LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice
Abstract
1. Introduction
2. Results
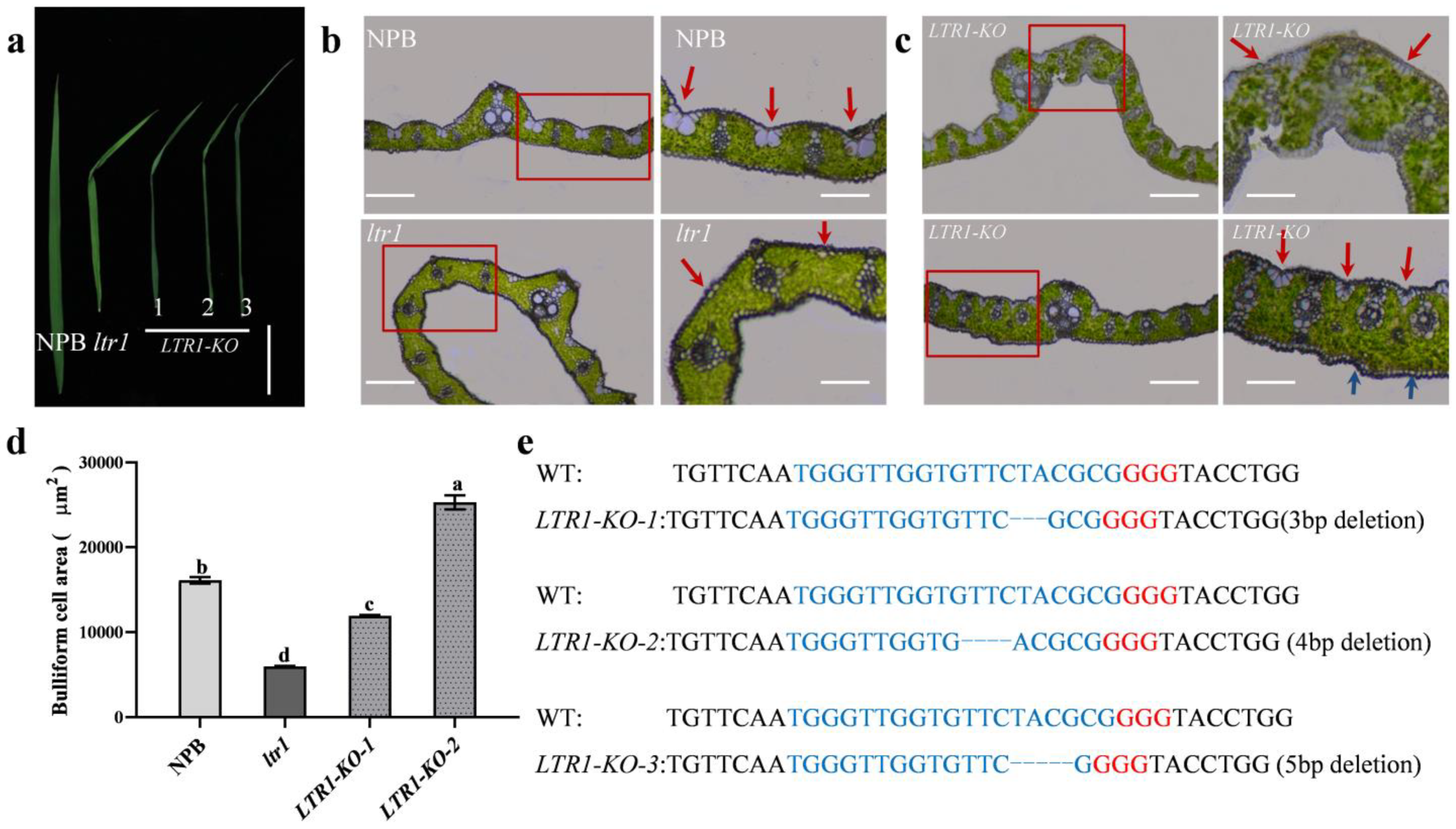
2.1. Identification of the ltr1 Mutant
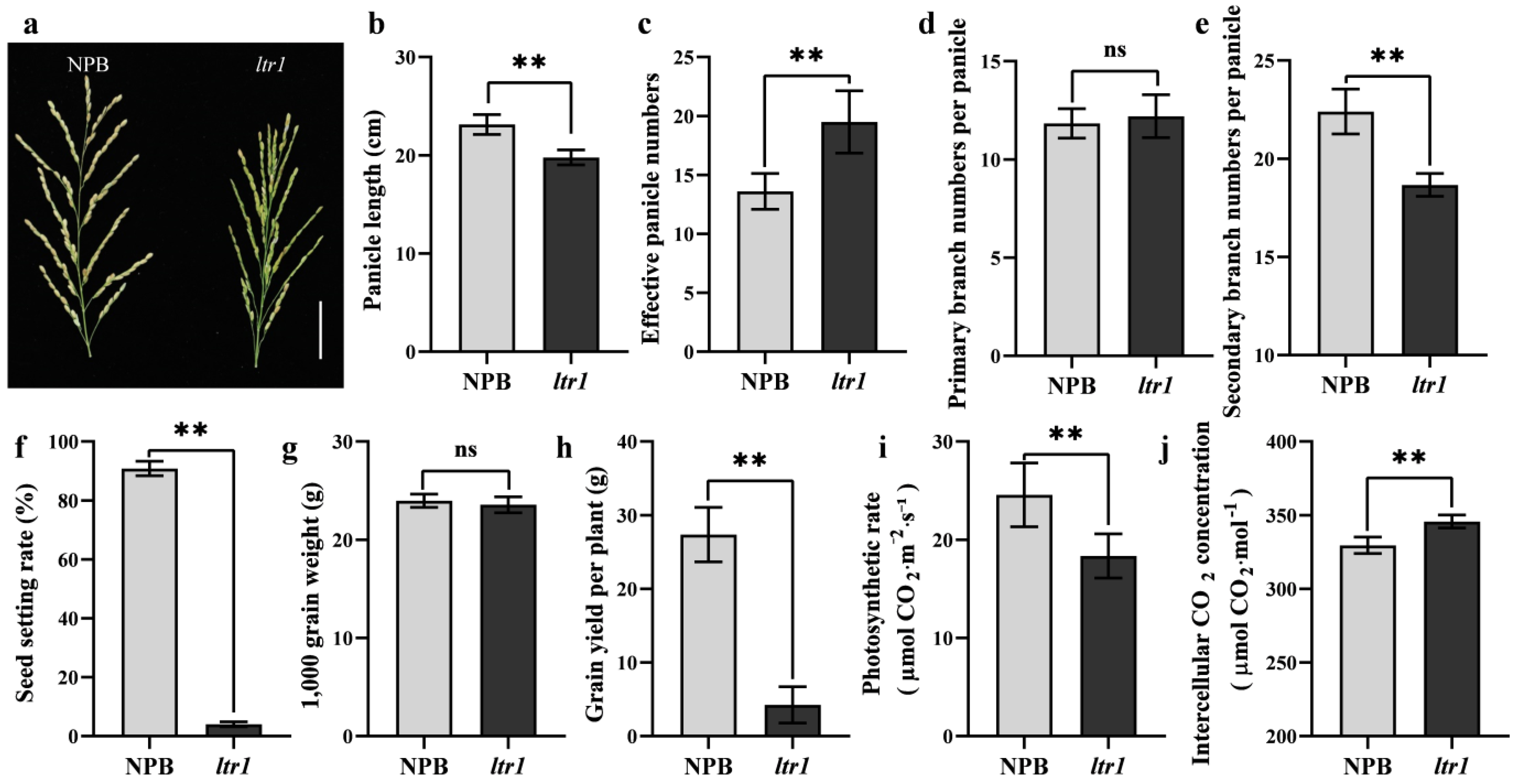
2.2. Effect of LTR1 on Photosynthetic Efficiency and Seed Setting Rate
2.3. Map-Based Cloning of LTR1
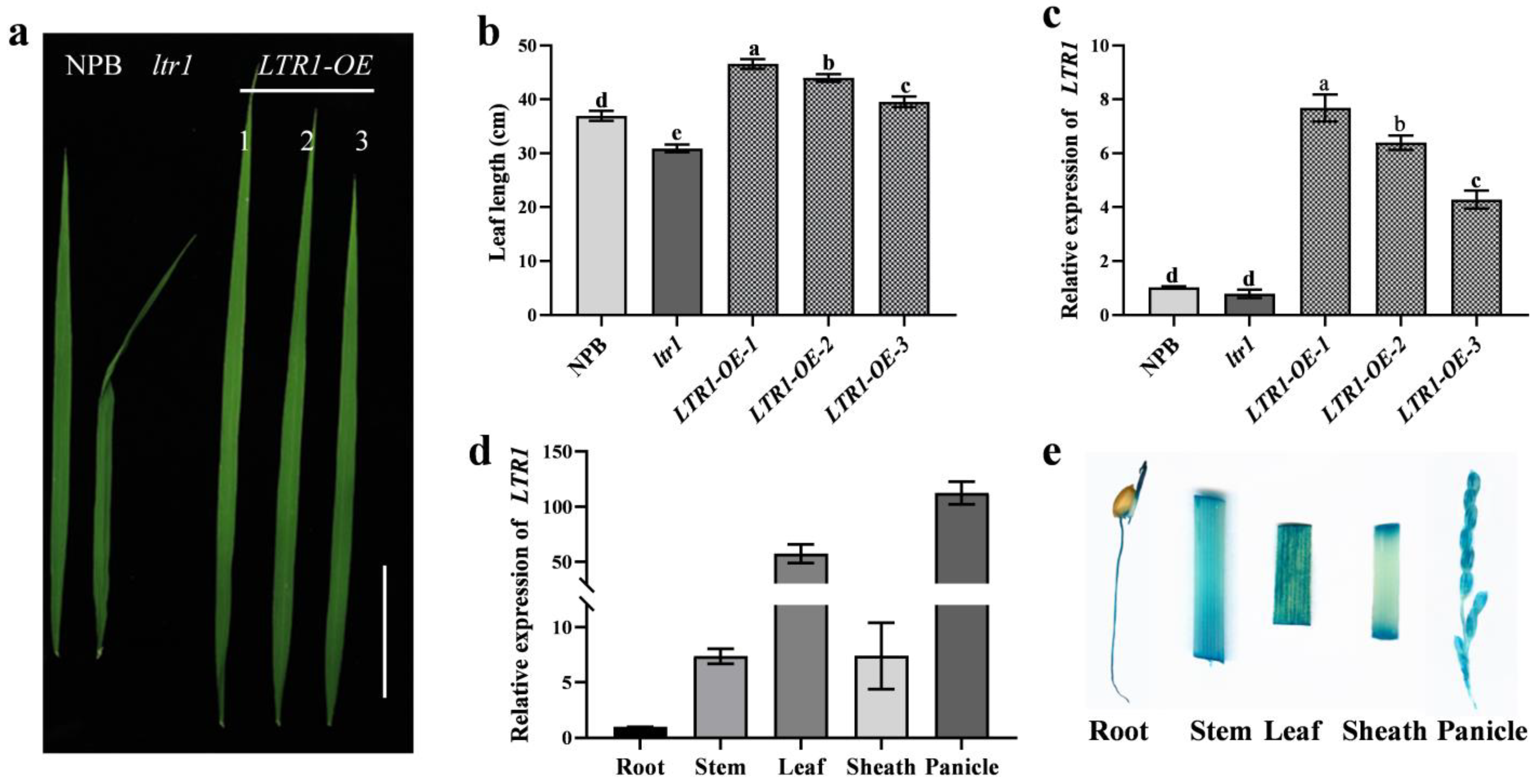
2.4. Overexpression and Targeted Deletion of LTR1
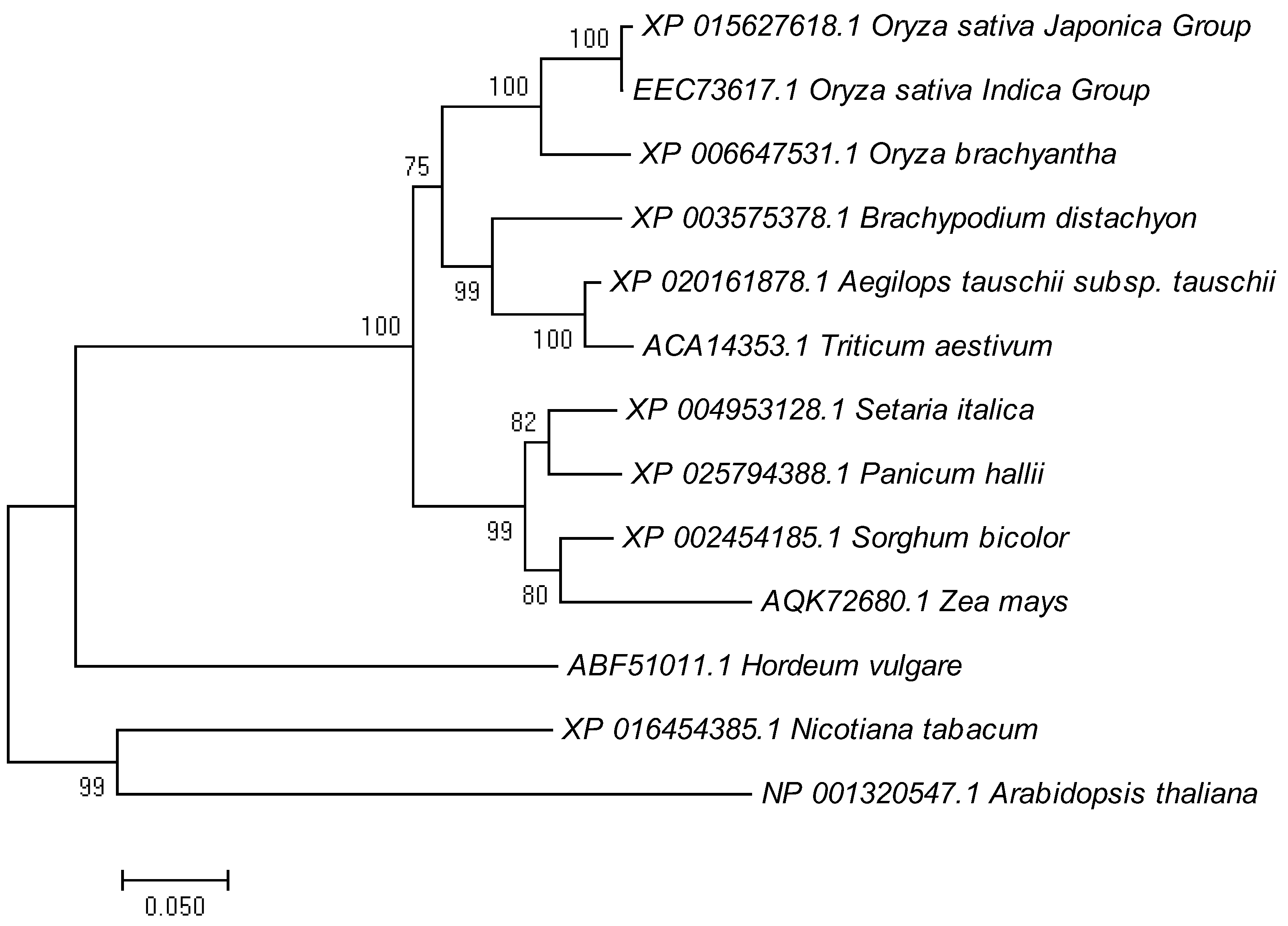
2.5. Phylogenetic Analysis of LTR1
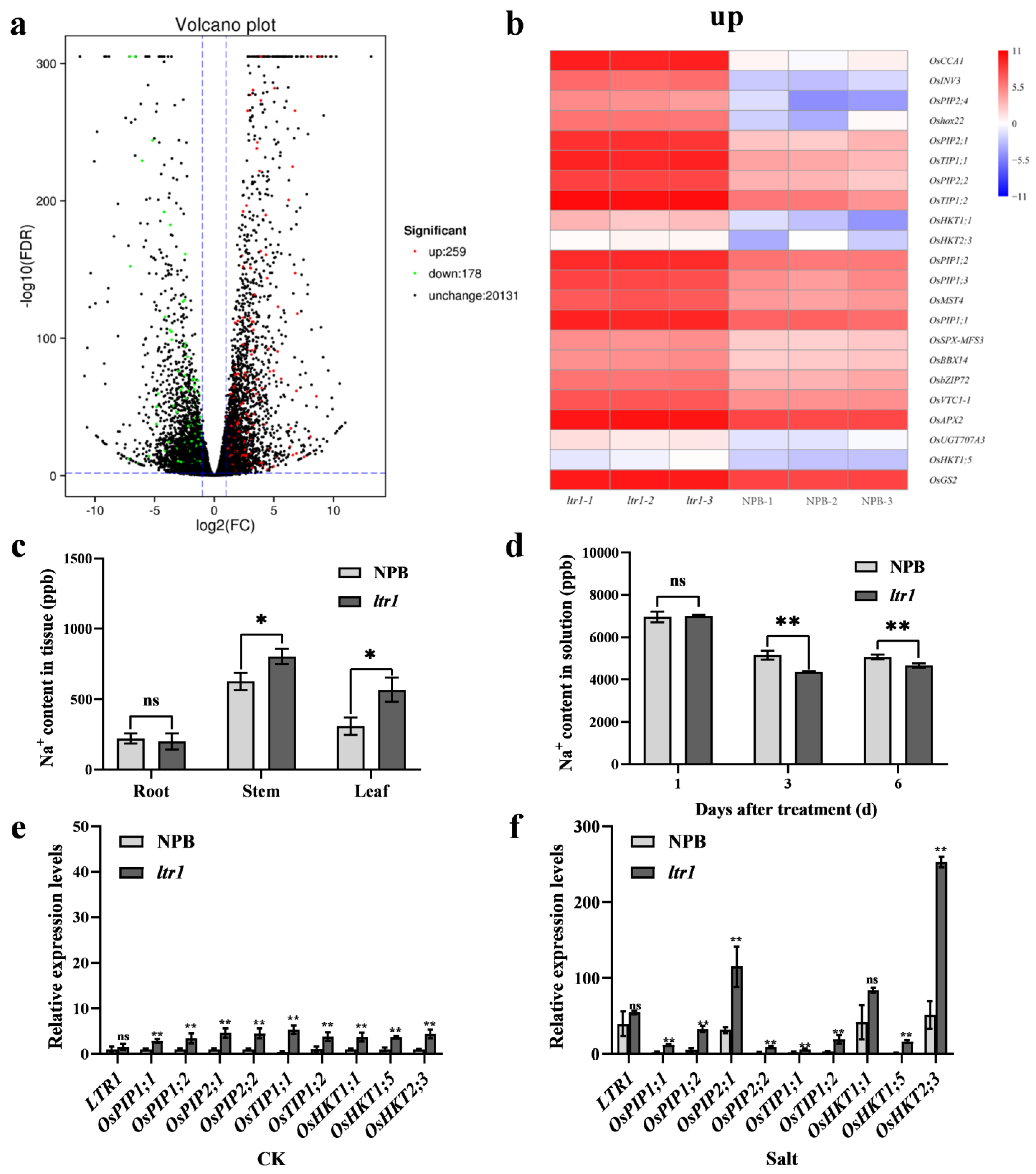
2.6. LTR1 Participates in Water Transport and Ion Homeostasis
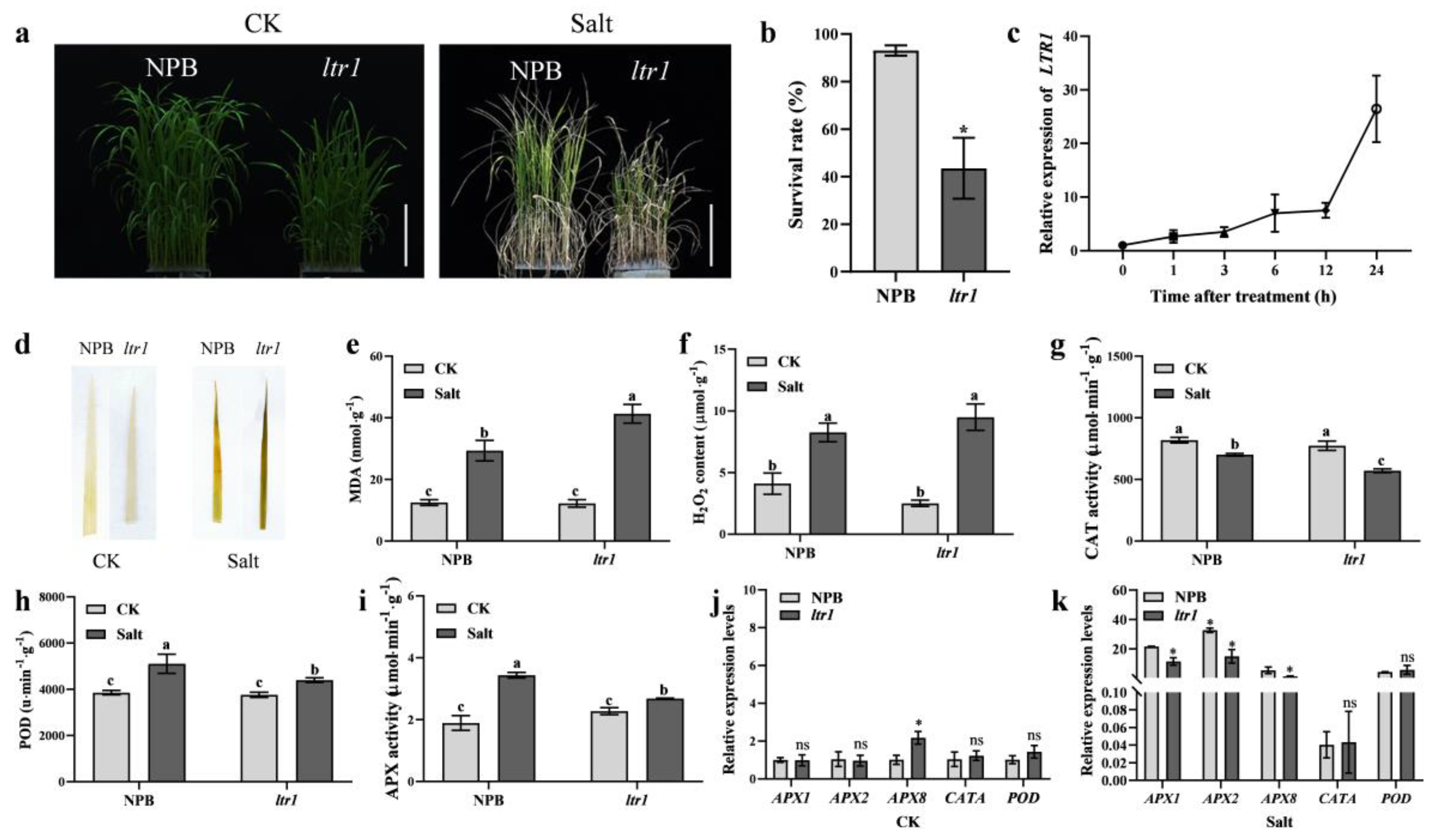
2.7. LTR1 Regulates Salt Tolerance in Rice
3. Discussion
3.1. LTR1 Encodes a Wax Synthesis Gene and Regulates Leaf Morphology
3.2. LTR1 Has Multiple Effects on Plant Growth and Development
3.3. LTR1 Regulated Salt Tolerance by Altering Plant Water Status and Ion Homeostasis
3.4. Prospects
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Phenotypicl Characterization and Histological Analysis
4.3. Measurements of Chlorophyll Content and Photosynthetic Parameters
4.4. Map-Based Cloning and Complementation Assay
4.5. Gene Editing and Overexpression
4.6. Histological GUS Assay
4.7. RNA-seq and Data Analysis
4.8. Determination of Stress-Related Physiological Index
4.9. Measurement of Na+ Content
4.10. RNA Extraction and Quantitative Real-Time PCR
4.11. Quantification and Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, G.H.; Xu, Q.; Zhu, X.D.; Qian, Q.; Xue, H.W. SHALLOT-LIKE1 is a KANADI transcription factor that modulates rice leaf rolling by regulating leaf abaxial cell development. Plant Cell 2009, 217, 19–35. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.Y.; Shen, A.; Xiong, W.; Sun, Q.L.; Luo, Q.; Song, T.; Li, Z.L.; Luan, W.J. Overexpression of OsHox32 results in pleiotropic effects on plant type architecture and leaf development in rice. Rice 2016, 94, 6. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.J.; Zhang, G.H.; Qian, Q.; Xue, H.W. Semi-rolled leaf1 encodes a putative glycosylphosphatidylinositol-anchored protein and modulates rice leaf rolling by regulating the formation of bulliform cells. Plant Physiol. 2012, 1591, 1488–1500. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.P.; Sun, X.H.; Zhang, Z.G.; Liu, P.; Wu, J.X.; Tian, C.J.; Qiu, J.L.; Lu, T.G. Leaf rolling controlled by the homeodomain leucine zipper class IV gene Roc5 in rice. Plant Physiol. 2011, 1561, 589–1602. [Google Scholar]
- Li, Y.; Yang, Y.; Liu, Y.; Li, D.; Zhao, Y.; Li, Z.; Liu, Y.; Jiang, D.; Li, J.; Zhou, H.; et al. Overexpression of OsAGO1b induces adaxially rolled leaves by affecting leaf abaxial sclerenchymatous cell development in rice. Rice 2019, 126, 60. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.; Cui, X.; Teng, S.; Kunnong, Z.; Wang, Y.; Chen, Z.; Sun, X.; Wu, J.; Ai, P.; Quick, W.P.; et al. HD-ZIP IV gene Roc8 regulates the size of bulliform cells and lignin content in rice. Plant Biotechnol. J. 2020, 182, 2559–2572. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, X.; Wang, L.; Xu, J.; Chen, J.; Fu, X.; Shen, N.; Nian, J.; Jiang, Z.; Hu, J.; et al. PHOTO-SENSITIVE LEAF ROLLING 1 encodes a polygalacturonase that modifies cell wall structure and drought tolerance in rice. New Phytol. 2021, 2298, 890–901. [Google Scholar] [CrossRef]
- Xu, P.; Ali, A.; Han, B.; Wu, X. Current advances in molecular basis and mechanisms regulating leaf morphology in rice. Front. Plant Sci. 2018, 91, 528. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.J.; Xu, J.; Qian, Q.; Zhang, G.H. Development of rice leaves: How histocytes modulate leaf polarity establishment. Rice Sci. 2020, 274, 468–479. [Google Scholar]
- Guo, W.; Chen, L.; Herrera-Estrella, L.; Cao, D.; Tran, L.P. Altering plant architecture to improve performance and resistance. Trends Plant Sci. 2020, 251, 1154–1170. [Google Scholar] [CrossRef] [PubMed]
- Springer, N. Shaping a better rice plant. Nat. Genet. 2010, 424, 475–476. [Google Scholar] [CrossRef]
- Lu, Z.; Yu, H.; Xiong, G.; Wang, J.; Jiao, Y.; Liu, G.; Jing, Y.; Meng, X.; Hu, X.; Qian, Q.; et al. Genome-wide binding analysis of the transcription activator ideal plant architecture1 reveals a complex network regulating rice plant architecture. Plant Cell 2013, 253, 3743–3759. [Google Scholar] [CrossRef]
- Wang, L.; Ming, L.; Liao, K.; Xia, C.; Sun, S.; Chang, Y.; Wang, H.; Fu, D.; Xu, C.; Wang, Z.; et al. Bract suppression regulated by the miR156/529-SPLs-NL1-PLA1 module is required for the transition from vegetative to reproductive branching in rice. Mol. Plant 2021, 141, 1168–1184. [Google Scholar] [CrossRef]
- Wang, J.; Zhou, L.; Shi, H.; Chern, M.; Yu, H.; Yi, H.; He, M.; Yin, J.; Zhu, X.; Li, Y.; et al. A single transcription factor promotes both yield and immunity in rice. Science 2018, 3611, 1026–1028. [Google Scholar] [CrossRef] [PubMed]
- Fujisawa, Y.; Kato, T.; Ohki, S.; Ishikawa, A.; Kitano, H.; Sasaki, T.; Asahi, T.; Iwasaki, Y. Suppression of the heterotrimeric G protein causes abnormal morphology, including dwarfism, in rice. Proc. Natl. Acad. Sci. USA 1999, 967, 7575–7580. [Google Scholar] [CrossRef] [PubMed]
- Jangam, A.P.; Pathak, R.R.; Raghuram, N. Microarray analysis of rice d1 (RGA1) mutant reveals the potential role of g-protein alpha subunit in regulating multiple abiotic stresses such as drought, salinity, heat, and cold. Front. Plant Sci. 2016, 71, 11. [Google Scholar] [CrossRef]
- Zhu, Y.; Li, T.; Xu, J.; Wang, J.; Wang, L.; Zou, W.; Zeng, D.; Zhu, L.; Chen, G.; Hu, J.; et al. Leaf width gene LW5/D1 affects plant architecture and yield in rice by regulating nitrogen utilization efficiency. Plant Physiol. Biochem. 2020, 1573, 359–369. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Zhang, C.; Zhang, G.; Fu, W.; Feng, B.; Chen, T.; Peng, S.; Tao, L.; Fu, G. Abscisic acid negatively modulates heat tolerance in rolled leaf rice by increasing leaf temperature and regulating energy homeostasis. Rice 2020, 131, 8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Chen, Z.; Cheng, J.; Lai, Y.; Wang, J.; Bao, Y.; Huang, J.; Zhang, H. QTL analysis of Na+ and K+ concentrations in roots and shoots under different levels of NaCl stress in rice (Oryza sativa L.). PLoS ONE 2012, 7, e51202. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Guo, Y. Unraveling salt stress signaling in plants. Integr. Plant Biol. 2018, 607, 796–804. [Google Scholar] [CrossRef] [PubMed]
- Deinlein, U.; Stephan, A.B.; Horie, T.; Luo, W.; Xu, G.; Schroeder, J.I. Plant salt-tolerance mechanisms. Trends Plant Sci. 2014, 193, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Tester, M.; Davenport, R. Na+ tolerance and Na+ transport in higher plants. Ann. Bot. 2003, 915, 503–527. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Salt and drought stress signal transduction in plants. Annu. Rev. Plant Biol. 2002, 532, 247–273. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Genetic analysis of plant salt tolerance using Arabidopsis. Plant Physiol. 2000, 1249, 941–948. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 714, 403–433. [Google Scholar] [CrossRef]
- Blumwald, E. Sodium transport and salt tolerance in plants. Curr. Opin. Cell Biol. 2000, 124, 431–434. [Google Scholar] [CrossRef]
- Long, L.M.; Patel, H.P.; Cory, W.C.; Stapleton, A.E. The maize epicuticular wax layer provides UV protection. Funct. Plant Biol. 2003, 307, 75–81. [Google Scholar] [CrossRef]
- Uppalapati, S.R.; Ishiga, Y.; Doraiswamy, V.; Bedair, M.; Mittal, S.; Chen, J.; Nakashima, J.; Tang, Y.; Tadege, M.; Ratet, P.; et al. Loss of abaxial leaf epicuticular wax in Medicago truncatula irg1/palm1 mutants results in reduced spore differentiation of anthracnose and nonhost rust pathogens. Plant Cell 2012, 243, 353–370. [Google Scholar] [CrossRef]
- Kan, Y.; Mu, X.R.; Zhang, H.; Gao, J.; Shan, J.X.; Ye, W.W.; Lin, H.X. TT2 controls rice thermotolerance through SCT1-dependent alteration of wax biosynthesis. Nat. Plants 2022, 85, 53–67. [Google Scholar] [CrossRef]
- Zhu, X.; Xiong, L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA 2013, 1101, 17790–17795. [Google Scholar] [CrossRef]
- Zhou, X.; Li, L.; Xiang, J.; Gao, G.; Xu, F.; Liu, A.; Zhang, X.; Peng, Y.; Chen, X.; Wan, X. OsGL1-3 is involved in cuticular wax biosynthesis and tolerance to water deficit in rice. PLoS ONE 2015, 10, e116676. [Google Scholar]
- Wang, Z.; Tian, X.; Zhao, Q.; Liu, Z.; Li, X.; Ren, Y.; Tang, J.; Fang, J.; Xu, Q.; Bu, Q. The E3 ligase DROUGHT HYPERSENSITIVE negatively regulates cuticular wax biosynthesis by promoting the degradation of transcription factor ROC4 in rice. Plant Cell 2018, 302, 228–244. [Google Scholar] [CrossRef] [PubMed]
- Wei, J.; Choi, H.; Jin, P.; Wu, Y.; Yoon, J.; Lee, Y.S.; Quan, T.; An, G. GL2-type homeobox gene Roc4 in rice promotes flowering time preferentially under long days by repressing Ghd7. Plant Sci. 2016, 2521, 133–143. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wan, L.; Zhang, L.; Zhang, Z.; Zhang, H.; Quan, R.; Zhou, S.; Huang, R. An ethylene response factor OsWR1 responsive to drought stress transcriptionally activates wax synthesis related genes and increases wax production in rice. Plant Mol. Biol. 2012, 782, 275–288. [Google Scholar] [CrossRef]
- Jenks, M.A.; Rashotte, A.M.; Tuttle, H.A.; Feldmann, K.A. Mutants in Arabidopsis thaliana altered in epicuticular wax and leaf morphology. Plant Physiol. 1996, 1103, 377–385. [Google Scholar] [CrossRef][Green Version]
- Chen, X.; Goodwin, S.M.; Boroff, V.L.; Liu, X.; Jenks, M.A. Cloning and characterization of the WAX2 gene of Arabidopsis involved in cuticle membrane and wax production. Plant Cell 2003, 151, 1170–1185. [Google Scholar] [CrossRef]
- Horton, P. Prospects for crop improvement through the genetic manipulation of photosynthesis: Morphological and biochemical aspects of light capture. J. Exp. Bot. 2000, 51, 475–485. [Google Scholar] [CrossRef]
- Sage, R.F. A model describing the regulation of ribulose-15, -bisphosphate carboxylase, electron transport, and triose phosphate use in response to light intensity and CO2 in C3 plants. Plant Physiol. 1990, 941, 1728–1734. [Google Scholar] [CrossRef]
- Qiu, Z.; Chen, D.; He, L.; Zhang, S.; Yang, Z.; Zhang, Y.; Wang, Z.; Ren, D.; Qian, Q.; Guo, L.; et al. The rice white green leaf 2 gene causes defects in chloroplast development and affects the plastid ribosomal protein S9. Rice 2018, 113, 9. [Google Scholar] [CrossRef]
- Schroeder, J.I.; Delhaize, E.; Frommer, W.B.; Guerinot, M.L.; Harrison, M.J.; Herrera-Estrella, L.; Horie, T.; Kochian, L.V.; Munns, R.; Nishizawa, N.K.; et al. Using membrane transporters to improve crops for sustainable food production. Nature 2013, 497, 60–66. [Google Scholar] [CrossRef]
- Dumanović, J.; Nepovimova, E.; Natić, M.; Kuča, K.; Jaćević, V. The significance of reactive oxygen species and antioxidant defense system in plants: A concise overview. Front. Plant Sci. 2020, 115, 52969. [Google Scholar] [CrossRef] [PubMed]
- Hasanuzzaman, M.; Bhuyan, M.; Zulfiqar, F.; Raza, A.; Mohsin, S.M.; Mahmud, J.A.; Fujita, M.; Fotopoulos, V. Reactive oxygen species and antioxidant defense in plants under abiotic stress: Revisiting the crucial role of a universal defense regulator. Antioxidants 2020, 9, 681. [Google Scholar] [CrossRef] [PubMed]
- Gill, S.S.; Tuteja, N. Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 2010, 489, 909–930. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Du, H.; Ning, J.; Ye, H.; Xiong, L. Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol. Biol. 2009, 704, 443–456. [Google Scholar] [CrossRef]
- Xu, J.; Wang, J.J.; Xue, H.W.; Zhang, G.H. Leaf direction: Lamina joint development and environmental responses. Plant Cell Environ. 2021, 442, 2441–2454. [Google Scholar] [CrossRef]
- Hong, Z.; Ueguchi-Tanaka, M.; Shimizu-Sato, S.; Inukai, Y.; Fujioka, S.; Shimada, Y.; Takatsuto, S.; Agetsuma, M.; Yoshida, S.; Watanabe, Y.; et al. Loss-of-function of a rice brassinosteroid biosynthetic enzyme, C-6 oxidase, prevents the organized arrangement and polar elongation of cells in the leaves and stem. Plant J. 2002, 324, 495–508. [Google Scholar] [CrossRef]
- Shen, C.; Wang, S.; Zhang, S.; Xu, Y.; Qian, Q.; Qi, Y.; Jiang, D. OsARF16, a transcription factor, is required for auxin and phosphate starvation response in rice (Oryza sativa L.). Plant Cell Environ. 2013, 366, 607–620. [Google Scholar] [CrossRef]
- Huang, G.; Hu, H.; van de Meene, A.; Zhang, J.; Dong, L.; Zheng, S.; Zhang, F.; Betts, N.S.; Liang, W.; Bennett, M.J.; et al. AUXIN RESPONSE FACTORS 6 and 17 control the flag leaf angle in rice by regulating secondary cell wall biosynthesis of lamina joints. Plant Cell 2021, 333, 3120–3133. [Google Scholar] [CrossRef]
- Toriba, T.; Suzaki, T.; Yamaguchi, T.; Ohmori, Y.; Tsukaya, H.; Hirano, H.Y. Distinct regulation of adaxial-abaxial polarity in anther patterning in rice. Plant Cell 2010, 221, 1452–1462. [Google Scholar] [CrossRef]
- Hasson, A.; Blein, T.; Laufs, P. Leaving the meristem behind: The genetic and molecular control of leaf patterning and morphogenesis. C. R. Biol. 2010, 3333, 350–360. [Google Scholar] [CrossRef]
- Shi, Z.; Wang, J.; Wan, X.; Shen, G.; Wang, X.; Zhang, J. Over-expression of rice OsAGO7 gene induces upward curling of the leaf blade that enhanced erect-leaf habit. Planta 2007, 2269, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Qin, B.X.; Tang, D.; Huang, J.; Li, M.; Wu, X.R.; Lu, L.L.; Wang, K.J.; Yu, H.X.; Chen, J.M.; Gu, M.H.; et al. Rice OsGL1-1 is involved in leaf cuticular wax and cuticle membrane. Mol. Plant 2011, 49, 985–995. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Ni, E.; Yang, J.; Zhou, H.; Liang, H.; Li, J.; Jiang, D.; Wang, Z.; Liu, Z.; Zhuang, C. Rice OsGL1-6 is involved in leaf cuticular wax accumulation and drought resistance. PLoS ONE 2013, 8, e65139. [Google Scholar] [CrossRef] [PubMed]
- Ali, A.; Wu, T.; Zhang, H.; Xu, P.; Zafar, S.A.; Liao, Y.; Chen, X.; Zhou, H.; Liu, Y.; Wang, W.; et al. A putative SUBTILISIN-LIKE SERINE PROTEASE 1 (SUBSrP1) regulates anther cuticle biosynthesis and panicle development in rice. J. Adv. Res. 2022. [Google Scholar] [CrossRef]
- Wang, X.; Guan, Y.; Zhang, D.; Dong, X.; Tian, L.; Qu, L.Q. A β-Ketoacyl-CoA Synthase Is Involved in Rice Leaf Cuticular Wax Synthesis and Requires a CER2-LIKE Protein as a Cofactor. Plant Physiol. 2017, 1739, 944–955. [Google Scholar] [CrossRef]
- Chen, H.; Zhang, Z.; Ni, E.; Lin, J.; Peng, G.; Huang, J.; Zhu, L.; Deng, L.; Yang, F.; Luo, Q.; et al. HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 2020, 2252, 2077–2093. [Google Scholar] [CrossRef]
- Yu, B.; Liu, L.; Wang, T. Deficiency of very long chain alkanes biosynthesis causes humidity-sensitive male sterility via affecting pollen adhesion and hydration in rice. Plant Cell Environ. 2019, 423, 3340–3354. [Google Scholar] [CrossRef]
- Maurel, C.; Boursiac, Y.; Luu, D.T.; Santoni, V.; Shahzad, Z.; Verdoucq, L. Aquaporins in Plants. Physiol. Rev. 2015, 951, 1321–1358. [Google Scholar] [CrossRef]
- Lian, H.L.; Yu, X.; Lane, D.; Sun, W.N.; Tang, Z.C.; Su, W.A. Upland rice and lowland rice exhibited different PIP expression under water deficit and ABA treatment. Cell Res. 2006, 166, 651–660. [Google Scholar] [CrossRef]
- Liu, C.; Fukumoto, T.; Matsumoto, T.; Gena, P.; Frascaria, D.; Kaneko, T.; Katsuhara, M.; Zhong, S.; Sun, X.; Zhu, Y.; et al. Aquaporin OsPIP1;1 promotes rice salt resistance and seed germination. Plant Physiol. Biochem. 2013, 631, 151–158. [Google Scholar] [CrossRef]
- Ren, Z.H.; Gao, J.P.; Li, L.G.; Cai, X.L.; Huang, W.; Chao, D.Y.; Zhu, M.Z.; Wang, Z.Y.; Luan, S.; Lin, H.X. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat. Genet. 2005, 371, 1141–1146. [Google Scholar] [CrossRef] [PubMed]
- Sunarpi; Horie, T.; Motoda, J.; Kubo, M.; Yang, H.; Yoda, K.; Horie, R.; Chan, W.Y.; Leung, H.Y.; Hattori, K.; et al. Enhanced salt tolerance mediated by AtHKT1 transporter-induced Na unloading from xylem vessels to xylem parenchyma cells. Plant J. 2005, 449, 928–938. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, J.I.; Ward, J.M.; Gassmann, W. Perspectives on the physiology and structure of inward-rectifying K+ channels in higher plants: Biophysical implications for K+ uptake. Annu. Rev. Biophys. Biomol. Struct. 1994, 234, 441–471. [Google Scholar] [CrossRef] [PubMed]
- Garciadeblás, B.; Senn, M.E.; Bañuelos, M.A.; Rodríguez-Navarro, A. Sodium transport and HKT transporters: The rice model. Plant J. 2003, 347, 788–801. [Google Scholar] [CrossRef] [PubMed]
- Shepherd, T.; Wynne Griffiths, D. The effects of stress on plant cuticular waxes. New Phytol. 2006, 1714, 469–499. [Google Scholar] [CrossRef]
- Lewandowska, M.; Keyl, A.; Feussner, I. Wax biosynthesis in response to danger: Its regulation upon abiotic and biotic stress. New Phytol. 2020, 2276, 698–713. [Google Scholar] [CrossRef] [PubMed]
- Sartory, D.P.; Grobbelaar, J.U. Extraction of chlorophyll a from freshwater phytoplankton for spectrophotometric analysis. Hydrobiologia 1984, 1141, 177–187. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y.; et al. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 81, 274–1284. [Google Scholar] [CrossRef]
- Jefferson, R.A.; Kavanagh, T.A.; Bevan, M.W. GUS fusions: Beta-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. Embo J. 1987, 63, 3901–3907. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Liu, Y.; Hu, S.; Xu, J.; Nian, J.; Cao, X.; Chen, M.; Cen, J.; Liu, X.; Zhang, Z.; et al. LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice. Int. J. Mol. Sci. 2022, 23, 8818. https://doi.org/10.3390/ijms23158818
Wang J, Liu Y, Hu S, Xu J, Nian J, Cao X, Chen M, Cen J, Liu X, Zhang Z, et al. LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice. International Journal of Molecular Sciences. 2022; 23(15):8818. https://doi.org/10.3390/ijms23158818
Chicago/Turabian StyleWang, Jiajia, Yiting Liu, Songping Hu, Jing Xu, Jinqiang Nian, Xiaoping Cao, Minmin Chen, Jiangsu Cen, Xiong Liu, Zhihai Zhang, and et al. 2022. "LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice" International Journal of Molecular Sciences 23, no. 15: 8818. https://doi.org/10.3390/ijms23158818
APA StyleWang, J., Liu, Y., Hu, S., Xu, J., Nian, J., Cao, X., Chen, M., Cen, J., Liu, X., Zhang, Z., Liu, D., Zhu, L., Hu, J., Ren, D., Gao, Z., Shen, L., Dong, G., Zhang, Q., Li, Q., ... Zhang, G. (2022). LEAF TIP RUMPLED 1 Regulates Leaf Morphology and Salt Tolerance in Rice. International Journal of Molecular Sciences, 23(15), 8818. https://doi.org/10.3390/ijms23158818









