New Advances in Using Virus-like Particles and Related Technologies for Eukaryotic Genome Editing Delivery
Abstract
1. A Brief Introduction to Designer Endonucleases
2. Background Knowledge for Developing VLPs as Genome Editing Delivery Vehicles
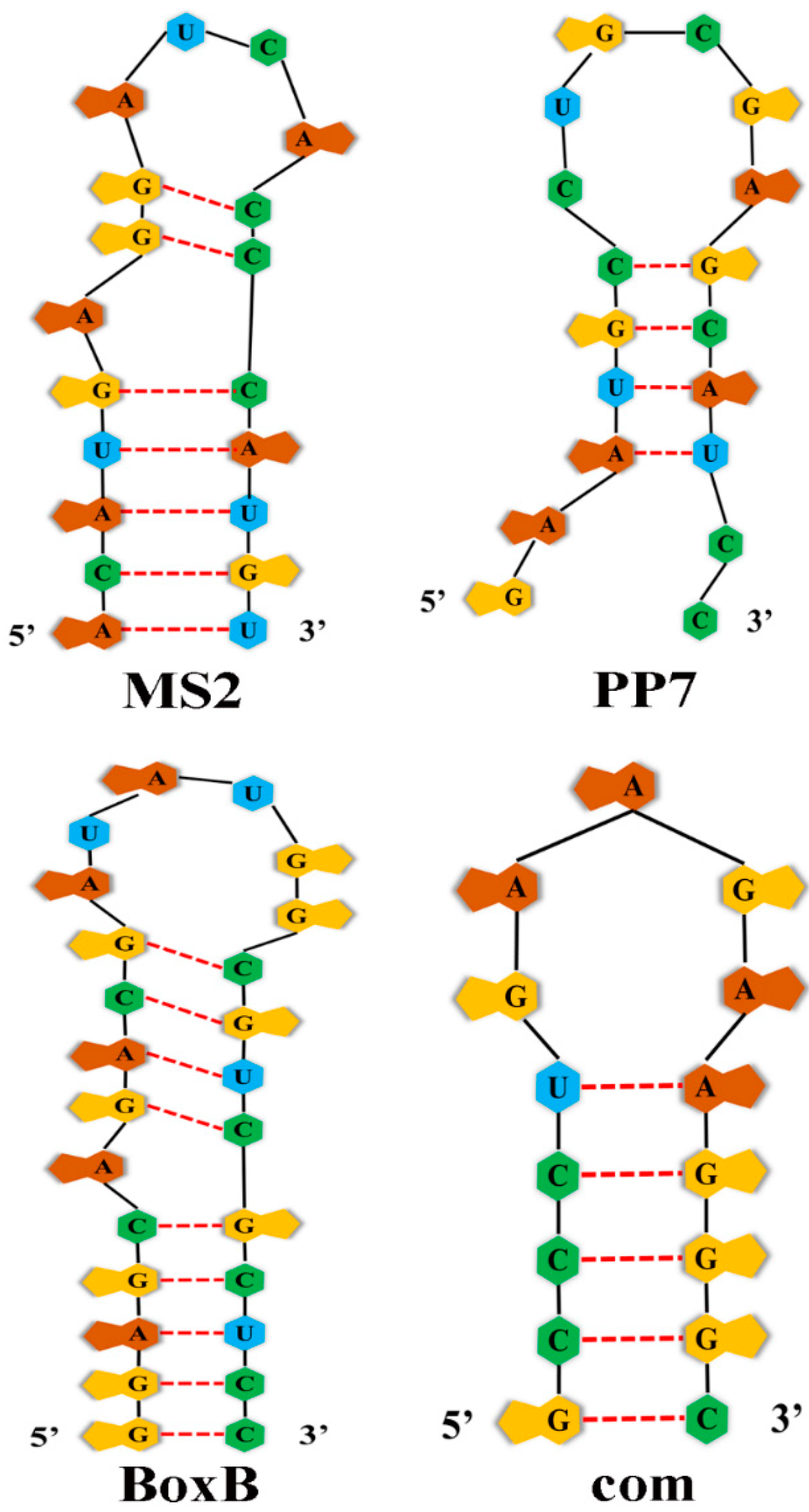
RNA Aptamers and Aptamer-Binding Proteins (ABPs)
3. Recently Developed VLPs as Safe Genome Editing Delivery Vehicles
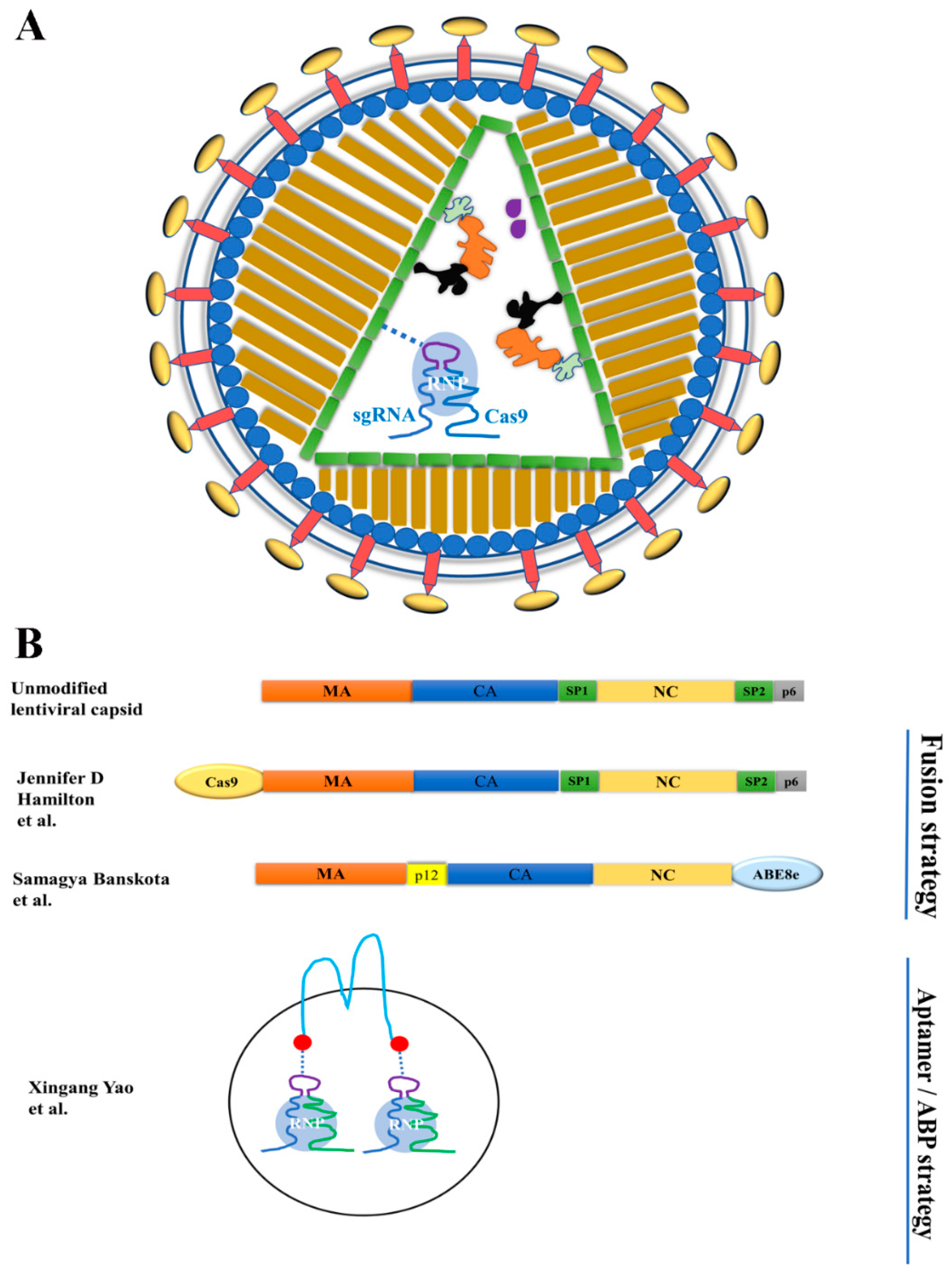
3.1. Using VLPs to Deliver Proteins or Ribonucleoproteinss
| Reference | Mechanism for Nuclease Recruitment | Capsid Type | Editing Effectors Delivered | Experiment Stage | Gene/Tissue | |
|---|---|---|---|---|---|---|
| The fusion strategy | Cai et al. [72] | Fusing editing effector to the N-terminus of Gag | LV | ZFN and TALEN | In vitro | GFP/CCR5/AAVS1—cell |
| Choi et al. [73] | Fusing Cas9 protein to the N-terminus of Gag | LV | SpCas9 | In vitro | LTR/CD4/CCR5—cell | |
| Mangeot et al. [74] | Fusing Cas9 to the C-terminus of MLV Gag | MLV | SpCas9 | In vivo | MYD88/DDX3/GFP/Hpd/Fto/Tyr/LoxP—cell | |
| Gee et al. [75] | Fusing FKBP12 to Gag, fusing FRB to SpCas9. FKBP12/AP21967/FRB interaction brings SpCas9 to Gag | LV | SpCas9 | In vivo | DMD—cell and mouse | |
| Indikova et al. [76] | Fusing Cas9 to the C-terminus of Vpr | LV | SpCas9 | In vitro | GFP/EMX1/FANCF/HEKs1/HEKs3—cell | |
| Hamilton et al. [10] | Fusing Cas9 to N-terminal Gag structural protein | LV | SpCas9 | In vitro | B2M/TRAC—cell | |
| Banskota et al. [12] | Fusing ABE8e to C-terminus of Gag | MLV | SpCas9 and ABE | In vivo | BCL11A/COL7A1—cell Pcsk 9/Dnmt 1—mouse | |
| The Aptamer/ABP Strategy | Lyu et al. [63,77] | Forming a three-component complex: NC-Com/aptamer-sgRNA/Cas9 protein | LV | SpCas9 and ABE | In vitro | IL2RG/HBB—cell |
| Lu et al. [78] | SaCas9 | |||||
| Yao et al. [5] | Forming a three-component complex: CD63-Com/aptamer-sgRNA/Cas9 protein | Exosome | SaCas9 SpCas9 ABE | In vivo | DMD/IL2RG/HBB—cell DMD—mouse |
3.1.1. VLP-Mediated Nuclease Delivery Using the Fusion Strategy
VLPs for Co-Delivery of Cas9 RNPs and LV Genomic RNA
Recent Applications of Previously Reported VLPs
VLPs for Delivering Base Editors
3.1.2. Delivering Cas9 and Base Editor RNPs by Exosomes Using the ABP/Aptamer Interaction Strategy
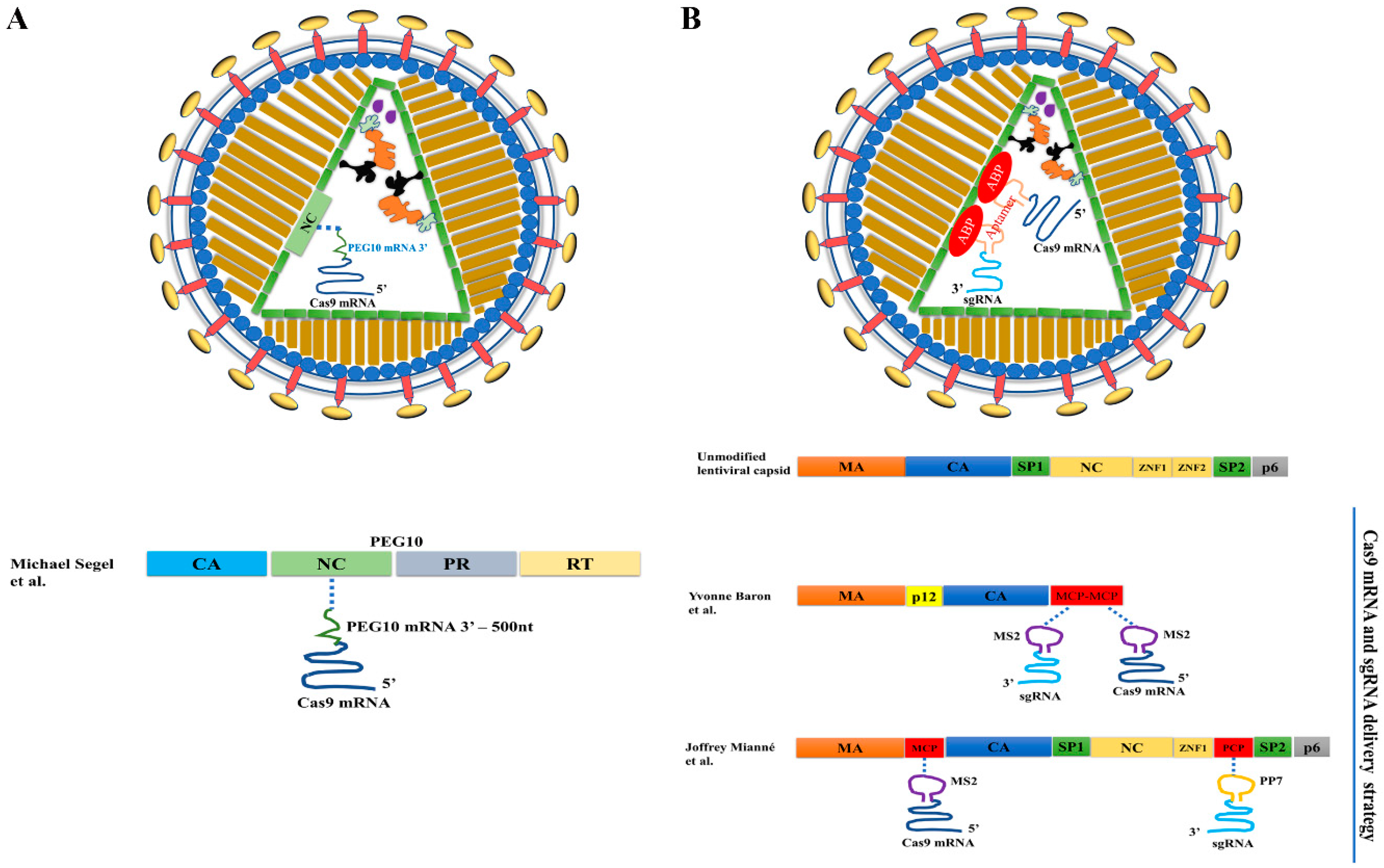
3.2. VLPs for Delivering Cas9 mRNAs
3.3. VLPs for Co-Delivery of Cas9 mRNA and sgRNA
| Reference | Virus Type | Capsid Modification | RNA Package | Copy Number | |
|---|---|---|---|---|---|
| mRNA | Mock et al. [83] | LV | Not modified | TALEN mRNA | 2 copies |
| Prel et al. [85] | LV | MCP replaced the second zinc finger domain of NC | SpCas9 mRNA | ~6 copies | |
| Lu et al. [64] | LV | MCP inserted after the second zinc finger domain of NC | SaCas9 mRNA | 50~100 copies | |
| Lindel et al. [84] | Foamy Viruses | Not modified | SpCas9 mRNA | 60 copies | |
| mRNA or mRNA & sgRNA | Segel et al. [11] | Endogenous retrotransposon | No modification of endogenous Gag homolog | SpCas9 mRNA (and sgRNA) | Not available |
| mRNA & sgRNA | Knopp et al. [14] | Murine Leukemia Virus or Rous sarcoma virus | Two copies of MCP replaced NC | SpCas9 mRNA and sgRNA | Not available |
| Baron et al. [15] | |||||
| Mianné et al. [16] | LV | MCP inserted the N terminus of CA and PCP replaced ZNF2 | SpCas9 mRNA and sgRNA | 1.43 copies | |
| hybrid mRNA & sgRNA | Ling et al. [6] | LV | MCP inserted the N terminus of Gag | SpCas9 mRNA and sgRNA | 3~4 copies |
| Yin et al. [7] | |||||
| Yadav et al. [13] | LV | MCP inserted after the second zinc finger domain of NC | SaCas9 mRNA and sgRNA | Not available |
3.3.1. Adding Aptamers to Cas9 mRNA and sgRNA for Co-Delivery
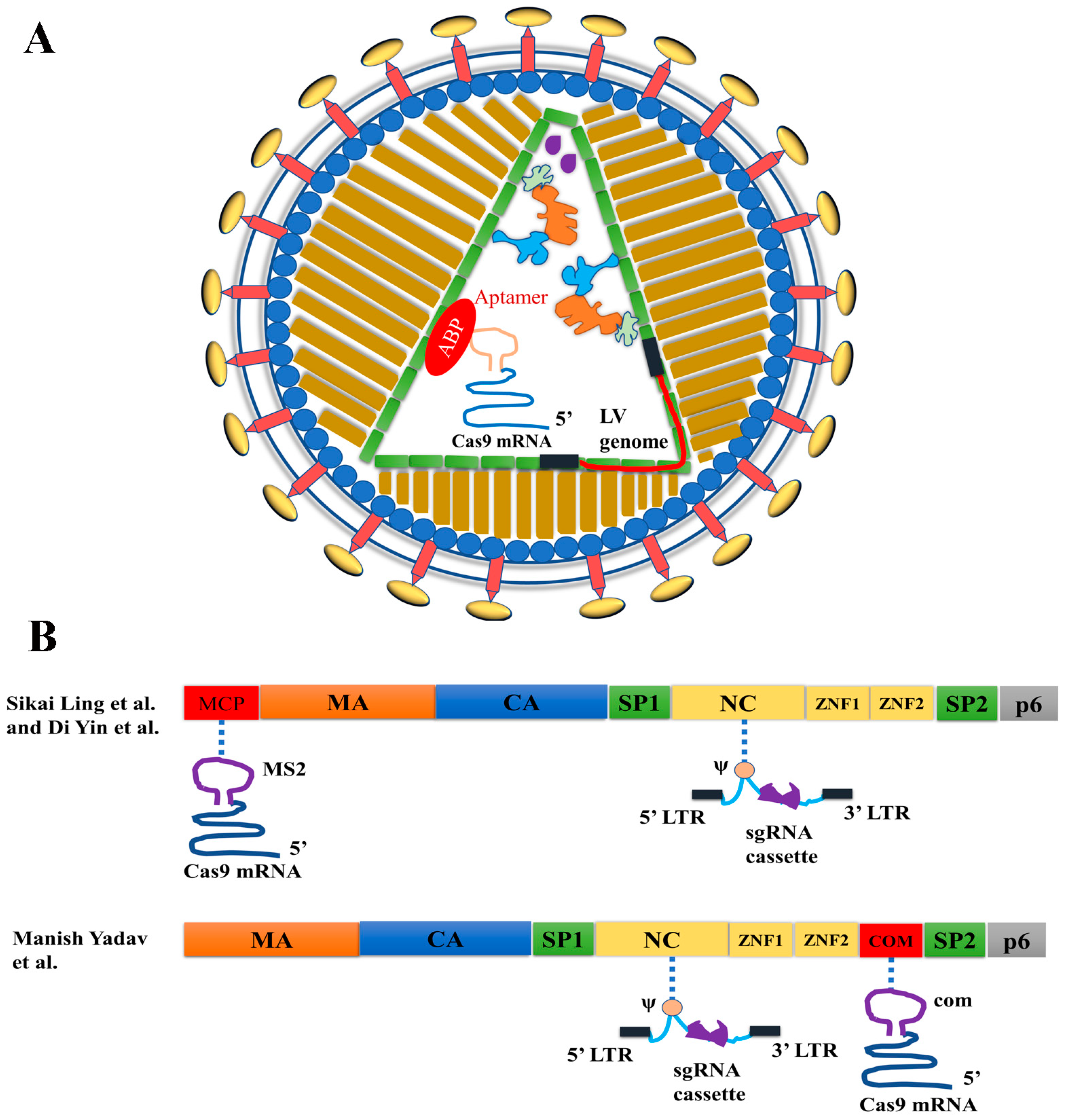
3.3.2. VLP and LV Hybrid Particles for Cas9 mRNA and sgRNA Co-Delivery
4. Applications of VLPs in Genome Editing
4.1. In Vitro Applications in Clinically Relevant Blood Cells
4.2. In Vivo Applications of VLPs in Mice
5. Summary
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| cDNA | Complementary DNA |
| RNA | Ribonucleic acid |
| gRNA | Guide RNA |
| sgRNA mRNA | Single guide RNA Messenger RNA |
| ZFN | Zinc finger endonuclease |
| TALEN | Transcription activator-like effector nuclease |
| CRISPR/Cas | Clustered Regularly Interspaced Short Palindromic Repeats/CRISPR-associated/CRISPR-associated |
| PAM | Protospacer adjacent motif |
| ABE | Adenine base editor |
| LV | Lentiviral vector |
| AAV | Adeno-associated virus-derived vector |
| IDLV | Integration-defective lentiviral vector |
| FMLV | Friend murine leukemia virus |
| HIV-1 | Human immunodeficiency virus type 1 |
| Gag | Group-specific antigen |
| MA | Matrix protein |
| CA | Capsid protein |
| NC | Nucleocapsid protein |
| RNP | Ribonucleoprotein |
| INDEL | Insertion and deletion |
| VLP | Virus-like particle |
| ABP | Aptamer binding protein |
| MCP | MS2 coating protein |
| PCP | PP7 coating protein |
References
- Woolf, T.M. Therapeutic repair of mutated nucleic acid sequences. Nat. Biotechnol. 1998, 16, 341–344. [Google Scholar] [CrossRef]
- Fu, Y.; Foden, J.A.; Khayter, C.; Maeder, M.L.; Reyon, D.; Joung, J.K.; Sander, J.D. High-frequency off-target mutagenesis induced by CRISPR-Cas nucleases in human cells. Nat. Biotechnol. 2013, 31, 822–826. [Google Scholar] [CrossRef]
- Nelson, C.E.; Wu, Y.; Gemberling, M.P.; Oliver, M.L.; Waller, M.A.; Bohning, J.D.; Robinson-Hamm, J.N.; Bulaklak, K.; Castellanos Rivera, R.M.; Collier, J.H.; et al. Long-term evaluation of AAV-CRISPR genome editing for Duchenne muscular dystrophy. Nat. Med. 2019, 25, 427–432. [Google Scholar] [CrossRef]
- Lyu, P.; Wang, L.; Lu, B. Virus-Like Particle Mediated CRISPR/Cas9 Delivery for Efficient and Safe Genome Editing. Life 2020, 10, 366. [Google Scholar] [CrossRef]
- Yao, X.; Lyu, P.; Yoo, K.; Yadav, M.K.; Singh, R.; Atala, A.; Lu, B. Engineered extracellular vesicles as versatile ribonucleoprotein delivery vehicles for efficient and safe CRISPR genome editing. J. Extracell. Vesicles 2021, 10, e12076. [Google Scholar] [CrossRef]
- Ling, S.; Yang, S.; Hu, X.; Yin, D.; Dai, Y.; Qian, X.; Wang, D.; Pan, X.; Hong, J.; Sun, X.; et al. Lentiviral delivery of co-packaged Cas9 mRNA and a Vegfa-targeting guide RNA prevents wet age-related macular degeneration in mice. Nat. Biomed. Eng. 2021, 5, 144–156. [Google Scholar] [CrossRef] [PubMed]
- Yin, D.; Ling, S.; Wang, D.; Dai, Y.; Jiang, H.; Zhou, X.; Paludan, S.R.; Hong, J.; Cai, Y. Targeting herpes simplex virus with CRISPR-Cas9 cures herpetic stromal keratitis in mice. Nat. Biotechnol. 2021, 39, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Fichter, C.; Aggarwal, A.; Wong, A.K.H.; McAllery, S.; Mathivanan, V.; Hao, B.; MacRae, H.; Churchill, M.J.; Gorry, P.R.; Roche, M.; et al. Modular Lentiviral Vectors for Highly Efficient Transgene Expression in Resting Immune Cells. Viruses 2021, 13, 1170. [Google Scholar] [CrossRef]
- Gutierrez-Guerrero, A.; Abrey Recalde, M.J.; Mangeot, P.E.; Costa, C.; Bernadin, O.; Perian, S.; Fusil, F.; Froment, G.; Martinez-Turtos, A.; Krug, A.; et al. Baboon Envelope Pseudotyped “Nanoblades” Carrying Cas9/gRNA Complexes Allow Efficient Genome Editing in Human T, B, and CD34(+) Cells and Knock-in of AAV6-Encoded Donor DNA in CD34(+) Cells. Front. Genome Ed. 2021, 3, 604371. [Google Scholar] [CrossRef] [PubMed]
- Hamilton, J.R.; Tsuchida, C.A.; Nguyen, D.N.; Shy, B.R.; McGarrigle, E.R.; Sandoval Espinoza, C.R.; Carr, D.; Blaeschke, F.; Marson, A.; Doudna, J.A. Targeted delivery of CRISPR-Cas9 and transgenes enables complex immune cell engineering. Cell Rep. 2021, 35, 109207. [Google Scholar] [CrossRef]
- Segel, M.; Lash, B.; Song, J.; Ladha, A.; Liu, C.C.; Jin, X.; Mekhedov, S.L.; Macrae, R.K.; Koonin, E.V.; Zhang, F. Mammalian retrovirus-like protein PEG10 packages its own mRNA and can be pseudotyped for mRNA delivery. Science 2021, 373, 882–889. [Google Scholar] [CrossRef] [PubMed]
- Banskota, S.; Raguram, A.; Suh, S.; Du, S.W.; Davis, J.R.; Choi, E.H.; Wang, X.; Nielsen, S.C.; Newby, G.A.; Randolph, P.B.; et al. Engineered virus-like particles for efficient in vivo delivery of therapeutic proteins. Cell 2022, 185, 250–265.e216. [Google Scholar] [CrossRef] [PubMed]
- Yadav, M.K.; Atala, A.; Lu, B. Developing all-in-one virus-like particles for Cas9 mRNA/single guide RNA co-delivery and aptamer-containing lentiviral vectors for improved gene expression. BioRxiv 2022. [Google Scholar] [CrossRef] [PubMed]
- Baron, Y.; Sens, J.; Lange, L.; Nassauer, L.; Klatt, D.; Hoffmann, D.; Kleppa, M.J.; Barbosa, P.V.; Keisker, M.; Steinberg, V.; et al. Improved alpharetrovirus-based Gag.MS2 particles for efficient and transient delivery of CRISPR-Cas9 into target cells. Mol. Ther. Nucleic Acids 2022, 27, 810–823. [Google Scholar] [CrossRef]
- Knopp, Y.; Geis, F.K.; Heckl, D.; Horn, S.; Neumann, T.; Kuehle, J.; Meyer, J.; Fehse, B.; Baum, C.; Morgan, M.; et al. Transient Retrovirus-Based CRISPR/Cas9 All-in-One Particles for Efficient, Targeted Gene Knockout. Mol. Ther. Nucleic Acids 2018, 13, 256–274. [Google Scholar] [CrossRef] [PubMed]
- Mianne, J.; Nasri, A.; Van, C.N.; Bourguignon, C.; Fieldes, M.; Ahmed, E.; Duthoit, C.; Martin, N.; Parrinello, H.; Louis, A.; et al. CRISPR/Cas9-mediated gene knockout and interallelic gene conversion in human induced pluripotent stem cells using non-integrative bacteriophage-chimeric retrovirus-like particles. BMC Biol. 2022, 20, 8. [Google Scholar] [CrossRef]
- Kim, Y.G.; Cha, J.; Chandrasegaran, S. Hybrid restriction enzymes: Zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. USA 1996, 93, 1156. [Google Scholar] [CrossRef]
- Li, T.; Huang, S.; Jiang, W.Z.; Wright, D.; Spalding, M.H.; Weeks, D.P.; Yang, B. TAL nucleases (TALNs): Hybrid proteins composed of TAL effectors and FokI DNA-cleavage domain. Nucleic Acids Res. 2011, 39, 359–372. [Google Scholar] [CrossRef]
- Moscou, M.J.; Bogdanove, A.J. A simple cipher governs DNA recognition by TAL effectors. Science 2009, 326, 1501. [Google Scholar] [CrossRef]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Kim, S.; Kim, J.M.; Kim, J.S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat. Biotechnol. 2013, 31, 230–232. [Google Scholar] [CrossRef] [PubMed]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; East, A.; Cheng, A.; Lin, S.; Ma, E.; Doudna, J. RNA-programmed genome editing in human cells. eLife 2013, 2, e00471. [Google Scholar] [CrossRef]
- Barrangou, R.; Fremaux, C.; Deveau, H.; Richards, M.; Boyaval, P.; Moineau, S.; Romero, D.A.; Horvath, P. CRISPR provides acquired resistance against viruses in prokaryotes. Science 2007, 315, 1709–1712. [Google Scholar] [CrossRef]
- Koonin, E.V.; Makarova, K.S.; Zhang, F. Diversity, classification and evolution of CRISPR-Cas systems. Curr. Opin. Microbiol. 2017, 37, 67–78. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. Genome editing. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Doench, J.G.; Hartenian, E.; Graham, D.B.; Tothova, Z.; Hegde, M.; Smith, I.; Sullender, M.; Ebert, B.L.; Xavier, R.J.; Root, D.E. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat. Biotechnol. 2014, 32, 1262–1267. [Google Scholar] [CrossRef]
- Nelson, C.E.; Hakim, C.H.; Ousterout, D.G.; Thakore, P.I.; Moreb, E.A.; Castellanos Rivera, R.M.; Madhavan, S.; Pan, X.; Ran, F.A.; Yan, W.X.; et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science 2016, 351, 403–407. [Google Scholar] [CrossRef]
- Tabebordbar, M.; Zhu, K.; Cheng, J.K.W.; Chew, W.L.; Widrick, J.J.; Yan, W.X.; Maesner, C.; Wu, E.Y.; Xiao, R.; Ran, F.A.; et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science 2016, 351, 407–411. [Google Scholar] [CrossRef]
- Long, C.; Amoasii, L.; Mireault, A.A.; McAnally, J.R.; Li, H.; Sanchez-Ortiz, E.; Bhattacharyya, S.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science 2016, 351, 400–403. [Google Scholar] [CrossRef] [PubMed]
- Mirgayazova, R.; Khadiullina, R.; Chasov, V.; Mingaleeva, R.; Miftakhova, R.; Rizvanov, A.; Bulatov, E. Therapeutic Editing of the TP53 Gene: Is CRISPR/Cas9 an Option? Genes 2020, 11, 704. [Google Scholar] [CrossRef] [PubMed]
- Gillmore, J.D.; Gane, E.; Taubel, J.; Kao, J.; Fontana, M.; Maitland, M.L.; Seitzer, J.; O’Connell, D.; Walsh, K.R.; Wood, K.; et al. CRISPR-Cas9 In Vivo Gene Editing for Transthyretin Amyloidosis. N. Engl. J. Med. 2021, 385, 493–502. [Google Scholar] [CrossRef]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef] [PubMed]
- Kaiser, J. CRISPR Helps a Blind Woman See, But Doesn’t Help All Patients. Available online: https://www.science.org/content/article/crispr-helps-blind-woman-see-doesn-t-help-all-patients (accessed on 29 September 2021).
- Stadtmauer, E.A.; Fraietta, J.A.; Davis, M.M.; Cohen, A.D.; Weber, K.L.; Lancaster, E.; Mangan, P.A.; Kulikovskaya, I.; Gupta, M.; Chen, F.; et al. CRISPR-engineered T cells in patients with refractory cancer. Science 2020, 367, eaba7365. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Bortesi, L.; Fischer, R. The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 2015, 33, 41–52. [Google Scholar] [CrossRef]
- Zhang, Z.T.; Jiménez-Bonilla, P.; Seo, S.O.; Lu, T.; Jin, Y.S.; Blaschek, H.P.; Wang, Y. Bacterial Genome Editing with CRISPR-Cas9: Taking Clostridium beijerinckii as an Example. Methods Mol. Biol. 2018, 1772, 297–325. [Google Scholar] [CrossRef]
- Qi, L.S.; Larson, M.H.; Gilbert, L.A.; Doudna, J.A.; Weissman, J.S.; Arkin, A.P.; Lim, W.A. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 2013, 152, 1173–1183. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Larson, M.H.; Morsut, L.; Liu, Z.; Brar, G.A.; Torres, S.E.; Stern-Ginossar, N.; Brandman, O.; Whitehead, E.H.; Doudna, J.A.; et al. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 2013, 154, 442–451. [Google Scholar] [CrossRef]
- Bikard, D.; Jiang, W.; Samai, P.; Hochschild, A.; Zhang, F.; Marraffini, L.A. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013, 41, 7429–7437. [Google Scholar] [CrossRef]
- Kim, S.; Kim, D.; Cho, S.W.; Kim, J.; Kim, J.S. Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 2014, 24, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Rees, H.A.; Komor, A.C.; Yeh, W.H.; Caetano-Lopes, J.; Warman, M.; Edge, A.S.B.; Liu, D.R. Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nat. Commun. 2017, 8, 15790. [Google Scholar] [CrossRef]
- Hsu, P.D.; Scott, D.A.; Weinstein, J.A.; Ran, F.A.; Konermann, S.; Agarwala, V.; Li, Y.; Fine, E.J.; Wu, X.; Shalem, O.; et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 2013, 31, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Shalem, O.; Sanjana, N.E.; Hartenian, E.; Shi, X.; Scott, D.A.; Mikkelson, T.; Heckl, D.; Ebert, B.L.; Root, D.E.; Doench, J.G.; et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 2014, 343, 84–87. [Google Scholar] [CrossRef] [PubMed]
- Cradick, T.J.; Fine, E.J.; Antico, C.J.; Bao, G. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013, 41, 9584–9592. [Google Scholar] [CrossRef]
- Maetzig, T.; Galla, M.; Baum, C.; Schambach, A. Gammaretroviral vectors: Biology, technology and application. Viruses 2011, 3, 677–713. [Google Scholar] [CrossRef]
- Kartikeyan, S.; Bharmal, R.N.; Tiwari, R.P.; Bisen, P.S. HIV and AIDS: Basic Elements and Priorities; Springer: Dordrecht, The Netherlands, 2007; pp. XIV–418. [Google Scholar]
- Ellington, A.D.; Szostak, J.W. In vitro selection of RNA molecules that bind specific ligands. Nature 1990, 346, 818–822. [Google Scholar] [CrossRef]
- Keefe, A.D.; Pai, S.; Ellington, A. Aptamers as therapeutics. Nat. Rev. Drug Discov. 2010, 9, 537–550. [Google Scholar] [CrossRef]
- Fouts, D.E.; True, H.L.; Celander, D.W. Functional recognition of fragmented operator sites by R17/MS2 coat protein, a translational repressor. Nucleic Acids Res. 1997, 25, 4464–4473. [Google Scholar] [CrossRef][Green Version]
- Lim, F.; Downey, T.P.; Peabody, D.S. Translational repression and specific RNA binding by the coat protein of the Pseudomonas phage PP7. J. Biol. Chem. 2001, 276, 22507–22513. [Google Scholar] [CrossRef] [PubMed]
- Wulczyn, F.G.; Kahmann, R. Translational stimulation: RNA sequence and structure requirements for binding of Com protein. Cell 1991, 65, 259–269. [Google Scholar] [CrossRef]
- Austin, R.J.; Xia, T.; Ren, J.; Takahashi, T.T.; Roberts, R.W. Designed arginine-rich RNA-binding peptides with picomolar affinity. J. Am. Chem. Soc. 2002, 124, 10966–10967. [Google Scholar] [CrossRef] [PubMed]
- Atkins, J.F.; Gesteland, R.F. The synthetase gene of the RNA phages R17, MS2 and f2 has a single UAG terminator codon. Mol. Gen. Genet. 1975, 139, 19–31. [Google Scholar] [CrossRef] [PubMed]
- Peabody, D.S.; Ely, K.R. Control of translational repression by protein-protein interactions. Nucleic Acids Res. 1992, 20, 1649–1655. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Mali, P.; Aach, J.; Stranges, P.B.; Esvelt, K.M.; Moosburner, M.; Kosuri, S.; Yang, L.; Church, G.M. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat. Biotechnol. 2013, 31, 833–838. [Google Scholar] [CrossRef]
- Zalatan, J.G.; Lee, M.E.; Almeida, R.; Gilbert, L.A.; Whitehead, E.H.; La Russa, M.; Tsai, J.C.; Weissman, J.S.; Dueber, J.E.; Qi, L.S.; et al. Engineering complex synthetic transcriptional programs with CRISPR RNA scaffolds. Cell 2015, 160, 339–350. [Google Scholar] [CrossRef]
- Konermann, S.; Brigham, M.D.; Trevino, A.E.; Joung, J.; Abudayyeh, O.O.; Barcena, C.; Hsu, P.D.; Habib, N.; Gootenberg, J.S.; Nishimasu, H.; et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 2015, 517, 583–588. [Google Scholar] [CrossRef]
- Ma, H.; Tu, L.C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. Multiplexed labeling of genomic loci with dCas9 and engineered sgRNAs using CRISPRainbow. Nat. Biotechnol. 2016, 34, 528–530. [Google Scholar] [CrossRef]
- Lyu, P.; Javidi-Parsijani, P.; Atala, A.; Lu, B. Delivering Cas9/sgRNA ribonucleoprotein (RNP) by lentiviral capsid-based bionanoparticles for efficient ‘hit-and-run’ genome editing. Nucleic Acids Res. 2019, 47, e99. [Google Scholar] [CrossRef]
- Lu, B.; Javidi-Parsijani, P.; Makani, V.; Mehraein-Ghomi, F.; Sarhan, W.M.; Sun, D.; Yoo, K.W.; Atala, Z.P.; Lyu, P.; Atala, A. Delivering SaCas9 mRNA by lentivirus-like bionanoparticles for transient expression and efficient genome editing. Nucleic Acids Res. 2019, 47, e44. [Google Scholar] [CrossRef]
- Larson, D.R.; Zenklusen, D.; Wu, B.; Chao, J.A.; Singer, R.H. Real-time observation of transcription initiation and elongation on an endogenous yeast gene. Science 2011, 332, 475–478. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Chao, J.A.; Singer, R.H. Fluorescence fluctuation spectroscopy enables quantitative imaging of single mRNAs in living cells. Biophys. J. 2012, 102, 2936–2944. [Google Scholar] [CrossRef] [PubMed]
- Lazinski, D.; Grzadzielska, E.; Das, A. Sequence-specific recognition of RNA hairpins by bacteriophage antiterminators requires a conserved arginine-rich motif. Cell 1989, 59, 207–218. [Google Scholar] [CrossRef]
- Wulczyn, F.G.; Bolker, M.; Kahmann, R. Translation of the bacteriophage Mu mom gene is positively regulated by the phage com gene product. Cell 1989, 57, 1201–1210. [Google Scholar] [CrossRef]
- Kaczmarczyk, S.J.; Sitaraman, K.; Young, H.A.; Hughes, S.H.; Chatterjee, D.K. Protein delivery using engineered virus-like particles. Proc. Natl. Acad. Sci. USA 2011, 108, 16998–17003. [Google Scholar] [CrossRef]
- Peretti, S.; Schiavoni, I.; Pugliese, K.; Federico, M. Cell death induced by the herpes simplex virus-1 thymidine kinase delivered by human immunodeficiency virus-1-based virus-like particles. Mol. Ther. 2005, 12, 1185–1196. [Google Scholar] [CrossRef]
- Joo, K.I.; Wang, P. Visualization of targeted transduction by engineered lentiviral vectors. Gene Ther. 2008, 15, 1384–1396. [Google Scholar] [CrossRef]
- Cai, Y.; Bak, R.O.; Mikkelsen, J.G. Targeted genome editing by lentiviral protein transduction of zinc-finger and TAL-effector nucleases. eLife 2014, 3, e01911. [Google Scholar] [CrossRef]
- Choi, J.G.; Dang, Y.; Abraham, S.; Ma, H.; Zhang, J.; Guo, H.; Cai, Y.; Mikkelsen, J.G.; Wu, H.; Shankar, P.; et al. Lentivirus pre-packed with Cas9 protein for safer gene editing. Gene Ther. 2016, 23, 627–633. [Google Scholar] [CrossRef]
- Mangeot, P.E.; Risson, V.; Fusil, F.; Marnef, A.; Laurent, E.; Blin, J.; Mournetas, V.; Massouridès, E.; Sohier, T.J.M.; Corbin, A.; et al. Genome editing in primary cells and in vivo using viral-derived Nanoblades loaded with Cas9-sgRNA ribonucleoproteins. Nat. Commun. 2019, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Gee, P.; Lung, M.S.Y.; Okuzaki, Y.; Sasakawa, N.; Iguchi, T.; Makita, Y.; Hozumi, H.; Miura, Y.; Yang, L.F.; Iwasaki, M.; et al. Extracellular nanovesicles for packaging of CRISPR-Cas9 protein and sgRNA to induce therapeutic exon skipping. Nat. Commun. 2020, 11, 1334. [Google Scholar] [CrossRef] [PubMed]
- Indikova, I.; Indik, S. Highly efficient ‘hit-and-run’ genome editing with unconcentrated lentivectors carrying Vpr.Prot.Cas9 protein produced from RRE-containing transcripts. Nucleic Acids Res. 2020, 48, 8178–8187. [Google Scholar] [CrossRef] [PubMed]
- Lyu, P.; Lu, Z.; Cho, S.I.; Yadav, M.; Yoo, K.; Atala, A.; Kim, J.S.; Lu, B. Adenine base editor ribonucleoproteins delivered by lentivirus-like particles show high on-target base editing and undetectable RNA off-target activities. CRISPR J. 2021, 4, 69–81. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Yao, X.; Lyu, P.; Yadav, M.; Yoo, K.; Atala, A.; Lu, B. Lentiviral Capsid-Mediated Streptococcus pyogenes Cas9 Ribonucleoprotein Delivery for Efficient and Safe Multiplex Genome Editing. CRISPR J. 2021, 4, 914–928. [Google Scholar] [CrossRef]
- Vindry, C.; Guillin, O.; Mangeot, P.E.; Ohlmann, T.; Chavatte, L. A Versatile Strategy to Reduce UGA-Selenocysteine Recoding Efficiency of the Ribosome Using CRISPR-Cas9-Viral-Like-Particles Targeting Selenocysteine-tRNA([Ser]Sec) Gene. Cells 2019, 8, 574. [Google Scholar] [CrossRef]
- Lyu, P.; Yoo, K.W.; Yadav, M.K.; Atala, A.; Aartsma-Rus, A.; Putten, M.V.; Duan, D.; Lu, B. Sensitive and reliable evaluation of single-cut sgRNAs to restore dystrophin by a GFP-reporter assay. PLoS ONE 2020, 15, e0239468. [Google Scholar] [CrossRef]
- Escola, J.M.; Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar] [CrossRef]
- Kobayashi, T.; Vischer, U.M.; Rosnoblet, C.; Lebrand, C.; Lindsay, M.; Parton, R.G.; Kruithof, E.K.; Gruenberg, J. The tetraspanin CD63/lamp3 cycles between endocytic and secretory compartments in human endothelial cells. Mol. Biol. Cell 2000, 11, 1829–1843. [Google Scholar] [CrossRef]
- Mock, U.; Riecken, K.; Berdien, B.; Qasim, W.; Chan, E.; Cathomen, T.; Fehse, B. Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci. Rep. 2014, 4, 6409. [Google Scholar] [CrossRef] [PubMed]
- Lindel, F.; Dodt, C.R.; Weidner, N.; Noll, M.; Bergemann, F.; Behrendt, R.; Fischer, S.; Dietrich, J.; Cartellieri, M.; Hamann, M.V.; et al. TraFo-CRISPR: Enhanced Genome Engineering by Transient Foamy Virus Vector-Mediated Delivery of CRISPR/Cas9 Components. Mol. Ther. Nucleic Acids 2019, 18, 708–726. [Google Scholar] [CrossRef] [PubMed]
- Prel, A.; Caval, V.; Gayon, R.; Ravassard, P.; Duthoit, C.; Payen, E.; Maouche-Chretien, L.; Creneguy, A.; Nguyen, T.H.; Martin, N.; et al. Highly efficient in vitro and in vivo delivery of functional RNAs using new versatile MS2-chimeric retrovirus-like particles. Mol. Ther. Methods Clin. Dev. 2015, 2, 15039. [Google Scholar] [CrossRef]
- Ma, H.; Tu, L.C.; Naseri, A.; Huisman, M.; Zhang, S.; Grunwald, D.; Pederson, T. CRISPR-Cas9 nuclear dynamics and target recognition in living cells. J. Cell Biol. 2016, 214, 529–537. [Google Scholar] [CrossRef] [PubMed]
- Hinz, J.M.; Laughery, M.F.; Wyrick, J.J. Nucleosomes Inhibit Cas9 Endonuclease Activity in Vitro. Biochemistry 2015, 54, 7063–7066. [Google Scholar] [CrossRef]
- Horlbeck, M.A.; Witkowsky, L.B.; Guglielmi, B.; Replogle, J.M.; Gilbert, L.A.; Villalta, J.E.; Torigoe, S.E.; Tjian, R.; Weissman, J.S. Nucleosomes impede Cas9 access to DNA in vivo and in vitro. eLife 2016, 5, e12677. [Google Scholar] [CrossRef]
- Isaac, R.S.; Jiang, F.; Doudna, J.A.; Lim, W.A.; Narlikar, G.J.; Almeida, R. Nucleosome breathing and remodeling constrain CRISPR-Cas9 function. eLife 2016, 5, e13450. [Google Scholar] [CrossRef]
- Kallimasioti-Pazi, E.M.; Thelakkad Chathoth, K.; Taylor, G.C.; Meynert, A.; Ballinger, T.; Kelder, M.J.E.; Lalevee, S.; Sanli, I.; Feil, R.; Wood, A.J. Heterochromatin delays CRISPR-Cas9 mutagenesis but does not influence the outcome of mutagenic DNA repair. PLoS Biol. 2018, 16, e2005595. [Google Scholar] [CrossRef]
- Hoffmann, M.; Wu, Y.J.; Gerber, M.; Berger-Rentsch, M.; Heimrich, B.; Schwemmle, M.; Zimmer, G. Fusion-active glycoprotein G mediates the cytotoxicity of vesicular stomatitis virus M mutants lacking host shut-off activity. J. Gen. Virol. 2010, 91, 2782–2793. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lyu, P.; Lu, B. New Advances in Using Virus-like Particles and Related Technologies for Eukaryotic Genome Editing Delivery. Int. J. Mol. Sci. 2022, 23, 8750. https://doi.org/10.3390/ijms23158750
Lyu P, Lu B. New Advances in Using Virus-like Particles and Related Technologies for Eukaryotic Genome Editing Delivery. International Journal of Molecular Sciences. 2022; 23(15):8750. https://doi.org/10.3390/ijms23158750
Chicago/Turabian StyleLyu, Pin, and Baisong Lu. 2022. "New Advances in Using Virus-like Particles and Related Technologies for Eukaryotic Genome Editing Delivery" International Journal of Molecular Sciences 23, no. 15: 8750. https://doi.org/10.3390/ijms23158750
APA StyleLyu, P., & Lu, B. (2022). New Advances in Using Virus-like Particles and Related Technologies for Eukaryotic Genome Editing Delivery. International Journal of Molecular Sciences, 23(15), 8750. https://doi.org/10.3390/ijms23158750






