokine profiles, TNF-α, IL-1β, IL-6, and IL-12. Pe LPS was unable to induce a significant response. TLR2 and TLR4 expressions were neither modified by Pg or Pe LPS. Pg LPS, but not Pe LPS, induced a macrophage M1 Profile.
1. Introduction
Apical periodontitis (AP) is a prevalent inflammatory disease caused by dental infection comprising the pulpal tissue of the root canal system following a carious lesion of the tooth, which naturally evolves to form an apical lesion of endodontic origin (ALEO), a hallmark of the disease [1]. AP represents the main cause of tooth loss and has been increasingly linked with low-grade systemic inflammation and, consequently, a higher risk of cardiovascular diseases and diabetes mellitus [2,3,4].
The development and progression of ALEO depends on the dynamic balance between the bacterial consortia, their respective virulence factors, and the host’s immune response [5,6,7]. Although endodontic infections have a polymicrobial nature, black-pigmented bacteria, such as Prevotella and Porphyromonas spp., have been recognized as central agents in the development and progression of the lesion [8,9,10]. Among them, Porphyromonas endodontalis (Pe) and Porphyromonas gingivalis (Pg), stand out because of their high prevalence and virulence factors in AP [3,11,12,13,14,15,16,17].
Lipopolysaccharide (LPS), a key virulence factor of Gram-negative bacteria, is a complex glycolipid from the outer membrane, composed of the following three covalently linked domains: lipid A or endotoxin, which is inserted in the outer membrane, the oligosaccharides core and the O-antigen (OAg), which is the outermost and most variable part of the molecule [18]. LPS from P. gingivalis has been widely reported to activate inflammatory host response and bone loss during periodontal inflammation [19,20,21]. Likewise, LPS from P. endodontalis reduced osteoblast viability, induced the expression of TNF-α and IL-6 via NF-κB, and stimulated osteoclastogenesis and bone resorption in the calvaria mouse model [20,22,23,24,25].
Despite the infectious etiology of AP, the nature, scope, and duration of the immune response against endodontic infection are the main determinants of apical tissue destruction [7,26,27,28]. Resident macrophages are often the first immune cell to recognize pathogen-associated molecular patterns (PAMPs), such as LPS released from Gram-negative bacteria, which is specifically recognized by Toll-like receptors (TLRs) [23]. Macrophages show broad plasticity and differentiation dynamics towards two main phenotypic profiles, pro-inflammatory M1 and anti-inflammatory/healing M2 [28]. M1 macrophages differentiate from the classical activation pathway triggered mainly by the IFN-γ/Jak–STAT1 pathway under the interplay with TLRs [29,30], playing an important role in fighting bacteria and intracellular pathogens, while M2 macrophages result from exposure to IL-4/IL-13 or IL-10 regulated by STAT6 [31]. Symptomatic AP has been associated with M1 over M2 predominance [32] and higher expression of TLR2 in association with its promoter’s hypomethylation [33,34].
Overall, since P. endodontalis and P. gingivalis coexist with high frequency in AP [35] it is important to decipher the individual and combined effects of P. endodontalis and P. gingivalis LPS over macrophage differentiation. This study aimed to characterize the polarization profiles and TLR2/4 expression of macrophages differentiated from THP-1 cells following P. gingivalis and/or P. endodontalis LPS stimulation from reference strain and clinical isolates.
2. Results
We characterized and visualized in silver-stained Tris-Tricine gels the electrophoretic profiles of bacterial lysates and their respective LPS extracts from reference strains P. gingivalis ATCC 33277, P. endodontalis ATCC 35406, and clinical isolates P. gingivalis (AC1Pg) and P. endodontalis (AC2Pe). The clinical isolates were previously obtained from patients with asymptomatic AP (AAP).
First, LPS from P. gingivalis ATCC 33277 and AC1Pg lysates consisted of an unsubstituted Lipid A-core region and a preferential length of lipid A-core substituted with more than eight repeating units of OAg. On the other hand, the electrophoretic profiles of the LPS obtained from both the reference strain and its respective clinical isolate (AC2Pe), showed a weaker unsubstituted lipid A-core region compared to that of P. gingivalis, and a preferential OAg substitution length of 8 to 14 repeated units (Figure 1a).
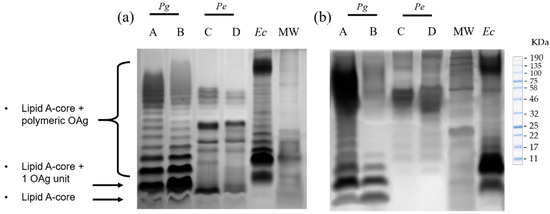
Figure 1.
Representative silver-stained Tris-Tricine gels of P. gingivalis and P. endodontalis LPS; (a) Bacterial lysates. (b) Purified LPS. A: P. gingivalis reference strain ATCC 33277; B: P. gingivalis from clinical isolate AC1Pg; C: P. endodontalis reference strain ATCC 35406; D: P. endodontalis from clinical isolate AC2Pe; Ec: commercial E. coli LPS (BioLabs, Ipswich, MA, USA). MW: Molecular Weight Ladder.
LPS purification with the modified TRIzol extraction protocol, consisting of an additional sonication step to ensure wall rupture, and treatments with DNase I, RNase, and proteinase K effectively removed nucleic acids and protein contaminants. The resultant P. gingivalis and P. endodontalis LPS retained their endotoxic activity (>85 EU/mL) and conserved their structural KDO, a component of the LPS core. The purified P. gingivalis LPS obtained from both the reference strain and its clinical isolate (AC1Pg), presented an electrophoretic profile similar to that observed in bacterial lysates, showing that the extraction protocol did not affect LPS integrity. In the case of the purified P. endodontalis LPS, the unsubstituted lipid A-core region was not visible (Figure 1b). However, both lysates and purified LPS exhibited a preferential length of 8 to 14 OAg repeated units.
Subsequently, we assessed P. gingivalis and P. endodontalis LPS capacity to induce polarization on THP-1 macrophages. For this purpose, cell cultures were stimulated for 24 h with LPS (10 µg/mL) from P. gingivalis ATCC 33277, P. gingivalis AC1Pg, P. endodontalis ATCC 35406, or P. endodontalis AC2Pe or co-stimulated with P. gingivalis ATCC 33277 LPS plus P. endodontalis ATCC 35406 LPS (10 μg/mL each). E. coli LPS stimulation (10 ng/mL) was used as a positive control.
Macrophage polarization was assessed as the percentage of double-positive cells for M1 macrophages (CD64+CD80+), or M2 (CD163+CD206+) and the M1/M2 ratio on THP-1 macrophage. The percentages of single-positive cells and the mean fluorescence intensity (MFI) of each surface marker (Figure 2) were also assessed.
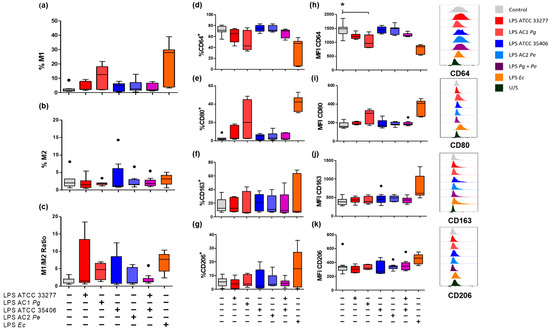
Figure 2.
Percentages of M1 and M2 macrophage populations in LPS-stimulated macrophages in vitro. Box and whisker plots represent the median and interquartile range of (a) Percentage of CD64+CD80+ for M1 macrophages. (b) CD163+CD206+ for M2 macrophages. (c) M1/M2 ratio. (d) CD64+, (e) CD80+ (f) CD163+ and (g) CD206+ cell percentage, respectively. (h) Mean fluorescence intensity (MFI) of CD64, (i) CD80, (j) CD163 and (k) CD206 respectively of THP-1 macrophages in vitro stimulated for 24 h with LPS. U/S Unstained. Independent experiments were performed in triplicate. Data were analyzed by the Kruskal-Wallis test. (●) Represents outlier data. (n = 3) * p < 0.05.
P. gingivalis LPS, specifically LPS from AC1Pg, showed a tendency to increase the M1 population over unstimulated macrophages by 12.7%. P. gingivalis LPS from ATCC 33277 and LPS from AC1Pg showed a tendency to increase the M1/M2 ratio to 1.5 and 4.7 respectively, although the changes were not significant (p > 0.05).
On the other hand, neither varieties of forms of P. endodontalis LPS did not induce changes in surface marker percentages, nor did co-stimulus with P. gingivalis ATCC 33277 LPS and P. endodontalis ATCC 35406 LPS. None of the evaluated LPS conditions influenced the percentage of double-positive M2 macrophages (Figure 2a–c).
Regarding single positive M1 macrophage populations on unstimulated THP-1 macrophages, 71.3% of the total population were CD64+, but only 2% were CD80+. E. coli LPS stimulus induced a decrease in the CD64+ population down to 47.3% but increased CD80+ macrophages up to 42.4% (Figure 2d,e). P. gingivalis LPS, P. endodontalis LPS, and co-stimuli did not significantly change the percentage of CD64+ nor CD80+ cell population.
CD64 surface levels (MFI) were reduced by P. gingivalis LPS, specifically AC1Pg LPS, below unstimulated control levels (p < 0.05). P. endodontalis LPS and the co-stimulus did not promote changes in CD64 nor CD80 surface levels (Figure 2h,i).
Evaluation of single positive M2 macrophage populations showed no significant changes in any of the evaluated conditions. P. gingivalis and/or P. endodontalis LPS did not change CD163 or CD206 surface levels (MFI), although E. coli LPS induced a CD163 increase above unstimulated control (Figure 2f,g,j,k).
In summary, neither P. gingivalis LPS nor P. endodontalis LPS induced changes in the percentage of cell surface macrophage markers in vitro. Surface levels of CD64 (MFI) were significantly reduced by LPS from clinical isolates of P. gingivalis.
Next, the cytokine profiles associated with M1 (TNF-α, IL-1β, IL-6, and IL-12) and anti-inflammatory M2 (IL-10 and IL-1RA) were evaluated in the supernatant of the stimulated THP-1 macrophages (Figure 3). P. gingivalis LPS stimulated the secretion levels of pro-inflammatory cytokines similarly to E. coli LPS, but in a minor amount, compared with the unstimulated control. Specifically, P. gingivalis ATCC 33277 LPS induced increases in TNF-α and IL-1β. Moreover, LPS from AC1Pg induced increases in TNF-α, IL-1β, IL-6, and IL-12 compared to unstimulated THP-1 macrophages (p < 0.05) (Figure 3). In the case of the anti-inflammatory cytokines, LPS from AC1Pg induced an increase of IL-10 compared to unstimulated cells (p < 0.05).
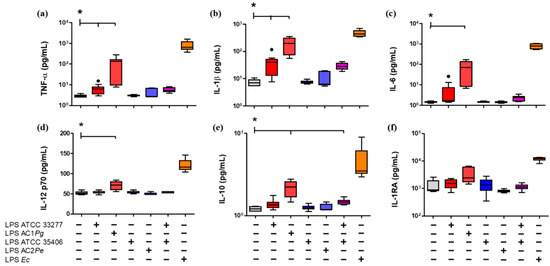
Figure 3.
Cytokine secretion LPS-stimulated macrophages in vitro. Box and whisker plots represent the median and interquartile range of pro-inflammatory cytokines (a) TNF-α, (b) IL-1β, (c) IL-6, and (d) IL-12p70, and anti-inflammatory cytokines (e) IL-10 and (f) IL-1RA, detected in the supernatant of THP-1 macrophages in vitro culture stimulated for 24 h with LPS. U/S Unstained. Experiments were performed in triplicate. Data were analyzed by the Kruskal-Wallis test. (●) Represents outlier data. (n = 3) * p < 0.05.
P. endodontalis LPS stimulation and the co-stimulus with P. gingivalis ATCC 33277/P. endodontalis ATCC 35406 LPS did not induce changes in the secretion of M1 or M2 cytokines levels compared with P. gingivalis ATCC 33277 LPS (Figure 3).
To evaluate the capability of P. gingivalis and P. endodontalis LPS to induce changes in the expression levels of TLR2 and TLR4, we assessed the percentage of positive macrophage populations for TLR2+ and TLR4+ and the MFI of TLR2 and TLR4 under the conditions previously described and, in addition, we included PAM3CSK4 stimulation, a TLR2 ligand, as a positive control. P. gingivalis and P. endodontalis LPS did not show significant differences in the percentages of TLR2+ and TLR4+ cells nor induced significant changes in TLR2 or TLR4 MFI compared with non-stimulated control (Figure 4). Although, both positive controls induced significant decrease of TLR2+ and TLR4+ populations, and TLR2 and TLR4 MFI decreased compared to the unstimulated control.
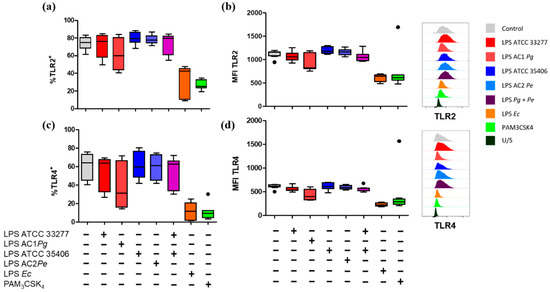
Figure 4.
Percentage of single stain and expression levels of TLR2 and TLR4 in LPS or lipopeptide-stimulated macrophages in vitro. Box and whisker plots represent the median and interquartile range of (a) TLR2+ and (b) TLR4+ percentage. (c) Mean fluorescence intensity (MFI) of TLR2 and (d) TLR4 of THP-1 macrophages in vitro stimulated for 24 h with LPS or PAM3CSK4. U/S Unstained. Experiments were performed in triplicate. Data were analyzed by the Kruskal-Wallis test. (●) Represents outlier data. (n = 3).
Correlation analysis between the macrophage surface markers, TLR2, TLR4, and the measured secreted cytokines revealed a strong positive correlation between CD64 and TLRs and a strong negative correlation between CD64 and pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 (Table 1).

Table 1.
Spearman’s correlation matrix of M1 and M2 surface markers, TLR2, TLR4, and secreted cytokines levels.
We finally evaluated the P. gingivalis and P. endodontalis LPS effects on TLR2 and TLR4 mRNA expression in THP-1 macrophages, using the DNA demethylating agent decitabine.
We showed that TLR2 or TLR4 mRNA levels were not affected by P. gingivalis and P. endodontalis LPS, nor did previous decitabine treatment (Figure 5).
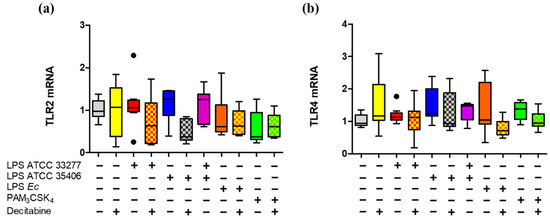
Figure 5.
TLR2 and TLR4 gene expression in LPS or lipopeptide-stimulated macrophages in vitro treated with decitabine. Box and whisker plots represent the median and interquartile range of relative gene expression of (a) TLR2 and (b) TLR4. THP-1 macrophages in vitro were treated with decitabine for 24 h and stimulated for 2 h with LPS or PAM3CSK4. Relative gene expression was normalized relative to 18S rRNA and expressed as the difference between 2–ΔΔCt. Experiments were performed in triplicate. Data were analyzed by the Kruskal-Wallis test. (●) Represents outlier data. (n = 3).
3. Discussion
Black-pigmented bacteria, such as P. gingivalis and P. endodontalis, are recognized as central agents in the etiopathogenesis and progression of AP, promoting pro-inflammatory and osteolytic responses due to their diversity of virulence factors [9,13,14,15,16,20]. LPS is a distinctive component of the outer membrane of Gram-negative bacteria and a potent inducer of cytokine secretion. In the case of P. gingivalis LPS, it has been described to induce an immune response by ligation of both TLR2 and TLR4 because of the heterogeneity of its lipid A structure [36].
In the present work, we purified P. gingivalis and P. endodontalis LPS from reference strains (ATCC) and clinical isolates and then we evaluated the polarization profiles in macrophages differentiated from THP-1 after stimulation with these LPSs. We observed that P. gingivalis LPS induced an M1 profile evidenced by reduction of CD64+ cells and increased pro-inflammatory cytokines secretion (TNF-α, IL-1β, IL-6, and IL-12), whereas LPS from P. endodontalis failed to induce a significant response.
The LPS purification was done using the extraction protocol described by Yi and Hackett, which is a fast, convenient, and low-scale extraction method that uses TRIzol as the main reagent [37]. Nevertheless, we found proteins and nucleic acid traces in our LPS extracts. For this reason, we incorporated enzymatic treatments with DNase, RNase, and proteinase K into the bacterial pellet as a previous step [38,39]. This modification allowed us to remove the contaminants, except for DNA/RNA traces visualized in GelRed-stained agarose gel as an electrophoretic band with a molecular weight smaller than 100 base pairs previously undescribed. Bacterial nucleic acids have immunogenic potential by ligating endosomal TLR7 and TLR9, triggering a response similar to TLR2. To eliminate any trace of nucleic acids, we incorporated the mechanical breakdown step by sonication of the bacterial pellet before the enzymatic treatment [38]. With these modifications to the Yi and Hackett protocol, we obtained P. gingivalis and P. endodontalis LPSs free of any detectable traces of proteins and nucleic acids. However, adding extra steps to the original extraction process diminished the performance extraction, fluctuating between 50–90% less dry weight LPS than initially obtained. Despite the latter, the presence of endotoxic activity was confirmed, which was greater than 85 EU/mL, well above that reported in commercial P. gingivalis LPS (~2 EU/mL) [40]. KDO, a structural component of the LPS inner-core, was also detected in the purified P. gingivalis and P. endodontalis LPS, showing that the structure of LPS was not affected by the extraction protocol.
Our P. gingivalis LPS electrophoretic profile exhibited a high mobility region of unsubstituted lipid A-core clearly defined in the gel and a preferential length of ≥8 OAg repeated units, with a molecular weight between 43 and 100 KDa, consistent with previous descriptions in the literature in other P. gingivalis strains [41,42,43,44]. On the other hand, compared to P. gingivalis LPS, P. endodontalis LPS exhibited a faint lipid A-core region not substituted by OAg and a preferential length of 8 to 14 OAg substitutions repeating units.
It has previously been reported that P. endodontalis LPS electrophoretic profile in Tris-Glycine gels exhibits a preferential chain length greater than 8 OAg repeated units [45]. In this study, we report that in Tris-Tricine gels, the P. endodontalis LPS exhibited a preferential chain length greater than 8 OAg repeated units but smaller than the preferential length of E. coli LPS. Although the electrophoretic profiles of P. endodontalis LPS previously presented by Park’s group are slightly different from ours, it is due to methodological differences, since Tris-Glycine gel provides higher resolution to bands with lower electrophoretic mobility while Tris-Tricine gel is suited for a higher resolution to bands with greater electrophoretic mobility [46].
During AP, macrophages exhibit a polarization switch towards M1, whereas disease exacerbation during symptomatic AP is associated with a reduced M2 differentiation profile, based on the reduced surface expression of CD163 along with higher IL-6 and IL-23 levels, which supports the role of the macrophages in the severity of apical lesions that can be triggered by endodontic bacterial antigens [32]. It has been previously reported that P. gingivalis LPS induces M1 cytokine secretion in bone marrow-derived macrophages once polarized to M1 or M2 [47], as well as M1 cytokine secretion in THP-1 differentiated to macrophages while being polarized with IFN-γ or IL-4 [48]. However, despite the above, the capacity of P. gingivalis and P. endodontalis LPS to induce macrophage polarization by analyzing their surface markers or cytokine secretion have not yet been described. In the present study, we phenotypically characterized M1 and M2 macrophage subpopulations based on the expression of their cell surface markers CD64 and CD80 for M1, as well as CD163 and CD206 for M2 [32,49,50,51].
Our positive control, E. coli LPS, induced an M1 macrophage phenotype with lower CD64 and higher CD80 surface levels in THP-1 macrophages. In the same way, P. gingivalis LPS induced a reduction of CD64 surface levels, although not on the same level. CD64 is an IgG Fc-γ receptor constitutively expressed on monocytes and macrophages and participates in cellular cytotoxicity by triggering phagocytosis, pro-inflammatory cytokine secretion, ROS production, and antigen presentation to T cells. A decrease in CD64 could suggest a decline in the phagocytic capacity of those macrophages [52,53,54]. In contrast, CD80 is expressed at low basal levels and, like CD86, is a co-stimulatory molecule that, combined with MHC/antigen complex, induces activation, proliferation, and differentiation of T cells [55,56]. For that reason, it might be plausible that macrophages differentiated from THP-1 and stimulated by LPS for 24 h decrease their phagocytic capacity but increase their ability to activate, proliferate and differentiate T cells.
Specifically, P. gingivalis LPS from our clinical isolate (AC1Pg) induced a decrease in CD64 surface levels and an increase in TNF-α, IL-1β, IL-6, and IL-12 cytokine secretion; while P. gingivalis LPS from ATCC 33277 strain only induced increased secretion of TNF-α and IL-1β in a statistically significant manner. The difference in magnitude response between E. coli LPS and P. gingivalis LPS is probably because P. gingivalis LPS is less immunologically active than E. coli LPS [57,58,59,60], which could be attributed to its lipid A heterogeneity. P. gingivalis LPS expresses two varieties of lipid A, a penta-acylated variety, capable of activating TLR4 in a similar way to E. coli LPS, and a tetra-acylated variety that antagonizes TLR4 by binding it but that does not trigger a response [36,57,61,62].
When comparing P. gingivalis LPS from different sources, the LPS from clinical isolate induced a more significant response than LPS from ATCC 33277 strain. It has been previously described that clinical isolates of P. gingivalis with low passages are more virulent than reference strains [63,64], presumably because P. gingivalis gradually loses the properties necessary to survive straight in vivo, in order to adapt to the in vitro culture conditions [65]. Therefore, since P. gingivalis from our clinical isolate was obtained from the root canal of a patient with AAP, we can attribute the different results of AC1Pg LPS compared to LPS of ATCC 33277 as being due to a higher virulence of P. gingivalis LPS from clinical isolates over the reference strains.
We also evaluated the immune response triggered by P. endodontalis LPS, and we observed that it did not induce changes in the secretion of the evaluated cytokines nor the surface levels of M1 or M2 markers. It would be plausible that P. endodontalis LPS possesses a lipid A heterogeneity similar to that described in P. gingivalis, but capable of inducing even lower immune responses, It cannot be ruled out that differences in the OAg region between P. endodontalis and P. gingivalis LPS may play a role in the low immune response induced in THP-1 macrophages [44,66,67].
Since AP is caused by a polymicrobial infection of the root canal system, we also studied the immune response of THP-1 macrophages co-stimulated with P. gingivalis and P. endodontalis LPS. Unexpectedly, LPS co-stimulation did not result in an increased immune response, suggesting that P. endodontalis LPS could attenuate the immune response induced by P. gingivalis LPS, by competing and blocking TLR4 and TLR2 [22,36,57], However, whether or not P. endodontalis LPS can attenuate the immune response in macrophages induced by P. gingivalis LPS or LPS from other Gram-negative bacteria needs further research. Altogether, these data suggest that macrophage polarization towards M1 during AP progression might be associated with the LPS burden of specific pathogens, such as P gingivalis.
Finally, we analyzed the TLR2 and TLR4 surface membrane levels on THP-1 cells differentiated into macrophages after stimulation 24 h with LPS. Surprisingly, both positive controls for ligands for TLR2 and TLR4 induced a reduction in TLR2+ and TLR4+ population in addition to reduced TLR2 and TLR4 surface levels. These results are difficult to interpret. Although increases in TLR2 and TLR4 levels have been described when stimulated with their respective ligands, other studies have reported a decrease. Specifically, an increase in surface expression of TLR2 and TLR4 was observed in THP-1 cells differentiated for 72 h with PMA (Phorbol 12-myristate 13-acetate) 17 nM and stimulated with P. gingivalis LPS or E. coli LPS 1 μg/mL for 24 h [68]. Similarly, THP-1 monocytes stimulated with P. gingivalis LPS or E. coli LPS 1 μg/mL for 24 h increased their TLR2+ or TLR4+ population respectively [69]. On the other hand, a decrease in surface expression of both TLR2 and TLR4, but not of TLR2 and TLR4 mRNA, was described after 24 h of stimulation with P. gingivalis LPS (1 μg/mL), PAM3CSK4 (1 μg/mL) or E. coli LPS (0.1 μg/mL) in human gingival fibroblasts primary cultures [70]. This result could suggest that the protein expression of TLR2 and TLR4 may depend on the concentration of LPS, the cell model, and the conditions of differentiation to macrophages (concentration and time of exposition to PMA). Additionally, the reduction of CD64 was directly correlated with TLR2 and TLR4. In this regard, a directly proportional expression between CD64, TLR2, and TLR4 has been described in monocytes differentiated into macrophages from peripheral blood [71] and neutrophils from tuberculosis patients [72], but not in vitro macrophages differentiated to a pro-inflammatory profile. These antecedents suggest that there might be a functional relationship between the two, which would help explain the reduction in TLRs observed on the surface of LPS-stimulated macrophages in our experimental model.
When E. coli LPS bonds with TLR4, a hexameric complex is formed, (TLR4/MD-2/LPS)2, which activates downstream signaling that induces endocytosis of the complex and, subsequently, transcriptional factor activation [73,74,75,76,77]. It has been reported that bone marrow-derived macrophages (BMDM) internalize TLR4 after 30 min of E. coli LPS stimulation [76]. In THP-1 differentiated to macrophages and stimulated with P. gingivalis or P. endodontalis LPS, or treated with decitabine, a demethylating agent, we did not observe changes in gene expression of TLR2 or TLR4. These results suggest that, in our model, DNA methylation would not play a predominant role in the reduction of TLR2 or TLR4 surface levels. On the other hand, it has been reported in THP-1 cells, that PAM3CSK4 induced both an increase of TLR2 and an increase of soluble TLR2 (sTLR2). The last one would be mediated by metalloproteinase activity and consequent proteolysis of TLR2 on the membrane surface [78], a process that could be stimulated by PMA and LPS [78,79,80]. These data suggest that the reduction in surface levels of both TLR2 and TLR4 by E. coli LPS and PAM3CSK4 could be explained, at least in part, by non-transcriptional mechanisms, such as recycling, lower export to membrane surface, or proteolysis of these receptors.
4. Materials and Methods
4.1. Bacterial Growth Conditions
P. gingivalis and P. endodontalis reference strains (ATCC 33277, ATCC 35406, respectively), and clinical isolates (AC1Pg, AC2Pe, respectively) previously obtained from AP patients, were inoculated on blood agar 5% supplemented with hemin-menadione (5 μg/mL) and cultured in an anaerobic chamber in an atmosphere of 80% N2, 10% CO2 and 10% H2 (Shel Lab, Cornelius, OR, USA) at 37 °C for 4 days. The bacteria were sub-cultured in 50 mL of brain-heart infusion (BHI) broth supplemented with hemin-menadione (5 µg/mL), grown to late exponential phase, centrifuged at 4500× g for 20 min, and stored at −80 °C. The clinical isolates P. gingivalis (AC1Pg) and P. endodontalis (AC2Pe) were identified and confirmed by sequencing of 763 bp fragments corresponding to the V3-V6 regions of the 16S RNA gene.
4.2. LPS Purification
LPS was purified by TRIzol extraction protocol [37], which was modified to remove nucleic acids and protein contaminants, as follows: The obtained bacterial pellets were sonicated with an ultrasonic processor, then incubated with DNase (0.1 mg/mL) (Sigma-Aldrich, St. Louis, MO, USA) and RNase (0.2 mg/mL) (Thermo Fisher, Waltham, MA, USA) at 37 °C overnight [38], and, subsequently, with Proteinase K (1 mg/mL) at 60 °C for 1.5 h. Then, they were homogenized in 2 mL TRIzol with 400 μL chloroform and centrifuged at 3900× g for 10 min at 4 °C. The upper aqueous phase was recovered and lyophilized overnight. The product was dissolved in 0.375 M MgCl2 in 95% ethanol, centrifuged at 4700× g for 5 min and the precipitate recovered. LPS purity was verified with a Bradford assay (protein detection limit 0.1 mg/mL) and GelRed stained agarose gel (detection limit 0.1 ng). Bacterial lysates were obtained from cell cultures grown to the exponential phase and adjusted to OD600 = 2.0 [81,82]. Purified LPS and bacterial lysates were visualized on silver-stained Tris-Tricine polyacrylamide gels (SDS-PAGE) [46,83] and compared with molecular weight standard #P7706S (BioLabs, Ipswich, MA, USA).
LPS endotoxic activity was determined with Pierce LAL Chromogenic Endotoxin Quantitation Kit (Thermo Fisher, Waltham, MA, USA), and 3-deoxy-D-manno-oct-2-ulosonic acid (KDO), a component of the LPS core, was determined by Purpald test (Sigma-Aldrich, St. Louis, MO, USA) [46].
4.3. Cell Culture
THP-1 cells (ATCC TIB-202) were cultured in supplemented RPMI 1640 medium (Corning, New York, NY, USA) with 10% fetal bovine serum (FBS) (Gibco, Thermo Fisher, Waltham, MA, USA) and maintained at 5% CO2, 37 °C. THP-1 cells were seeded in triplicates at 1 × 106 cells per well in six-well plates at a concentration of 5 × 105 cells/mL and differentiated to macrophages with 10 nM phorbol 12-myristate 13-acetate (PMA) (Abcam, UK, Cambridge, UK) for 24 h in supplemented RPMI medium with 10% FBS. Non-adherent cells were removed, and adherent cells were washed and incubated for 24 h with RPMI medium with 10% FBS.
THP-1 cells differentiated to macrophages were stimulated with LPS from P. gingivalis, ATCC 33277 or AC1Pg (10 μg/mL), LPS from P. endodontalis ATCC 35406 or AC2Pe (10 μg/mL) or the combination of LPS from P. gingivalis ATCC 33277 (10 μg/mL) plus LPS from P. endodontalis ATCC 35406 (10 μg/mL) in RPMI medium without FBS. Cells were stimulated for 2 h to measure TLR2 and TLR4 transcriptional levels or 24 h to assess cell surface proteins levels or cytokine secretion levels [84,85]. Unstimulated cells were used as a negative control. LPS from E. coli O127: B8 (10 ng/mL) (Sigma-Aldrich, St. Louis, MO, USA) or PAM3CSK4 (10 ng/mL) (InvivoGen, San Diego, CA, USA) were used as positive controls. Also, a group of cells treated with the demethylation agent 5-aza-2’-deoxycytidine (Decitabine, 500 nM; Sigma-Aldrich, St. Louis, MO, USA) was included to explore whether DNA methylation changes influenced P. gingivalis or P. endodontalis LPS-induced TLR2 and TLR4 gene expression.
4.4. Flow Cytometry
Cells were placed in a V bottom 96-well plate in PBS for complete staining. Macrophages were stained with Fixable Viability Dye eFluor 780 (Invitrogen, Thermo Fisher, Waltham, MA, USA) 1:1000 in PBS for 30 min at 4 °C in the dark to discard the dead cells. Later, the cells were incubated with a mixture of all fluorochrome-conjugated antibodies at 1:25 each in PBS 2% FBS, for 30 min at 4 °C in the dark. The cells were single-stained under the same conditions as controls. After washing with PBS, the cells were fixed with 2% paraformaldehyde (Invitrogen, Thermo Fisher, Waltham, MA, USA) and analyzed by flow cytometry (LSR Fortessa X-20®; Becton Dickinson Immunocytometry Systems, Franklin Lakes, NJ, USA). Flow cytometric data analysis was performed with Flow Jo v10 software (TreeStar, Ashland, OR, USA). The following mouse monoclonal anti-human antibodies conjugated with their respective fluorochromes were used: CD64 conjugated with Brilliant Violet 510 (BV510) and CD80 conjugated with fluorescein isothiocyanate (FITC) were used to identify M1 macrophage subpopulations; CD163 conjugated with PE/Cy7 and CD206 conjugated with Brilliant Violet 421 (BV421) were used to identify M2 macrophages subpopulations; TLR2 conjugated with APC and TLR4 conjugated with PE were also used (Biolegend, San Diego, CA, USA). Fixable Viability Dye (FVD) eFluor 780 (Invitrogen, Thermo Fisher, Waltham, MA, USA).
THP-1 macrophages were first gated according to their forward- and side-scatter profiles. Negative FVD cells were gated to discard dead cells. For the determination of the frequencies of M1 and M2 macrophage subpopulations, cells were gated over CD64+CD80+ or CD163+CD206+. The threshold in each case was determined by the comparison with unstained samples. The M1/M2 ratio was calculated as the frequency of M1 double-positive macrophages divided by the frequency of the M2 double-positive macrophages in each sample. The intensity of expression of each specific cell surface marker was expressed as MFI.
4.5. Cytokine Secretion
Supernatant from stimulated cells was collected and stored at −80 °C. Cytokines quantification was performed using a multiplex bead-based immunoassay (Human Magnetic Luminex Assay®, R&D Systems, Minneapolis, MN, USA) for TNF-α, IL-1β, IL-6, IL-12p70, IL-10, and IL-1RA, following the manufacturer instructions. Data from the multiplex panel were read using a dedicated platform (Magpix®, Millipore, MO, USA) and software (Milliplex AnalystR®, Viagene Tech, Minneapolis, MN, USA), according to the manufacturer instructions.
4.6. RNA Expression
TLR2 and TLR4 relative mRNA levels were determined by qRT-PCR, and cDNA was amplified using their appropriate 5′-3′ forward primer and 5′-3′ reverse primer sets, respectively, as follows: CTCAACACGGGAAACCTCAC and CGTCCACCACTAAGAACG for 18S rRNA, CTCTCGGTGTCGGAATGTC and AGGATCAGCAGGAACAGAGC for TLR2 and CCCTCCCCTGTACCCTTC and TCCCTGCCTTGAATACCTTC for TLR4. All primer sets and reagents (KAPA™SYBR® Fast; KAPA Biosystems, Woburn, MA, USA) were used in qPCR equipment (StepOnePlus®; Applied Biosystems, Singapore) as follows: 95 °C for 3 min and 40 cycles of 95 °C for 30 s and 60 °C for 30 s. To detect nonspecific product formation and false-positive amplification, a melt curve of 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s was performed. Relative quantification of mRNA was analyzed using the 2–ΔΔCT method. The normalization of gene expression was done against the 18S rRNA.
4.7. Data Analysis
The Shapiro-Wilk test was used to determine the distribution of the continuous data. Inferential analyses were performed using the Kruskal-Wallis test for non-normally distributed data. The statistical analysis was performed using STATA 12® (StataCorpLP, College Station, TX, USA). The figures were performed in Flow Jo v10 software (TreeStar, Ashland, OR, USA) and Graph Prism 5 (GraphPad Software, Inc., San Diego, CA, USA).
5. Conclusions
In summary, P. gingivalis LPS induces a macrophage polarization profile predominantly towards M1, similarly, although less evidently, than that generated by E. coli LPS, based on their cell surface markers and secreted cytokines. On the other hand, P. endodontalis LPS did not cause a clear immune response in THP-1 macrophages. P. gingivalis and P. endodontalis LPS stimulation did not induce TLR2 or TLR4 levels changes in THP-1 macrophages. These data suggest that macrophage polarization towards M1 during AP progression might be associated with the LPS burden of specific pathogens, such as P gingivalis.
Author Contributions
Conceptualization and funding acquisition, M.H.; Formal analysis, P.V., M.H., A.H. and A.E.; Investigation, P.V., A.C., J.A., A.F. and D.G.-Q.; Methodology, P.V., A.C. and D.G.-Q.; Project administration, Resources, Supervision, Writing—review & editing, M.H., A.H. and A.E.; Validation J.A.; Visualization; P.V.; Writing—original draft, P.V. and D.G.-Q. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by FONDECYT, grant numbers 1160741 and 1200098.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
Pablo Veloso thanks the fellowship CONICYT 21171640, from the Chilean Government and B. Pesce for supporting flow cytometric procedures at the MED.UCHILE-FACS Laboratory from Biomedical Sciences Institute, School of Medicine of the University of Chile, Santiago de Chile.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hargreaves, K.M.; Goodis, H.E.; Tay, F.R. Seltzer and Bender’s Dental Pulp; Quintessence Pub.: Batavia, IL, USA, 2012. [Google Scholar]
- Gomes, M.S.; Blattner, T.C.; Sant’ Ana Filho, M.; Grecca, F.S.; Hugo, F.N.; Fouad, A.F.; Reynolds, M.A. Can apical periodontitis modify systemic levels of inflammatory markers? A systematic review and meta-analysis. J. Endod. 2013, 39, 1205–1217. [Google Scholar] [CrossRef] [PubMed]
- Bordagaray, M.J.; Fernández, A.; Garrido, M.; Astorga, J.; Hoare, A.; Hernández, M. Systemic and Extraradicular Bacterial Translocation in Apical Periodontitis. Front. Cell Infect. Microbiol. 2021, 2011, 649925. [Google Scholar] [CrossRef] [PubMed]
- Segura-Egea, J.J.; Martin-Gonzalez, J.; Cabanillas-Balsera, D.; Fouad, A.F.; Velasco-Ortega, E.; Lopez-Lopez, J. Association between diabetes and the prevalence of radiolucent periapical lesions in root-filled teeth: Systematic review and meta-analysis. Clin. Oral Investig. 2016, 20, 1133–1141. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, J.L.; Baumgartner, J.C.; Gluskin, A.H.; Hartwell, G.R.; Walton, R.E. Identify and define all diagnostic terms for periapical/periradicular health and disease states. J. Endod. 2009, 35, 1658–1674. [Google Scholar] [CrossRef]
- Hsiao, W.W.; Li, K.L.; Liu, Z.; Jones, C.; Fraser-Liggett, C.M.; Fouad, A.F. Microbial transformation from normal oral microbiota to acute endodontic infections. BMC Genom. 2012, 13, 345. [Google Scholar] [CrossRef]
- Buonavoglia, A.; Latronico, F.; Pirani, C.; Greco, M.F.; Corrente, M.; Prati, C. Symptomatic and asymptomatic apical periodontitis associated with red complex bacteria: Clinical and microbiological evaluation. Odontology 2013, 101, 84–88. [Google Scholar] [CrossRef]
- Paster, B.J.; Olsen, I.; Aas, J.A.; Dewhirst, F.E. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontology 2000 2006, 42, 80–87. [Google Scholar] [CrossRef]
- Seol, J.H.; Cho, B.H.; Chung, C.P.; Bae, K.S. Multiplex polymerase chain reaction detection of black-pigmented bacteria in infections of endodontic origin. J. Endod. 2006, 32, 110–114. [Google Scholar] [CrossRef]
- Marchesan, J.; Jiao, Y.; Schaff, R.A.; Hao, J.; Morelli, T.; Kinney, J.S.; Gerow, E.; Sheridan, R.; Rodrigues, V.; Paster, B.J.; et al. TLR4, NOD1 and NOD2 mediate immune recognition of putative newly identified periodontal pathogens. Mol. Oral Microbiol. 2016, 31, 243–258. [Google Scholar] [CrossRef]
- Silva, N.; Dutzan, N.; Hernandez, M.; Dezerega, A.; Rivera, O.; Aguillon, J.C.; Aravena, O.; Lastres, P.; Pozo, P.; Vernal, R.; et al. Characterization of progressive periodontal lesions in chronic periodontitis patients: Levels of chemokines, cytokines, matrix metalloproteinase-13, periodontal pathogens and inflammatory cells. J. Clin. Periodontol. 2008, 35, 206–214. [Google Scholar] [CrossRef]
- Rôças, I.N.; Siqueira, J.F., Jr.; Debelian, G.J. Analysis of symptomatic and asymptomatic primary root canal infections in adult Norwegian patients. J. Endod. 2011, 37, 1206–1212. [Google Scholar] [CrossRef]
- Colombo, A.P.; Bennet, S.; Cotton, S.L.; Goodson, J.M.; Kent, R.; Haffajee, A.D.; Socransky, S.S.; Hasturk, H.; Van Dyke, T.E.; Dewhirst, F.E.; et al. Impact of periodontal therapy on the subgingival microbiota of severe periodontitis: Comparison between good responders and individuals with refractory periodontitis using the human oral microbe identification microarray. J. Periodontol. 2012, 83, 1279–1287. [Google Scholar] [CrossRef]
- Montagner, F.; Jacinto, R.C.; Signoretti, F.G.; Sanches, P.F.; Gomes, B.P. Clustering behavior in microbial communities from acute endodontic infections. J. Endod. 2012, 38, 158–162. [Google Scholar] [CrossRef]
- Belstrom, D.; Fiehn, N.E.; Nielsen, C.H.; Kirkby, N.; Twetman, S.; Klepac-Ceraj, V.; Paster, B.J.; Holmstrup, P. Differences in bacterial saliva profile between periodontitis patients and a control cohort. J. Clin. Periodontol. 2014, 41, 104–112. [Google Scholar] [CrossRef]
- Perez-Chaparro, P.J.; Goncalves, C.; Figueiredo, L.C.; Faveri, M.; Lobao, E.; Tamashiro, N.; Duarte, P.; Feres, M. Newly identified pathogens associated with periodontitis: A systematic review. J. Dent. Res. 2014, 93, 846–858. [Google Scholar] [CrossRef]
- Jiménez, C.; Carvajal, D.; Hernández, M.; Valenzuela, F.; Astorga, J.; Fernández, A. Levels of the interleukins 17A, 22, and 23 and the S100 protein family in the gingival crevicular fluid of psoriatic patients with or without periodontitis. An. Bras. Dermatol. 2021, 96, 163–170. [Google Scholar] [CrossRef]
- Silhavy, T.J.; Kahne, D.; Walker, S. The bacterial cell envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000414. [Google Scholar] [CrossRef]
- Kato, H.; Taguchi, Y.; Tominaga, K.; Umeda, M.; Tanaka, A. Porphyromonas gingivalis LPS inhibits osteoblastic differentiation and promotes pro-inflammatory cytokine production in human periodontal ligament stem cells. Arch. Oral Biol. 2014, 59, 167–175. [Google Scholar] [CrossRef]
- Mirucki, C.S.; Abedi, M.; Jiang, J.; Zhu, Q.; Wang, Y.H.; Safavi, K.E.; Clark, R.B.; Nichols, F.C. Biologic activity of Porphyromonas endodontalis complex lipids. J. Endod. 2014, 40, 1342–1348. [Google Scholar] [CrossRef][Green Version]
- Herath, T.D.; Darveau, R.P.; Seneviratne, C.J.; Wang, C.Y.; Wang, Y.; Jin, L. Heterogeneous Porphyromonas gingivalis LPS modulates immuno-inflammatory response, antioxidant defense and cytoskeletal dynamics in human gingival fibroblasts. Sci. Rep. 2016, 6, 29829. [Google Scholar] [CrossRef]
- Tang, Y.; Sun, F.; Li, X.; Zhou, Y.; Yin, S.; Zhou, X. Porphyromonas endodontalis lipopolysaccharides induce RANKL by mouse osteoblast in a way different from that of Escherichia coli lipopolysaccharide. J. Endod. 2011, 37, 1653–1658. [Google Scholar] [CrossRef] [PubMed]
- Papadopoulos, G.; Weinberg, E.O.; Massari, P.; Gibson, F.C., 3rd; Wetzler, L.M.; Morgan, E.F.; Genco, C.A. Macrophage-specific TLR2 signaling mediates pathogen-induced TNF-dependent inflammatory oral bone loss. J. Immunol. 2013, 190, 1148–1157. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, D.; Okamura, H.; Teramachi, J.; Ochiai, K.; Qiu, L.; Haneji, T. Calcium hydroxide suppresses Porphyromonas endodontalis lipopolysaccharide-induced bone destruction. J. Dent. Res. 2014, 93, 508–513. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Qiu, L.; Guo, J.; Qu, L.; Xu, L.; Zhong, M. Effect of lipopolysaccharides from Porphyromonas endodontalis on the expression of macrophage colony stimulating factor in mouse osteoblasts. Zhonghua Kou Qiang Yi Xue Za Zhi 2014, 49, 535–539. [Google Scholar]
- Azuma, M. Fundamental mechanisms of host immune.e responses to infection. J. Periodontal. Res. 2006, 41, 361–373. [Google Scholar] [CrossRef]
- Gazivoda, D.; Dzopalic, T.; Bozic, B.; Tatomirovic, Z.; Brkic, Z.; Colic, M. Production of proinflammatory and immunoregulatory cytokines by inflammatory cells from periapical lesions in culture. J. Oral Pathol. Med. 2009, 38, 605–611. [Google Scholar] [CrossRef]
- Sima, C.; Glogauer, M. Macrophage subsets and osteoimmunology: Tuning of the immunological recognition and effector systems that maintain alveolar bone. Periodontolology 2000 2013, 63, 80–101. [Google Scholar] [CrossRef]
- Derlindati, E.; Dei Cas, A.; Montanini, B.; Spigoni, V.; Curella, V.; Aldigeri, R.; Ardigo, D.; Zavaroni, I.; Bonadonna, R.C. Transcriptomic analysis of human polarized macrophages: More than one role of alternative activation? PLoS ONE 2015, 10, e0119751. [Google Scholar]
- Murray, P.J. Macrophage Polarization. Annu. Rev. Physiol. 2017, 10, 541–566. [Google Scholar] [CrossRef]
- Zhou, D.; Huang, C.; Lin, Z.; Zhan, S.; Kong, L.; Fang, C.; Li, J. Macrophage polarization and function with emphasis on the evolving roles of coordinated regulation of cellular signaling pathways. Cell Signal. 2014, 26, 192–197. [Google Scholar] [CrossRef]
- Veloso, P.; Fernandez, A.; Terraza-Aguirre, C.; Alvarez, C.; Vernal, R.; Escobar, A.; Hernandez, M. Macrophages skew towards M1 profile through reduced CD163 expression in symptomatic apical periodontitis. Clin. Oral Investig. 2020, 24, 4571–4581. [Google Scholar] [CrossRef]
- Fernández, A.; Veloso, P.; Astorga, J.; Rodriguez, C.; Torres, V.A.; Valdes, M.; Garrido, M.; Gebicke-Haerter, P.J.; Hernandez, M. Epigenetic regulation of TLR2-mediated periapical inflammation. Int. Endod. J. 2020, 53, 1229–1237. [Google Scholar] [CrossRef]
- Fernández, A.; Cardenas, A.M.; Astorga, J.; Veloso, P.; Alvarado, A.; Merino, P.; Pino, D.; Reyes-Court, D.; Hernandez, M. Expression of Toll-like receptors 2 and 4 and its association with matrix metalloproteinases in symptomatic and asymptomatic apical periodontitis. Clin. Oral Investig. 2019, 23, 4205–4212. [Google Scholar] [CrossRef]
- Pereira, C.V.; Stipp, R.N.; Fonseca, D.C.; Pereira, L.J.; Hofling, J.F. Detection and clonal analysis of anaerobic bacteria associated to endodontic-periodontal lesions. J. Periodontol. 2011, 82, 1767–1775. [Google Scholar] [CrossRef]
- Reife, R.A.; Coats, S.R.; Al-Qutub, M.; Dixon, D.M.; Braham, P.A.; Billharz, R.J.; Howald, W.N.; Darveau, R.P. Porphyromonas gingivalis lipopolysaccharide lipid A heterogeneity: Differential activities of tetra- and penta-acylated lipid A structures on E-selectin expression and TLR4 recognition. Cell Microbiol. 2006, 8, 857–868. [Google Scholar] [CrossRef]
- Yi, E.C.; Hackett, M. Rapid isolation method for lipopolysaccharide and lipid A from gram-negative bacteria. Analyst 2000, 125, 651–656. [Google Scholar] [CrossRef]
- Rezania, S.; Amirmozaffari, N.; Tabarraei, B.; Jeddi-Tehrani, M.; Zarei, O.; Alizadeh, R.; Masjedian, F.; Zarnani, A.H. Extraction, Purification and Characterization of Lipopolysaccharide from Escherichia coli and Salmonella typhi. Avicenna J. Med. Biotechnol. 2011, 3, 3–9. [Google Scholar]
- Al-Agha, A.G.M. Extraction and Purification of Lipopolysaccharide of Klebsiella pneumoniae Isolates. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 90–100. [Google Scholar] [CrossRef]
- Nativel, B.; Couret, D.; Giraud, P.; Meilhac, O.; d’Hellencourt, C.L.; Viranaicken, W.; Da Silva, C.R. Porphyromonas gingivalis lipopolysaccharides act exclusively through TLR4 with a resilience between mouse and human. Sci. Rep. 2017, 7, 15789. [Google Scholar] [CrossRef]
- Shapira, L.; Champagne, C.; Van Dyke, T.E.; Amar, S. Strain-dependent activation of monocytes and inflammatory macrophages by lipopolysaccharide of Porphyromonas gingivalis. Infect. Immun. 1998, 66, 2736–2742. [Google Scholar] [CrossRef]
- Paramonov, N.; Rangarajan, M.; Hashim, A.; Gallagher, A.; Aduse-Opoku, J.; Slaney, J.M.; Hounsell, E.; Curtis, M.A. Structural analysis of a novel anionic polysaccharide from Porphyromonas gingivalis strain W50 related to Arg-gingipain glycans. Mol. Microbiol. 2005, 58, 847–863. [Google Scholar] [CrossRef] [PubMed]
- Rangarajan, M.; Aduse-Opoku, J.; Paramonov, N.; Hashim, A.; Bostanci, N.; Fraser, O.P.; Tarelli, E.; Curtis, M.A. Identification of a second lipopolysaccharide in Porphyromonas gingivalis W50. J. Bacteriol. 2008, 190, 2920–2932. [Google Scholar] [CrossRef] [PubMed]
- Diaz, L.; Hoare, A.; Soto, C.; Bugueno, I.; Silva, N.; Dutzan, N.; Venegas, D.; Salinas, D.; Perez-Donoso, J.M.; Gamonal, J.; et al. Changes in lipopolysaccharide profile of Porphyromonas gingivalis clinical isolates correlate with changes in colony morphology and polymyxin B resistance. Anaerobe 2015, 33, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Park, C.J.; Park, D.S.; Yoo, H.M.; Oh, T.S.; Lim, S.S. IL-1 and TNF-alpha release in human polymorphonuclear leukocytes after exposure to calcium hydroxide treated Porphyromonas endodontalis lipopolysaccharide. J. Korean Acad. Conserv. Dent. 2002, 5, 463–472. [Google Scholar] [CrossRef]
- Marolda, C.L.; Lahiry, P.; Vines, E.; Saldias, S.; Valvano, M.A. Micromethods for the characterization of lipid A-core and O-antigen lipopolysaccharide. Methods Mol. Biol 2006, 347, 237–252. [Google Scholar]
- Holden, J.A.; Attard, T.J.; Laughton, K.M.; Mansell, A.; O’Brien-Simpson, N.M.; Reynolds, E.C. Porphyromonas gingivalis lipopolysaccharide weakly activates M1 and M2 polarized mouse macrophages but induces inflammatory cytokines. Infect. Immun. 2014, 82, 4190–4203. [Google Scholar] [CrossRef]
- Belfield, L.A.; Bennett, J.H.; Abate, W.; Jackson, S.K. Exposure to Porphyromonas gingivalis LPS during macrophage polarisation leads to diminished inflammatory cytokine production. Arch. Oral Biol. 2017, 81, 41–47. [Google Scholar] [CrossRef]
- Ambarus, C.A.; Krausz, S.; van Eijk, M.; Hamann, J.; Radstake, T.R.; Reedquist, K.A.; Tak, P.P.; Baeten, D.L. Systematic validation of specific phenotypic markers for in vitro polarized human macrophages. J. Immunol. Methods 2012, 375, 196–206. [Google Scholar] [CrossRef]
- Barros, M.H.; Hauck, F.; Dreyer, J.H.; Kempkes, B.; Niedobitek, G. Macrophage polarisation: An immunohistochemical approach for identifying M1 and M2 macrophages. PLoS ONE 2013, 8, e80908. [Google Scholar] [CrossRef]
- Ortiz, M.C.; Lefimil, C.; Rodas, P.I.; Vernal, R.; Lopez, M.; Acuna-Castillo, C.; Imarai, M.; Escobar, A. Neisseria gonorrhoeae Modulates Immunity by Polarizing Human Macrophages to a M2 Profile. PLoS ONE 2015, 10, e0130713. [Google Scholar] [CrossRef]
- Hulett, M.D.; Hogarth, P.M. The second and third extracellular domains of FcgammaRI (CD64) confer the unique high affinity binding of IgG2a. Mol. Immunol. 1998, 35, 989–996. [Google Scholar] [CrossRef]
- Dugast, A.S.; Tonelli, A.; Berger, C.T.; Ackerman, M.E.; Sciaranghella, G.; Liu, Q.; Sips, M.; Toth, I.; Piechocka-Trocha, A.; Ghebremichael, M.; et al. Decreased Fc receptor expression on innate immune cells is associated with impaired antibody-mediated cellular phagocytic activity in chronically HIV-1 infected individuals. Virology 2011, 415, 160–167. [Google Scholar] [CrossRef]
- Hogarth, P.M.; Pietersz, G.A. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat. Rev. Drug Discov. 2012, 11, 311–331. [Google Scholar] [CrossRef]
- Bugeon, L.; Hargreaves, R.E.; Crompton, T.; Outram, S.; Rahemtulla, A.; Porter, A.C.; Dallman, M.J. Selective silencing of full-length CD80 but not IgV-CD80 leads to impaired clonal deletion of self-reactive T cells and altered regulation of immune responses. Eur. J. Immunol. 2001, 31, 118–127. [Google Scholar] [CrossRef]
- Carreno, B.M.; Collins, M. The B7 family of ligands and its receptors: New pathways for costimulation and inhibition of immune responses. Annu. Rev. Immunol. 2002, 20, 29–53. [Google Scholar] [CrossRef]
- Coats, S.R.; Reife, R.A.; Bainbridge, B.W.; Pham, T.T.; Darveau, R.P. Porphyromonas gingivalis lipopolysaccharide antagonizes Escherichia coli lipopolysaccharide at toll-like receptor 4 in human endothelial cells. Infect. Immun. 2003, 71, 6799–6807. [Google Scholar] [CrossRef]
- Liu, R.; Desta, T.; Raptis, M.; Darveau, R.P.; Graves, D.T.P. gingivalis and E. coli lipopolysaccharides exhibit different systemic but similar local induction of inflammatory markers. J. Periodontol. 2008, 79, 1241–1247. [Google Scholar] [CrossRef]
- Zhang, Y.; Gaekwad, J.; Wolfert, M.A.; Boons, G.J. Synthetic tetra-acylated derivatives of lipid A from Porphyromonas gingivalis are antagonists of human TLR4. Org. Biomol. Chem. 2008, 6, 3371–3381. [Google Scholar] [CrossRef] [PubMed]
- Herath, T.D.; Darveau, R.P.; Seneviratne, C.J.; Wang, C.Y.; Wang, Y.; Jin, L. Tetra- and penta-acylated lipid A structures of Porphyromonas gingivalis LPS differentially activate TLR4-mediated NF-kappaB signal transduction cascade and immuno-inflammatory response in human gingival fibroblasts. PLoS ONE 2013, 8, e58496. [Google Scholar] [CrossRef]
- Coats, S.R.; Pham, T.T.; Bainbridge, B.W.; Reife, R.A.; Darveau, R.P. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J. Immunol. 2005, 175, 4490–4498. [Google Scholar] [CrossRef]
- Hajishengallis, G.; Liang, S.; Payne, M.A.; Hashim, A.; Jotwani, R.; Eskan, M.A.; McIntosh, M.L.; Alsam, A.; Kirkwood, K.L.; Lambris, J.D.; et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host. Microbe 2011, 10, 497–506. [Google Scholar] [CrossRef] [PubMed]
- Lamont, R.J.; Chan, A.; Belton, C.M.; Izutsu, K.T.; Vasel, D.; Weinberg, A. Porphyromonas gingivalis invasion of gingival epithelial cells. Infect. Immun. 1995, 63, 3878–3885. [Google Scholar] [CrossRef] [PubMed]
- Mendez, K.N.; Hoare, A.; Soto, C.; Bugueno, I.; Olivera, M.; Meneses, C.; Perez-Donoso, J.M.; Castro-Nallar, E.; Bravo, D. Variability in Genomic and Virulent Properties of Porphyromonas gingivalis Strains Isolated from Healthy and Severe Chronic Periodontitis Individuals. Front. Cell Infect. Microbiol. 2019, 9, 246. [Google Scholar] [CrossRef] [PubMed]
- Kiyama-Kishikawa, M.; Hiratsuka, K.; Abiko, Y. Gene expression profiling and characterization under hemin limitation in Porphyromonas gingivalis. J. Oral Sci. 2005, 47, 191–197. [Google Scholar] [CrossRef][Green Version]
- Ortega, X.; Hunt, T.A.; Loutet, S.; Vinion-Dubiel, A.D.; Datta, A.; Choudhury, B.; Goldberg, J.B.; Carlson, R.; Valvano, M.A. Reconstitution of O-specific lipopolysaccharide expression in Burkholderia cenocepacia strain J2315, which is associated with transmissible infections in patients with cystic fibrosis. J. Bacteriol. 2005, 187, 1324–1333. [Google Scholar] [CrossRef]
- Paterson, G.K.; Cone, D.B.; Peters, S.E.; Maskell, D.J. The enzyme phosphoglucomutase (Pgm) is required by Salmonella enterica serovar Typhimurium for O-antigen production, resistance to antimicrobial peptides and in vivo fitness. Microbiology 2009, 155 Pt 10, 3403–3410. [Google Scholar] [CrossRef]
- Martin, M.; Katz, J.; Vogel, S.N.; Michalek, S.M. Differential induction of endotoxin tolerance by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli. J. Immunol. 2001, 167, 5278–5285. [Google Scholar] [CrossRef]
- Sun, Y.; Li, H.; Sun, M.J.; Zheng, Y.Y.; Gong, D.J.; Xu, Y. Endotoxin tolerance induced by lipopolysaccharides derived from Porphyromonas gingivalis and Escherichia coli: Alternations in Toll-like receptor 2 and 4 signaling pathway. Inflammation 2014, 37, 268–276. [Google Scholar] [CrossRef]
- Andrukhov, O.; Ertlschweiger, S.; Moritz, A.; Bantleon, H.P.; Rausch-Fan, X. Different effects of P. gingivalis LPS and E. coli LPS on the expression of interleukin-6 in human gingival fibroblasts. Acta Odontol. Scand. 2014, 72, 337–345. [Google Scholar] [CrossRef]
- Krutzik, S.R.; Tan, B.; Li, H.; Ochoa, M.T.; Liu, P.T.; Sharfstein, S.E.; Graeber, T.G.; Sieling, P.A.; Liu, Y.J.; Rea, T.H.; et al. TLR activation triggers the rapid differentiation of monocytes into macrophages and dendritic cells. Nat. Med. 2005, 11, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Hilda, J.N.; Selvaraj, A.; Das, S.D. Mycobacterium tuberculosis H37Rv is more effective compared to vaccine strains in modulating neutrophil functions: An in vitro study. FEMS Immunol. Med. Microbiol. 2018, 66, 372–381. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Hoshino, K.; Kaisho, T.; Sanjo, H.; Takeuchi, O.; Sugiyama, M.; Okabe, M.; Takeda, K.; et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science 2003, 301, 640–643. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, M.; Sato, S.; Hemmi, H.; Uematsu, S.; Hoshino, K.; Kaisho, T.; Takeuchi, O.; Takeda, K.; Akira, S. TRAM is specifically involved in the Toll-like receptor 4-mediated MyD88-independent signaling pathway. Nat. Immunol. 2003, 4, 1144–1150. [Google Scholar] [CrossRef] [PubMed]
- Kagan, J.C.; Su, T.; Horng, T.; Chow, A.; Akira, S.; Medzhitov, R. TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 2008, 9, 361–368. [Google Scholar] [CrossRef]
- Rosadini, C.V.; Kagan, J.C. Early innate immune responses to bacterial LPS. Curr. Opin. Immunol. 2017, 44, 14–19. [Google Scholar] [CrossRef]
- Stephens, M.; Liao, S.; von der Weid, P. Ultra-purification of Lipopolysacharides reveals species-specific signalling bias of TLR4: Importance in macrophage function. Sci. Rep. 2021, 11, 1335. [Google Scholar] [CrossRef]
- Langjahr, P.; Diaz-Jimenez, D.; De la Fuente, M.; Rubio, E.; Golenbock, D.; Bronfman, F.C.; Quera, R.; Gonzalez, M.J.; Hermoso, M.A. Metalloproteinase-dependent TLR2 ectodomain shedding is involved in soluble toll-like receptor 2 (sTLR2) production. PLoS ONE 2014, 9, e104624. [Google Scholar] [CrossRef]
- Gebbia, J.A.; Coleman, J.L.; Benach, J.L. Selective induction of matrix metalloproteinases by Borrelia burgdorferi via toll-like receptor 2 in monocytes. J. Infect. Dis. 2004, 189, 113–119. [Google Scholar] [CrossRef][Green Version]
- Roomi, M.W.; Monterrey, J.C.; Kalinovsky, T.; Rath, M.; Niedzwiecki, A. In vitro modulation of MMP-2 and MMP-9 in human cervical and ovarian cancer cell lines by cytokines, inducers and inhibitors. Oncol. Rep. 2010, 23, 605–614. [Google Scholar]
- Hoare, A.; Bravo, D.; Martinic, M.; Valvano, M.A.; Contreras, I.; Alvarez, S.A. The normal chain length distribution of the O antigen is required for the interaction of Shigella flexneri 2a with polarized Caco-2 cells. Biol. Res. 2012, 45, 21–26. [Google Scholar] [CrossRef]
- Bravo, D.; Silva, C.; Carter, J.A.; Hoare, A.; Alvarez, S.A.; Blondel, C.J.; Zaldivar, M.; Valvano, M.A.; Contreras, I. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J. Med. Microbiol. 2008, 57 Pt 8, 938–946. [Google Scholar] [CrossRef]
- Tsai, C.M.; Frasch, C.E. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 1982, 119, 115–119. [Google Scholar] [CrossRef]
- Takashiba, S.; Van Dyke, T.E.; Amar, S.; Murayama, Y.; Soskolne, A.W.; Shapira, L. Differentiation of monocytes to macrophages primes cells for lipopolysaccharide stimulation via accumulation of cytoplasmic nuclear factor kappaB. Infect. Immun. 1999, 67, 5573–5578. [Google Scholar] [CrossRef]
- Maess, M.B.; Wittig, B.; Cignarella, A.; Lorkowski, S. Reduced PMA enhances the responsiveness of transfected THP-1 macrophages to polarizing stimuli. J. Immunol. Methods 2013, 402, 76–81. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).