Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A
Abstract
1. Introduction
2. Results and Discussion
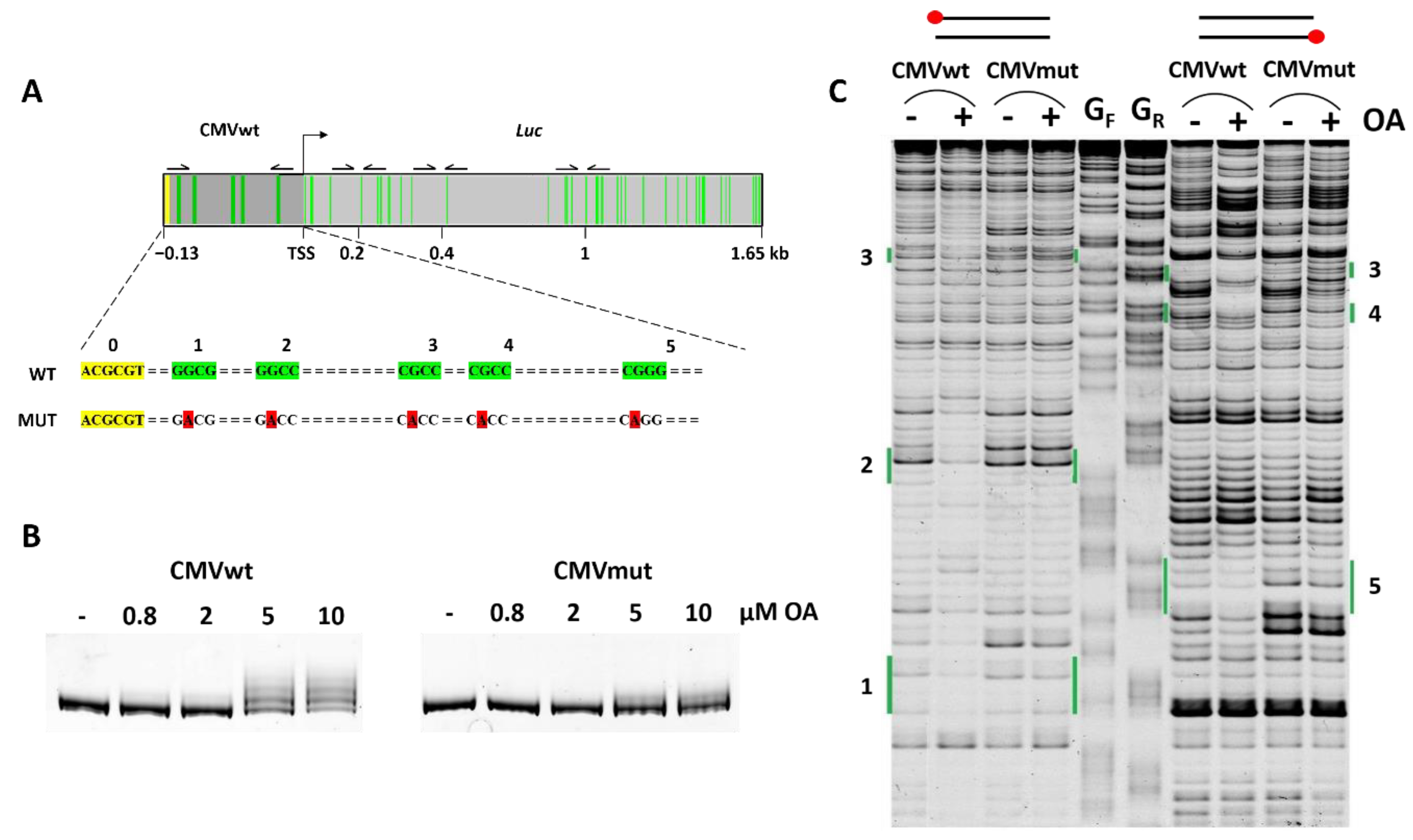
2.1. GC Tetrads in the Cytomegalovirus Promoter Are Critical for OA–DNA Complex Formation: An In Vitro Study
2.2. OA–DNA Complex Formation Impedes Transcription Driven by CMVwt but Not Cmvmut
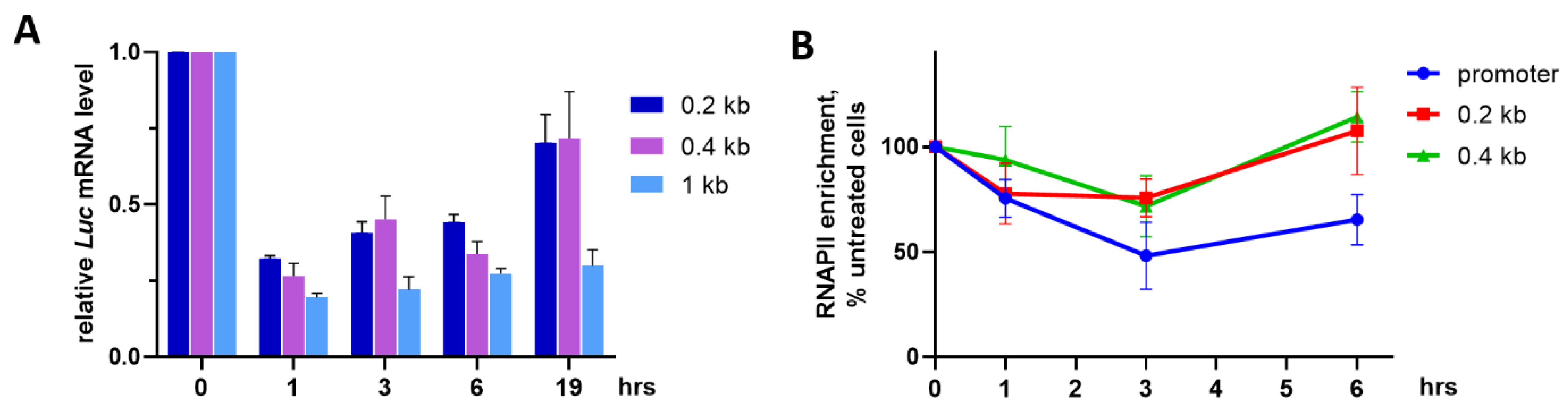
2.3. Reversibility of OA-Induced Transcriptional Attenuation
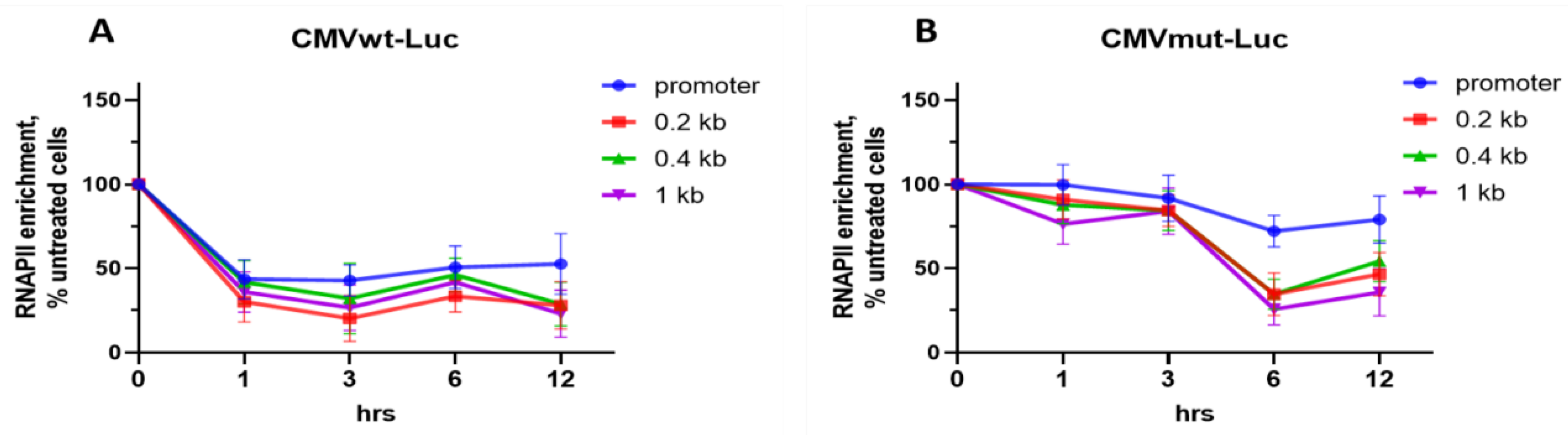
2.4. Differential Effects of OA on Gene Occupancy by Total RNAPII
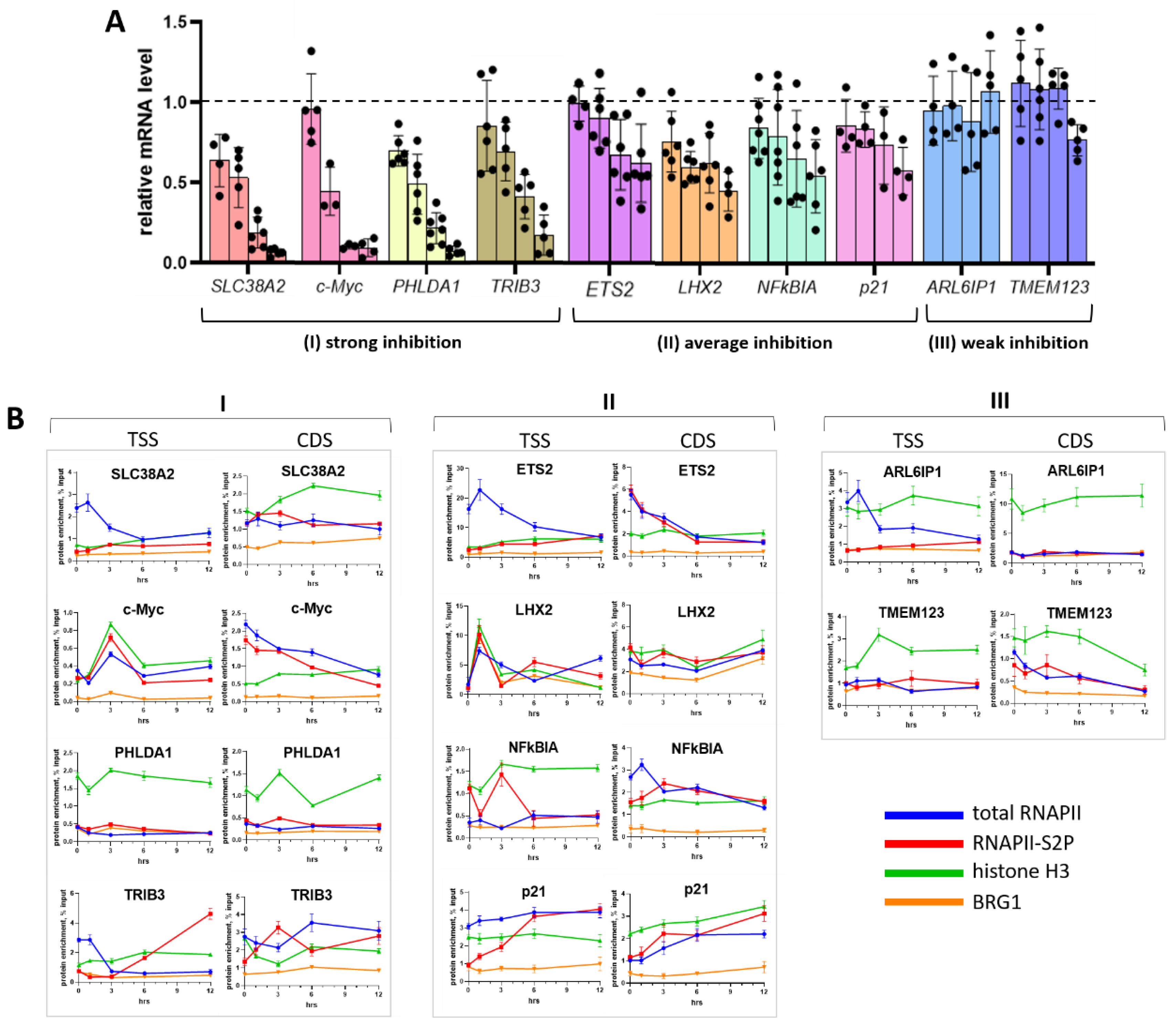
2.5. Does the GC Context Matter?
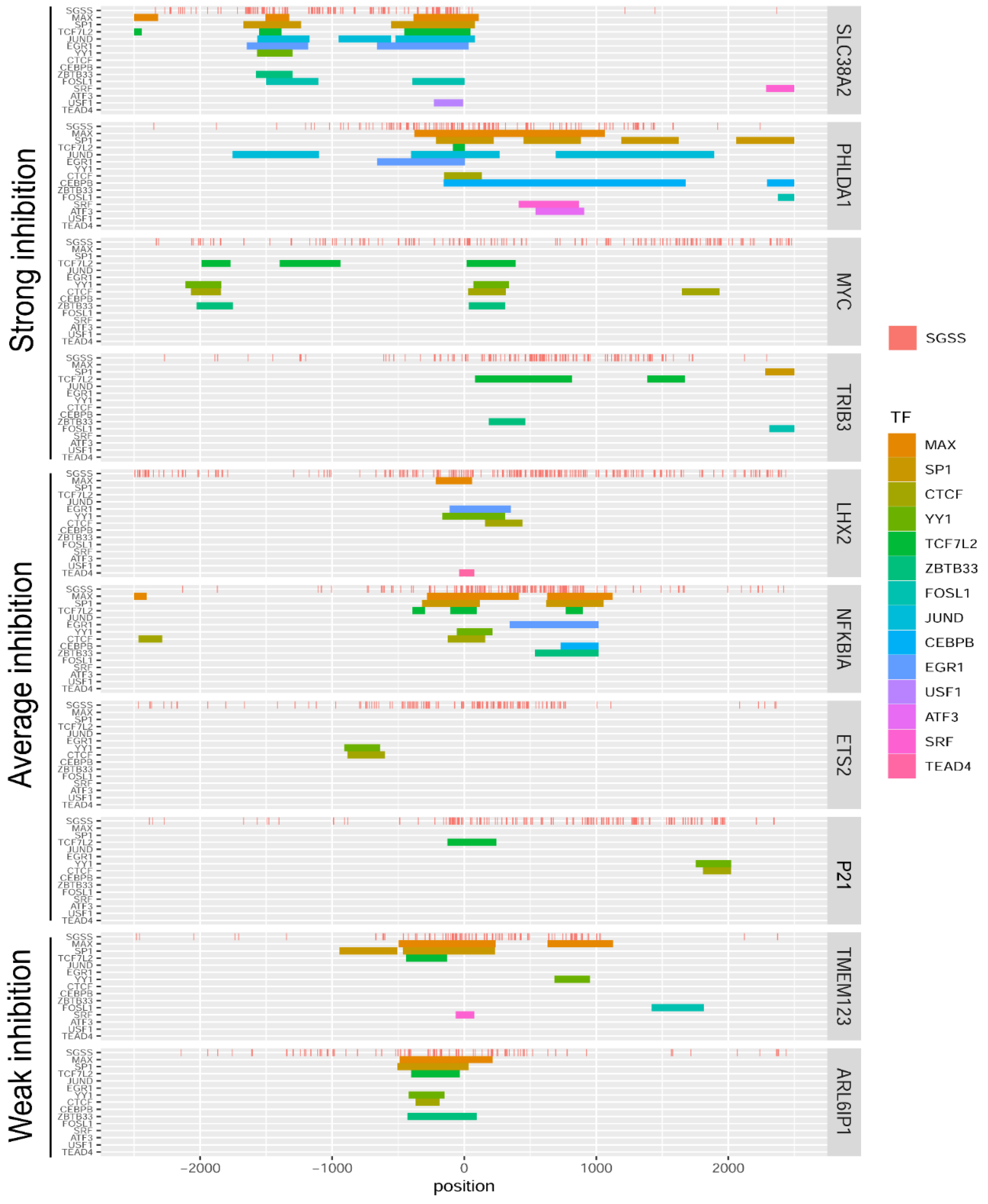
2.6. Sensitivity of Individual Genes to OA Is Independent of GC Tetrad Location
3. Materials and Methods
3.1. Cell Lines and Treatment
3.2. Antibodies
3.3. Plasmids, Cloning, and Cell Transfection
3.4. Real-Time qPCR
3.5. Chromatin Immunoprecipitation
3.6. EMSA
3.7. DNAse I Protection Assays
3.8. Mapping GC Tetrads and TF Consensus Binding Sites
3.9. Statistical Analysis
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- González, A.; Fillat, M.F.; Lanas, Á. Transcriptional regulators: Valuable targets for novel antibacterial strategies. Future Med. Chem. 2018, 10, 541–560. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, D.; Scarlato, V.; Vannini, A. Targeting of regulators as a promising approach in the search for novel antimicrobial agents. Microorganisms 2022, 10, 185. [Google Scholar] [CrossRef] [PubMed]
- Klein, K. Bromodomain protein inhibition: A novel therapeutic strategy in rheumatic diseases. RMD Open 2018, 4, e000744. [Google Scholar] [CrossRef]
- Franklin, R.M. The inhibition of ribonucleic acid synthesis in mammalian cells by actinomycin D. Biochim. Biophys. Acta 1963, 72, 555–565. [Google Scholar] [CrossRef]
- Ji, M.; Jiang, S.; Zhao, J.; Wan, X.; Feng, F.; Ren, T.; Yang, J.; Xiang, Y. Efficacies of FAEV and EMA/CO regimens as primary treatment for gestational trophoblastic neoplasia. Br. J. Cancer 2022, 127, 524–530. [Google Scholar] [CrossRef] [PubMed]
- Laham-Karam, N.; Pinto, G.P.; Poso, A.; Kokkonen, P. Transcription and translation inhibitors in cancer treatment. Front. Chem. 2020, 8, 276. [Google Scholar] [CrossRef]
- Kulkarni, K.K.; Bankar, K.G.; Shukla, R.N.; Das, C.; Banerjee, A.; Dasgupta, D.; Vasudevan, M. Global gene expression profiling data analysis reveals key gene families and biological processes inhibited by mithramycin in sarcoma cell lines. Genom. Data 2015, 3, 8–14. [Google Scholar] [CrossRef]
- Lombó, F.; Menéndez, N.; Salas, J.A.; Méndez, C. The aureolic acid family of antitumor compounds: Structure, mode of action, biosynthesis, and novel derivatives. Appl. Microbiol. Biotechnol. 2006, 73, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Tevyashova, A.N.; Olsufyeva, E.N.; Turchin, K.F.; Balzarini, J.; Bykov, E.E.; Dezhenkova, L.G.; Shtil, A.A.; Preobrazhenskaya, M.N. Reaction of the antitumor antibiotic olivomycin I with aryl diazonium salts. Synthesis, cytotoxic and antiretroviral potency of 5-aryldiazenyl-6-O-deglycosyl derivatives of olivomycin I. Bioorg. Med. Chem. 2009, 17, 4961–4967. [Google Scholar] [CrossRef]
- Bianchi, N.; Rutigliano, C.; Passadore, M.; Tomassetti, M.; Pippo, L.; Mischiati, C.; Feriotto, G.; Gambari, R. Targeting of the HIV-1 long terminal repeat with chromomycin potentiates the inhibitory effects of a triplex-forming oligonucleotide on Sp1-DNA interactions and in vitro transcription. Biochem. J. 1997, 326, 919–927. [Google Scholar] [CrossRef][Green Version]
- Osada, N.; Kosuge, Y.; Ishige, K.; Ito, Y. Mithramycin, an agent for developing new therapeutic drugs for neurodegenerative diseases. J. Pharmacol. Sci. 2013, 122, 251–256. [Google Scholar] [CrossRef]
- Stack, E.C.; Del Signore, S.J.; Luthi-Carter, R.; Soh, B.Y.; Goldstein, D.R.; Matson, S.; Goodrich, S.; Markey, A.L.; Cormier, K.; Hagerty, S.W.; et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Gen. 2007, 16, 1164–1175. [Google Scholar] [CrossRef] [PubMed]
- Kersten, W.; Kersten, H.; Szybalski, W.; Fiandt, M. Physicochemical properties of complexes between deoxyribonucleic acid and antibiotics which affect ribonucleic acid synthesis (actinomycin, daunomycin, cinerubin, nogalamycin, chromomycin, mithramycin, and olivomycin). Biochemistry 1966, 5, 236–244. [Google Scholar] [CrossRef] [PubMed]
- Gause, G.F. Olivomycin, chromomycin, and mithramycin. In Mechanism of Action of Antimicrobial and Antitumor Agents; Antibiotics; Corcoran, J.W., Hahn, F.E., Snell, J.F., Arora, K.L., Eds.; Springer: Berlin/Heidelberg, Germany, 1975; Volume 3, pp. 197–202. [Google Scholar]
- Keniry, M.A.; Banville, D.L.; Simmonds, P.M.; Shafer, R. Nuclear magnetic resonance comparison of the binding sites of mithramycin and chromomycin on the self-complementary oligonucleotide d(ACCCGGGT)2. Evidence that the saccharide chains have a role in sequence specificity. J. Mol. Biol. 1993, 231, 753–767. [Google Scholar] [CrossRef] [PubMed]
- Hou, M.H.; Robinson, H.; Gao, Y.G.; Wang, A.H. Crystal structure of the [Mg2+-(chromomycin A3)2]-d(TTGGCCAA)2 complex reveals GGCC binding specificity of the drug dimer chelated by a metal ion. Nucleic Acids Res. 2004, 32, 2214–2222. [Google Scholar] [CrossRef]
- Beniaminov, A.D.; Chashchina, G.V.; Livshits, M.A.; Kechko, O.I.; Mitkevich, V.A.; Mamaeva, O.K.; Tevyashova, A.N.; Shtil, A.A.; Shchyolkina, A.K.; Kaluzhny, D.N. Discrimination between G/C binding sites by olivomycin a is determined by kinetics of the drug-DNA Interaction. Int. J. Mol. Sci. 2020, 21, 5299. [Google Scholar] [CrossRef]
- Grohar, P.J.; Woldemichael, G.M.; Griffin, L.B.; Mendoza, A.; Chen, Q.R.; Yeung, C.; Currier, D.G.; Davis, S.; Khanna, C.; Khan, J.; et al. Identification of an inhibitor of the EWS-FLI1 oncogenic transcription factor by high-throughput screening. J. Nat. Cancer Inst. 2011, 103, 962–978. [Google Scholar] [CrossRef]
- Guillon, N.; Tirode, F.; Boeva, V.; Zynovyev, A.; Barillot, E.; Delattre, O. The oncogenic EWS-FLI1 protein binds in vivo GGAA microsatellite sequences with potential transcriptional activation function. PLoS ONE 2009, 4, e4932. [Google Scholar] [CrossRef]
- Snyder, R.C.; Ray, R.; Blume, S.; Miller, D.M. Mithramycin blocks transcriptional initiation of the c-myc P1 and P2 promoters. Biochemistry 1991, 30, 4290–4297. [Google Scholar] [CrossRef]
- Sleiman, S.F.; Langley, B.C.; Basso, M.; Berlin, J.; Xia, L.; Payappilly, J.B.; Kharel, M.K.; Guo, H.; Marsh, J.L.; Thompson, L.M.; et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 2011, 31, 6858–6870. [Google Scholar] [CrossRef]
- Tornin, J.; Martinez-Cruzado, L.; Santos, L.; Rodriguez, A.; Núñez, L.; Oro, P.; Ana, H.M.; Allonca, E.; Teresa, F.-G.M.; Astudillo, A.; et al. Inhibition of SP1 by the mithramycin analog EC-8042 efficiently targets tumor initiating cells in sarcoma. Oncotarget 2016, 7, 30935–30950. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.-S.; Nam, J.-S.; Jung, J.-Y.; Cho, N.-P.; Cho, S.-D. Modulation of specificity protein 1 by mithramycin A as a novel therapeutic strategy for cervical cancer. Sci. Rep. 2014, 4, 7162. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, R. Chemotherapy for the Brain: The antitumor antibiotic mithramycin prolongs survival in a mouse model of Huntington’s disease. J. Neurosci. 2004, 24, 10335–10342. [Google Scholar] [CrossRef] [PubMed]
- Lin, R.; Hsu, C.; Wang, Y. Mithramycin A inhibits DNA methyltransferase and metastasis potential of lung cancer cells. Anticancer Drugs 2007, 18, 1157–1164. [Google Scholar] [CrossRef] [PubMed]
- Federico, A.; Steinfass, T.; Larribère, L.; Novak, D.; Morís, F.; Núñez, L.E.; Umansky, V.; Utikal, J. Mithramycin A and mithralog EC-8042 inhibit SETDB1 expression and its oncogenic activity in malignant melanoma. Mol. Ther. Oncolytics 2020, 18, 83–99. [Google Scholar] [CrossRef]
- Chasse, M.H.; Johnson, B.K.; Boguslawski, E.A.; Sorensen, K.M.; Rosien, J.E.; Kang, M.H.; Reynolds, C.P.; Heo, L.; Madaj, Z.B.; Beddows, I.; et al. Mithramycin induces promoter reprogramming and differentiation of rhabdoid tumor. EMBO Mol. Med. 2021, 13, 12640. [Google Scholar] [CrossRef]
- Sergeev, A.V.; Tevyashova, A.N.; Vorobyov, A.P.; Gromova, E.S. The effect of antitumor antibiotic olivomycin A and its new semi-synthetic derivative olivamide on the activity of murine DNA methyltransferase Dnmt3a. Biochemistry 2019, 84, 62–70. [Google Scholar] [CrossRef]
- Mir, M.A.; Dasgupta, D. Interaction of antitumor drug, mithramycin, with chromatin. Biochem. Biophys. Res. Commun. 2001, 280, 68–74. [Google Scholar] [CrossRef]
- Das, S.; Dasgupta, D. Binding of (MTR)2Zn2+ complex to chromatin: A comparison with (MTR)2Mg2+ complex. J. Inorg. Biochem. 2005, 99, 707–715. [Google Scholar] [CrossRef]
- Kubosaki, A.; Tomaru, Y.; Tagami, M.; Arner, E.; Miura, H.; Suzuki, T.; Suzuki, M.; Suzuki, H.; Hayashizaki, Y. Genome-wide investigation of in vivo EGR-1 binding sites in monocytic differentiation. Genome Biol. 2009, 10, R41. [Google Scholar] [CrossRef]
- Verheul, T.C.J.; van Hijfte, L.; Perenthaler, E.; Barakat, T.S. The Why of YY1: Mechanisms of transcriptional regulation by Yin Yang 1. Front. Cell Dev. Biol. 2020, 8, 592164. [Google Scholar] [CrossRef]
- Isagulieva, A.K.; Soshnikova, N.V.; Shtil, A.A. Inhibition of the c-Myc oncogene by the aureolic acid group antibiotics. Dokl. Biochem. Biophys. 2021, 500, 308–311. [Google Scholar] [CrossRef]
- Fox, K.R.; Howarth, N.R. Investigations into the sequence-selective binding of mithramycin and related ligands to DNA. Nucl. Acids Res. 1985, 13, 8695–8714. [Google Scholar] [CrossRef]
- Carpenter, M.L.; Marks, J.N.; Fox, K.R. DNA-sequence binding preference of the GC-selective ligand mithramycin. Deoxyribonuclease-I/deoxyribonuclease-II and hydroxy-radical footprinting at CCCG, CCGC, CGGC, GCCC and GGGG flanked by (AT)n and AnTn. Eur. J. Biochem. 1993, 215, 561–566. [Google Scholar] [CrossRef]
- Albertini, V. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer Streptomyces argillaceus exhibits improved transcriptional repressor activity: Implications for cancer therapy. Nucl. Acids Res. 2006, 34, 1721–1734. [Google Scholar] [CrossRef] [PubMed]
- Cheglakov, I.B.; Tevyashova, A.N.; Kurbatov, L.K.; Tatarsky, V.V., Jr.; Samusenko, A.V.; Preobrazhenskaya, M.N.; Shtil, A.A. Altered transcription and replication are the mechanisms of cytotoxicity of antitumor antibiotic olivomycin A. Dokl. Biochem. Biophys. 2010, 435, 320–322. [Google Scholar] [CrossRef]
- Simonova, V.S.; Samusenko, A.V.; Filippova, N.A.; Tevyashova, A.N.; Lyniv, L.S.; Kulik, G.I.; Chekhun, V.F.; Shtil, A.A. Olivomycin induces tumor cell apoptosis and suppresses p53-induced transcription. Dokl. Biochem. Biophys. 2005, 139, 455–459. [Google Scholar] [CrossRef]
- Collas, P. The current state of chromatin immunoprecipitation. Mol. Biotechnol. 2010, 45, 87–100. [Google Scholar] [CrossRef]
- Bowman, E.A.; Kelly, W.G. RNA Polymerase II transcription elongation and Pol II CTD Ser2 phosphorylation. Nucleus 2014, 5, 224–236. [Google Scholar] [CrossRef]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Rec. Sign. 2008, 6, e004. [Google Scholar] [CrossRef]
- Ross, J.; Bernstein, P.; Prokipcak, R.D.; Herrick, D.J. Regulation of c-myc mRNA half-life by an RNA-binding protein. In Tumor Biology; NATO ASI Series; Tsiftsoglou, A.S., Sartorelli, A.C., Housman, D.E., Dexter, T.M., Eds.; Springer: Berlin/Heidelberg, Germany, 1996; Volume 99, pp. 257–272. [Google Scholar]
- Tevyashova, A.N.; Durandin, N.A.; Vinogradov, A.M.; Zbarsky, V.B.; Reznikova, M.I.; Dezhenkova, L.G.; Bykov, E.E.; Olsufyeva, E.N.; Kuzmin, V.A.; Shtil, A.A.; et al. Role of the acyl groups in carbohydrate chains in cytotoxic properties of olivomycin A. J. Antibiot. 2013, 66, 523–530. [Google Scholar] [CrossRef] [PubMed]
- Brechalov, A.V.; Georgieva, S.G.; Soshnikova, N.V. Mammalian cells contain two functionally distinct PBAF complexes incorporating different isoforms of PHF10 signature subunit. Cell Cycle 2014, 13, 1970–1979. [Google Scholar] [CrossRef] [PubMed]
- Soshnikova, N.V.; Tatarskiy, E.V.; Tatarskiy, V.V.; Klimenko, N.S.; Shtil, A.A.; Nikiforov, M.A.; Georgieva, S.G. PHF10 subunit of PBAF complex mediates transcriptional activation by MYC. Oncogene 2021, 40, 6071–6080. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Isagulieva, A.K.; Kaluzhny, D.N.; Beniaminov, A.D.; Soshnikova, N.V.; Shtil, A.A. Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A. Int. J. Mol. Sci. 2022, 23, 8871. https://doi.org/10.3390/ijms23168871
Isagulieva AK, Kaluzhny DN, Beniaminov AD, Soshnikova NV, Shtil AA. Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A. International Journal of Molecular Sciences. 2022; 23(16):8871. https://doi.org/10.3390/ijms23168871
Chicago/Turabian StyleIsagulieva, Alexandra K., Dmitry N. Kaluzhny, Artemy D. Beniaminov, Nataliya V. Soshnikova, and Alexander A. Shtil. 2022. "Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A" International Journal of Molecular Sciences 23, no. 16: 8871. https://doi.org/10.3390/ijms23168871
APA StyleIsagulieva, A. K., Kaluzhny, D. N., Beniaminov, A. D., Soshnikova, N. V., & Shtil, A. A. (2022). Differential Impact of Random GC Tetrad Binding and Chromatin Events on Transcriptional Inhibition by Olivomycin A. International Journal of Molecular Sciences, 23(16), 8871. https://doi.org/10.3390/ijms23168871





