A Multiscale Molecular Dynamic Analysis Reveals the Effect of Sialylation on EGFR Clustering in a CRISPR/Cas9-Derived Model
Abstract
1. Introduction
2. Results
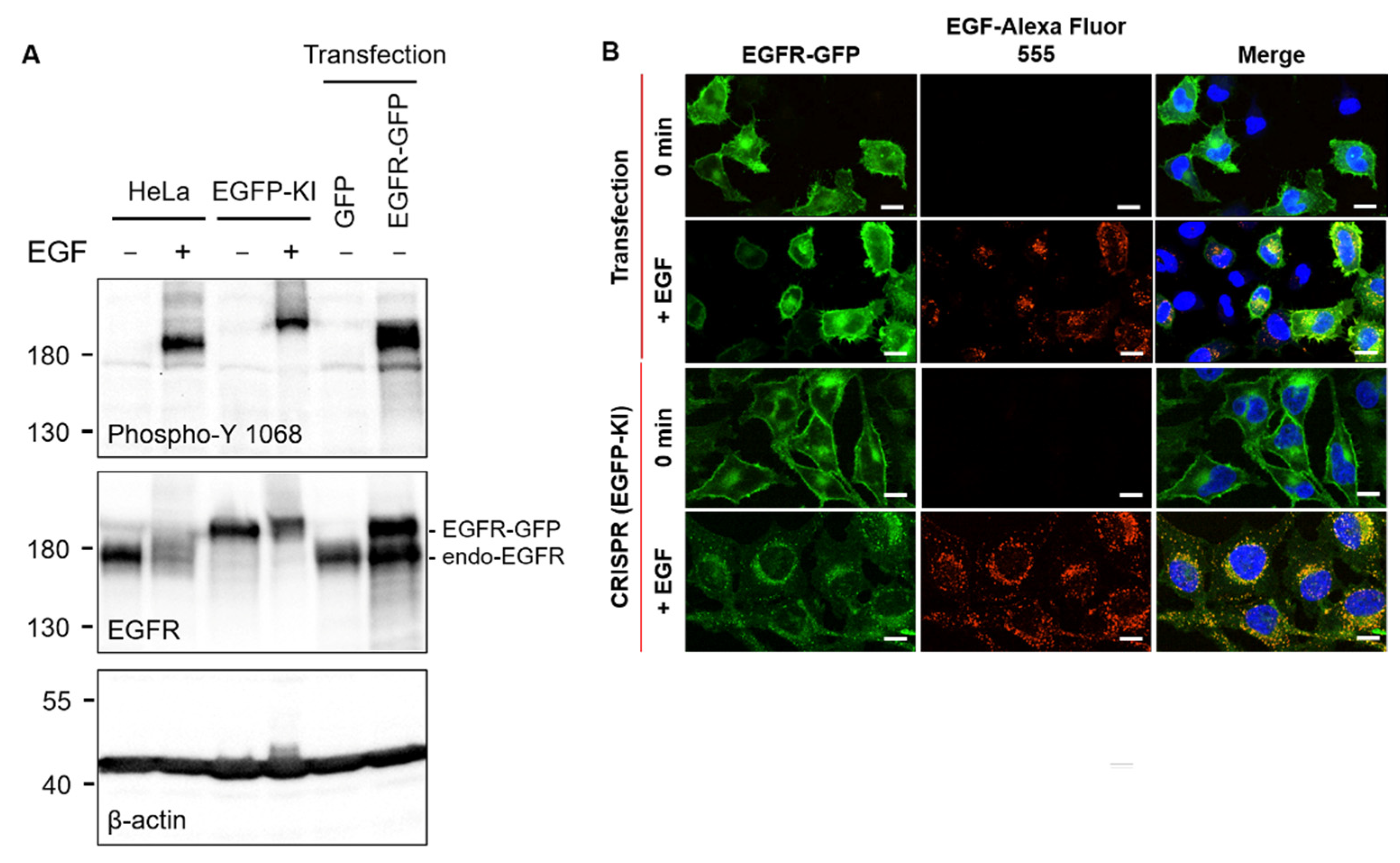
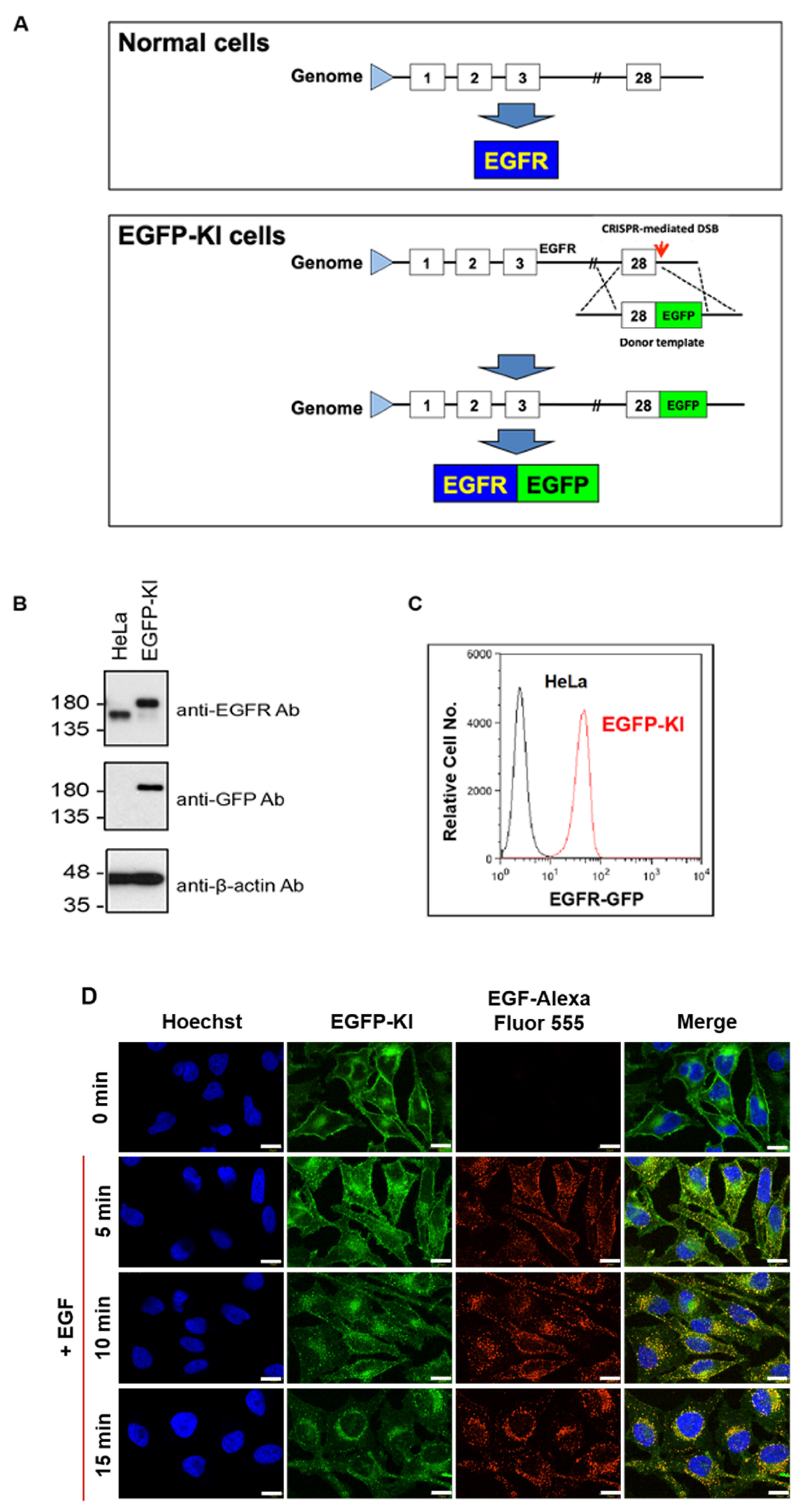
2.1. CRISPR/Cas9 Gene Editing for Constructing EGFP-KI HeLa Cells
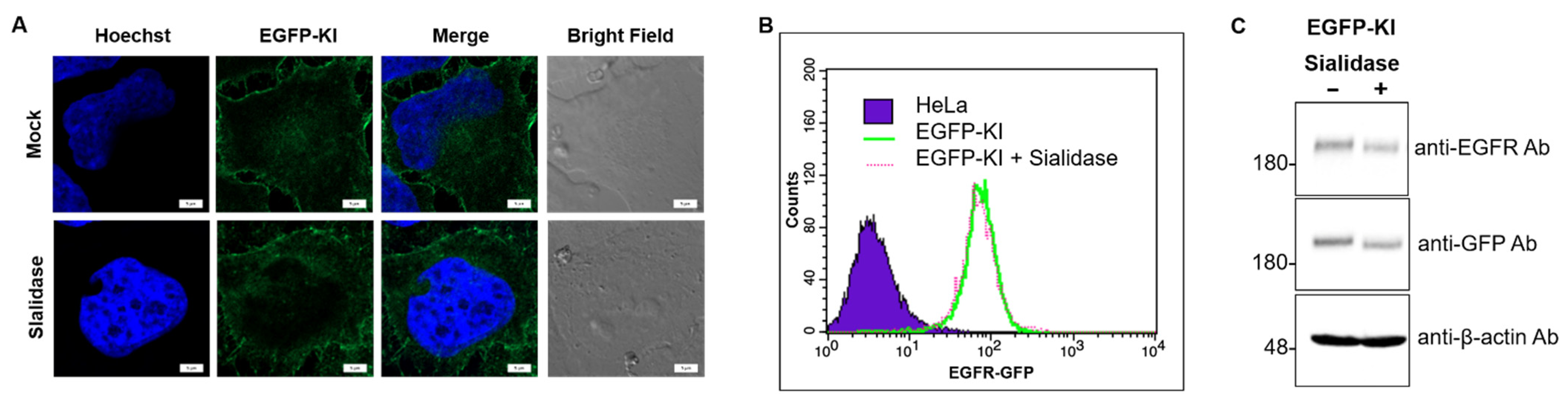
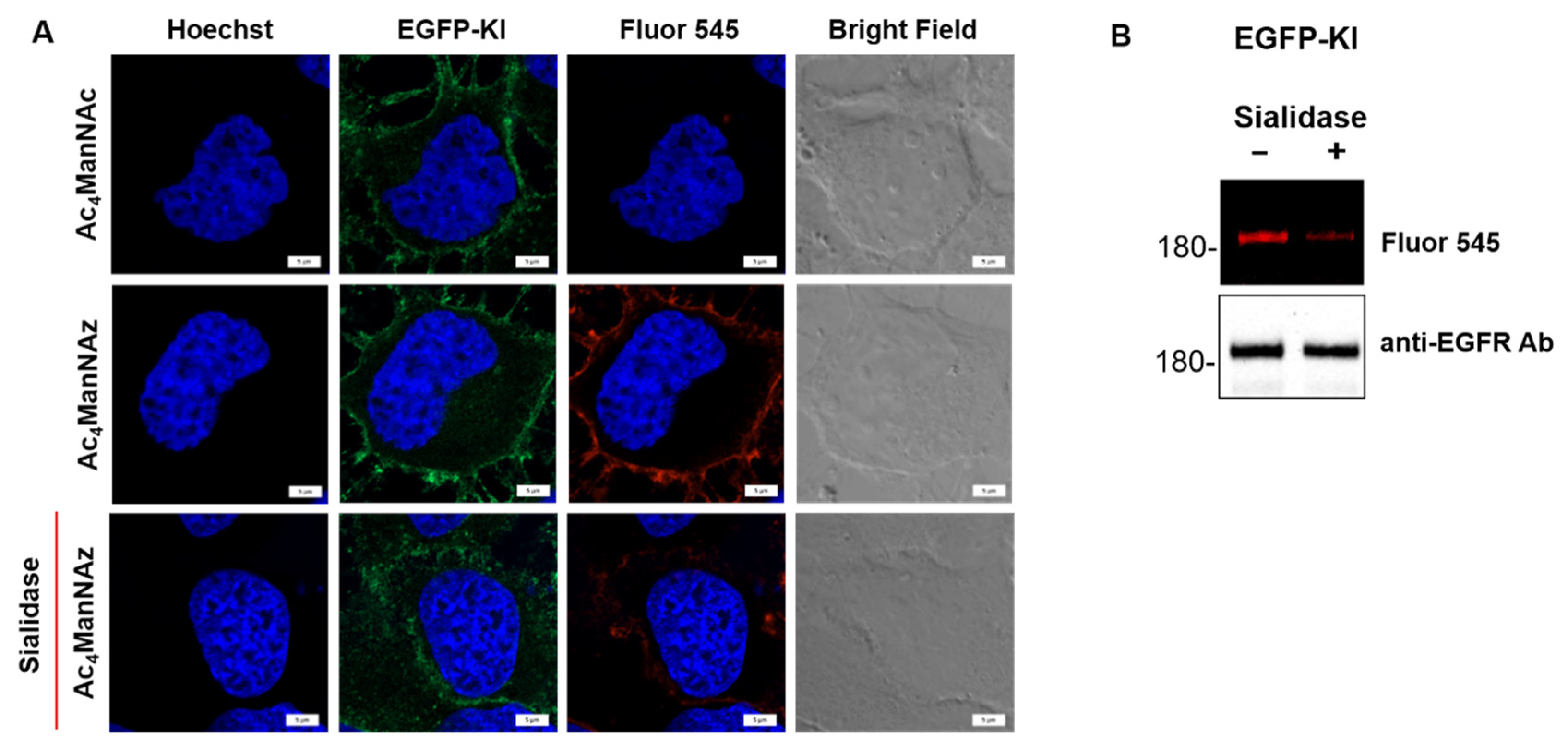
2.2. Metabolic Labeling of Sialic Acid by Strain-Promoted Azide-Alkyne Cycloaddition (SPAAC) to Confirm EGFR Desialylation by Sialidase
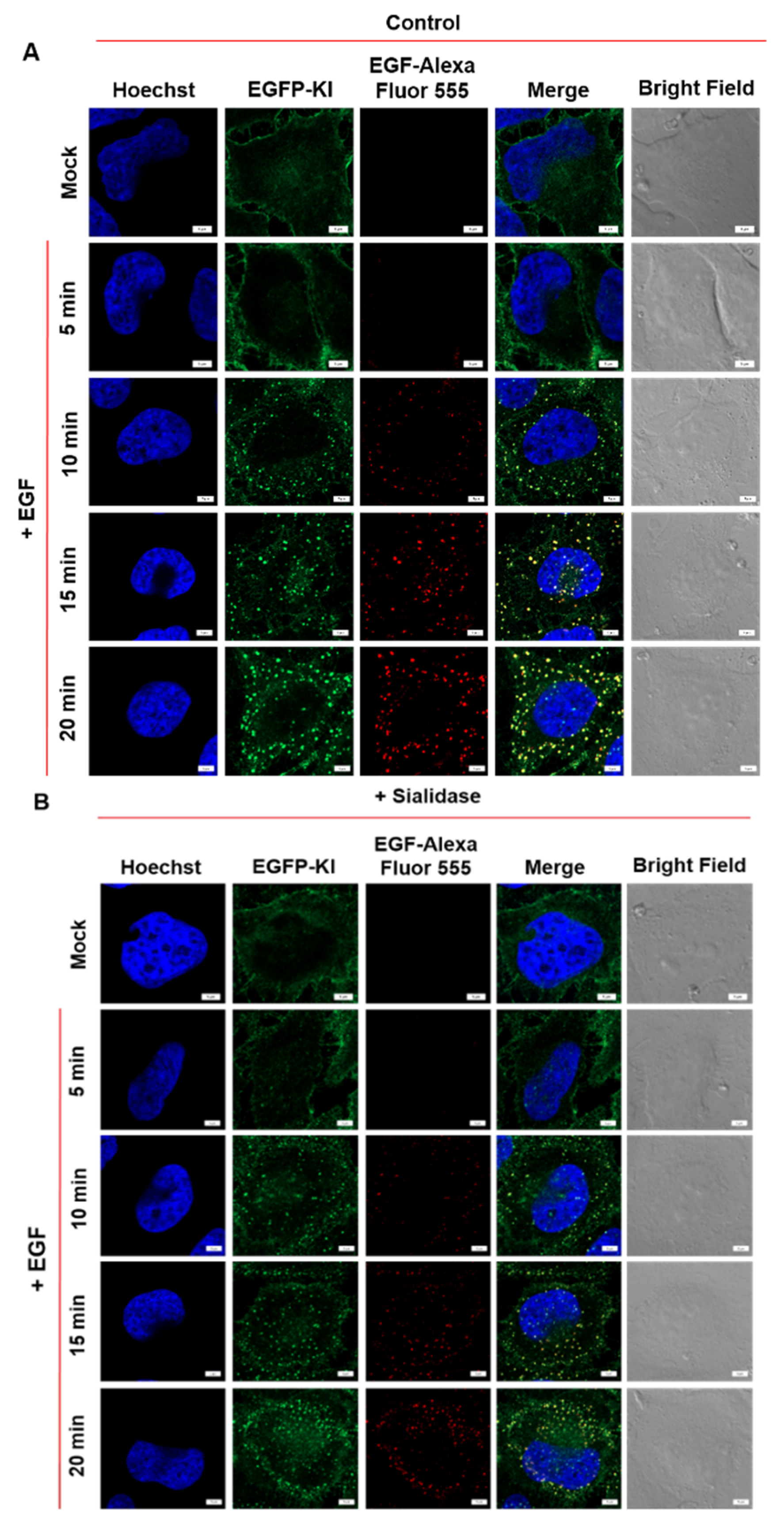
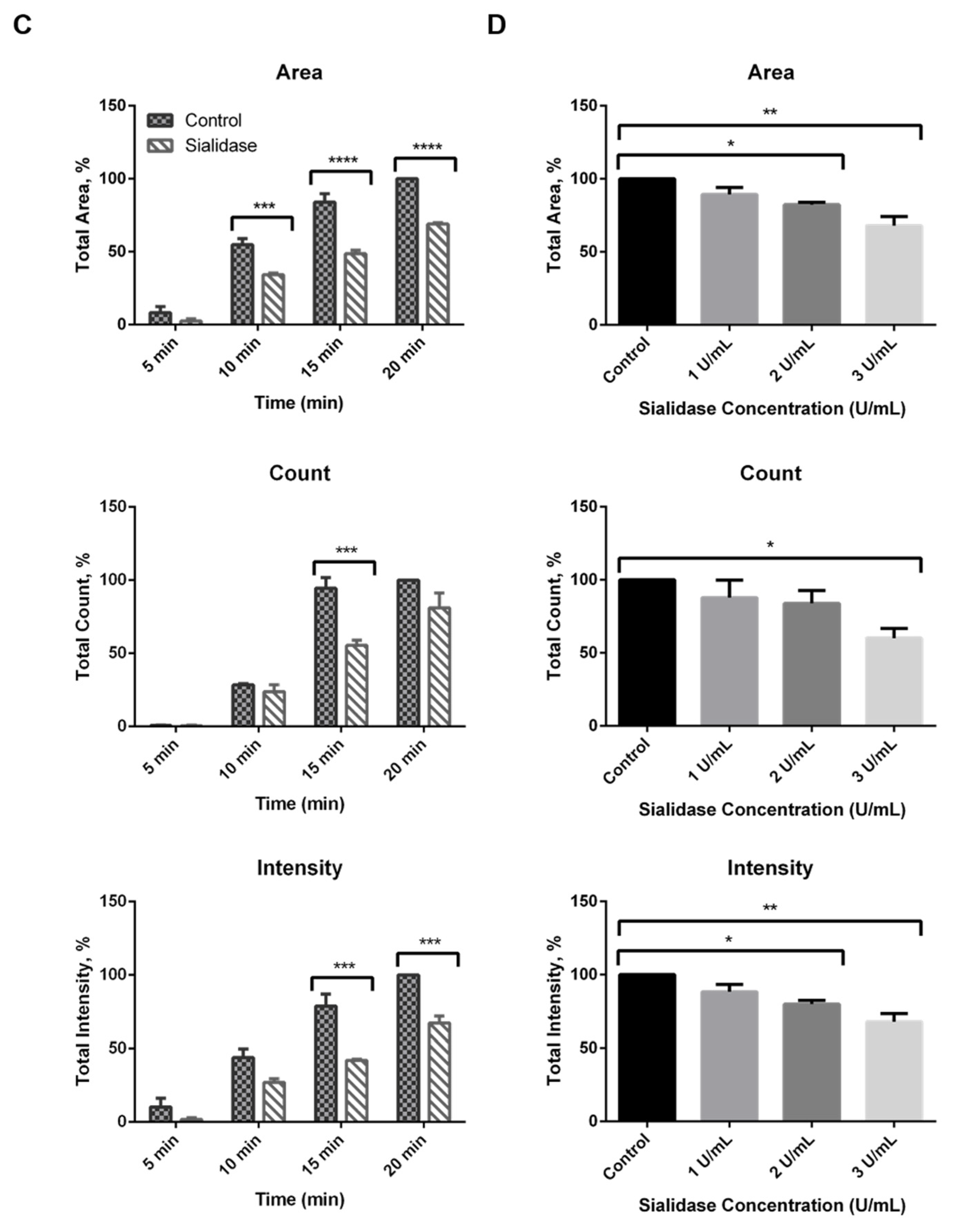
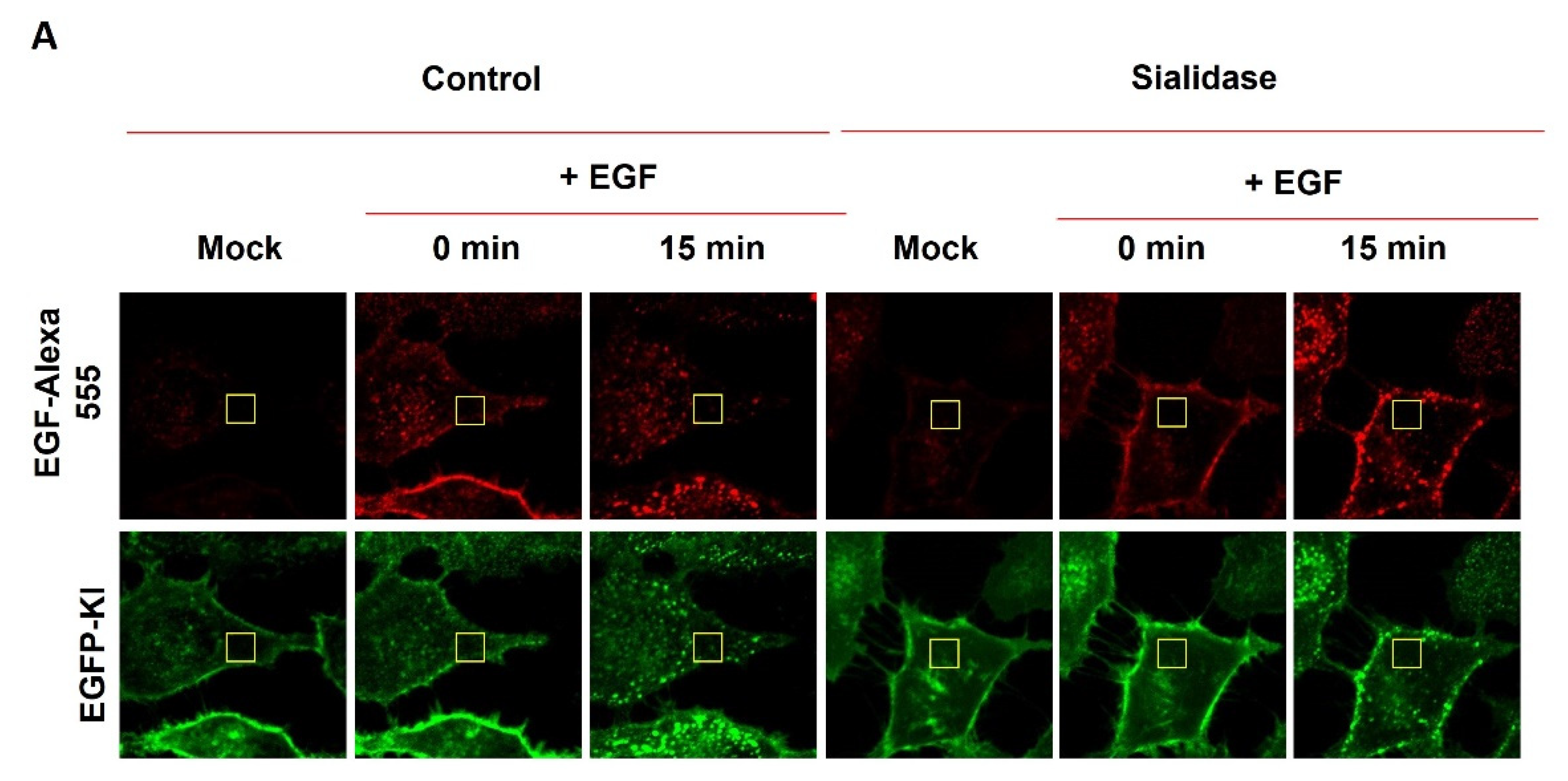
2.3. Effect of Sialidase on EGF-Induced Internalization of EGFR Clusters
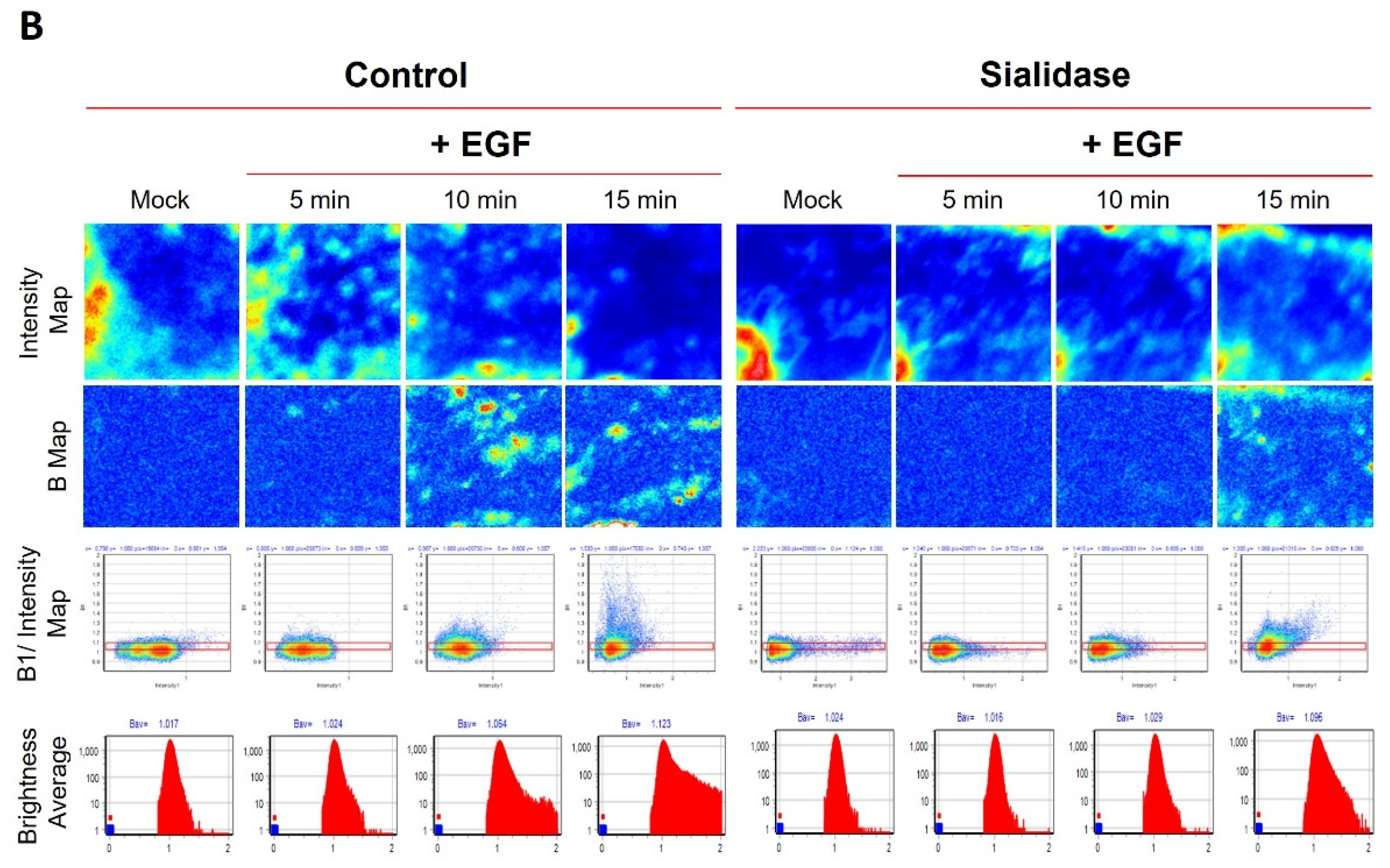
2.4. Evaluation of the Temperature-Dependent EGFR Clustering Induced by EGF Using N&B Analysis
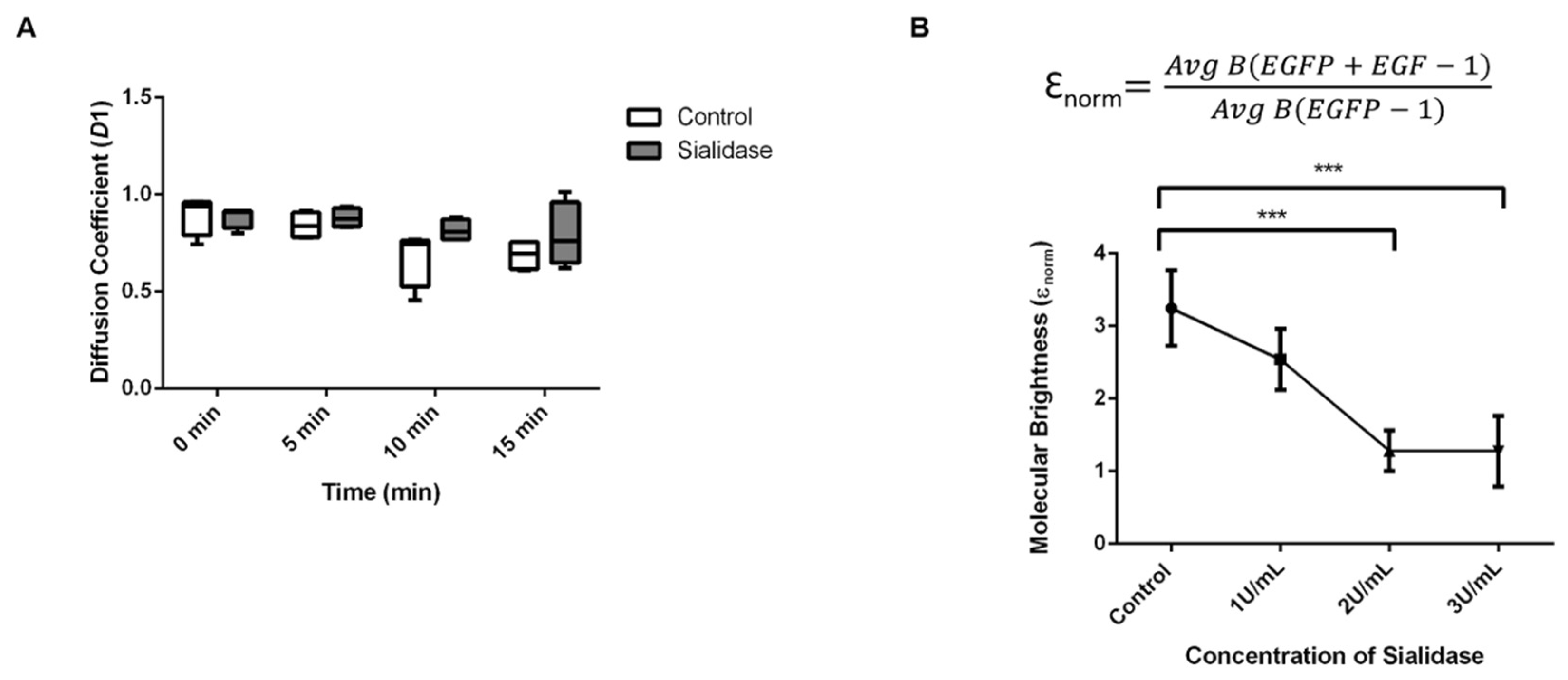
2.5. Residual Sialic Acid Mediates EGF-Induced EGFR Clustering
2.6. Diffusion Coefficient of EGFR-GFP in Desialylated EGFP-KI Cells
3. Discussion
4. Materials and Methods
4.1. Chemicals and Materials
4.2. Antibodies and Plasmids
4.3. Cell Cultures
4.4. Constructs and sgRNA/Cas9 Plasmid
4.5. Construction of EGFP-KI Donor Vectors
4.6. Establishment of the EGFP-KI Cells
4.7. Validation of EGFP-KI Cells Using Immunoblotting, Flow Cytometry and Confocal Microscopy
4.8. EGFR-GFP-Expressing Cells (Transfection and CRISPR/Cas9), Western Blotting and Fluorescence Confocal Microscopy Imaging
4.9. Determination of Sialic Acid Cell Surface Labeling
4.10. EGF Ligand-Induced Kinetic Studies after Treatment with Sialidase
4.11. High-Content Analysis of the Count, Area, and Intensity of EGFP-KI Cells
4.12. Calibration and Instrumentation Setup
4.13. Sample Preparation for RICS Imaging
4.14. Data and Acquisition of RICS Images
4.15. Analysis of N&B Measurements
4.16. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Gschwind, A.; Fischer, O.M.; Ullrich, A. The discovery of receptor tyrosine kinases: Targets for cancer therapy. Nat. Rev. Cancer 2004, 4, 361–370. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Cell Signaling by Receptor Tyrosine Kinases. Cell 2000, 103, 211–225. [Google Scholar] [CrossRef]
- Grandis, J.R.; Sok, J.C. Signaling through the epidermal growth factor receptor during the development of malignancy. Pharmacol. Ther. 2004, 102, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Schlessinger, J. Ligand-Induced, Receptor-Mediated Dimerization and Activation of EGF Receptor. Cell 2002, 110, 669–672. [Google Scholar] [CrossRef]
- Yarden, Y.; Schlessinger, J. Epidermal growth factor induces rapid, reversible aggregation of the purified epidermal growth factor receptor. Biochemistry 1987, 26, 1443–1451. [Google Scholar] [CrossRef]
- Stray, S.J.; Cummings, R.D.; Air, G.M. Influenza virus infection of desialylated cells. Glycobiology 2000, 10, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Varki, A.; Gagneux, P. Multifarious roles of sialic acids in immunity. Ann. N. Y. Acad. Sci. 2012, 1253, 16–36. [Google Scholar] [CrossRef]
- Gao, Y.; Luan, X.; Melamed, J.; Brockhausen, I. Role of Glycans on Key Cell Surface Receptors That Regulate Cell Proliferation and Cell Death. Cells 2021, 10, 1252. [Google Scholar] [CrossRef]
- Wang, X.; Huong, S.-M.; Chiu, M.L.; Raab-Traub, N.; Huang, E.-S. Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 2003, 424, 456–461. [Google Scholar] [CrossRef]
- Wang, X.; Huang, D.Y.; Huong, S.-M.; Huang, E.-S. Integrin αvβ3 is a coreceptor for human cytomegalovirus. Nat. Med. 2005, 11, 515–521. [Google Scholar] [CrossRef]
- Eierhoff, T.; Hrincius, E.R.; Rescher, U.; Ludwig, S.; Ehrhardt, C. The Epidermal Growth Factor Receptor (EGFR) Promotes Uptake of Influenza A Viruses (IAV) into Host Cells. PLoS Pathog. 2010, 6, e1001099. [Google Scholar] [CrossRef] [PubMed]
- Sieben, C.; Sezgin, E.; Eggeling, C.; Manley, S. Influenza A viruses use multivalent sialic acid clusters for cell binding and receptor activation. PLoS Pathog. 2020, 16, e1008656. [Google Scholar] [CrossRef] [PubMed]
- Wen, F.; Wan, X.-F. Influenza Neuraminidase: Underrated Role in Receptor Binding. Trends Microbiol. 2019, 27, 477–479. [Google Scholar] [CrossRef] [PubMed]
- Lau, K.S.; Partridge, E.A.; Grigorian, A.; Silvescu, C.I.; Reinhold, V.N.; Demetriou, M.; Dennis, J.W. Complex N-Glycan Number and Degree of Branching Cooperate to Regulate Cell Proliferation and Differentiation. Cell 2007, 129, 123–134. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, M.; Yokoe, S.; Asahi, M.; Lee, S.H.; Li, W.; Osumi, D.; Miyoshi, E.; Taniguchi, N. N-glycan of ErbB family plays a crucial role in dimer formation and tumor promotion. Biochim. Biophys. Acta (BBA)-Gen. Subj. 2008, 1780, 520–524. [Google Scholar] [CrossRef] [PubMed]
- Britain, C.M.; Holdbrooks, A.T.; Anderson, J.C.; Willey, C.D.; Bellis, S.L. Sialylation of EGFR by the ST6Gal-I sialyltransferase promotes EGFR activation and resistance to gefitinib-mediated cell death. J. Ovarian Res. 2018, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Zhu, M.; Linhai, Y.; Song, Y.; Gui, X.; Tan, G.; Li, J.; Liu, Y.; Deng, Z.; Chen, X.; et al. Increasing HER2 α2,6 sialylation facilitates gastric cancer progression and resistance via the Akt and ERK pathways. Oncol. Rep. 2018, 40, 2997–3005. [Google Scholar] [CrossRef]
- Liu, Y.-C.; Yen, H.-Y.; Chen, C.-Y.; Chen, C.-H.; Cheng, P.-F.; Juan, Y.-H.; Chen, C.-H.; Khoo, K.-H.; Yu, C.-J.; Yang, P.-C.; et al. Sialylation and fucosylation of epidermal growth factor receptor suppress its dimerization and activation in lung cancer cells. Proc. Natl. Acad. Sci. USA 2011, 108, 11332–11337. [Google Scholar] [CrossRef]
- Park, J.-J.; Yi, J.Y.; Jin, Y.B.; Lee, Y.-J.; Lee, J.-S.; Lee, Y.-S.; Ko, Y.-G.; Lee, M. Sialylation of epidermal growth factor receptor regulates receptor activity and chemosensitivity to gefitinib in colon cancer cells. Biochem. Pharmacol. 2012, 83, 849–857. [Google Scholar] [CrossRef]
- Yen, H.-Y.; Liu, Y.-C.; Chen, N.-Y.; Tsai, C.-F.; Wang, Y.-T.; Chen, Y.-J.; Hsu, T.-L.; Yang, P.-C.; Wong, C.-H. Effect of sialylation on EGFR phosphorylation and resistance to tyrosine kinase inhibition. Proc. Natl. Acad. Sci. USA 2015, 112, 6955–6960. [Google Scholar] [CrossRef]
- Ratz, M.; Testa, I.; Hell, S.W.; Jakobs, S. CRISPR/Cas9-mediated endogenous protein tagging for RESOLFT super-resolution microscopy of living human cells. Sci. Rep. 2015, 5, 9592. [Google Scholar] [CrossRef] [PubMed]
- Tian, X.; Gu, T.; Patel, S.; Bode, A.M.; Lee, M.-H.; Dong, Z. CRISPR/Cas9—An evolving biological tool kit for cancer biology and oncology. NPJ Precis. Oncol. 2019, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Koch, B.; Nijmeijer, B.; Kueblbeck, M.; Cai, Y.; Walther, N.; Ellenberg, J. Generation and validation of homozygous fluorescent knock-in cells using CRISPR–Cas9 genome editing. Nat. Protoc. 2018, 13, 1465–1487. [Google Scholar] [CrossRef] [PubMed]
- Digman, M.A.; Gratton, E. Analysis of diffusion and binding in cells using the RICS approach. Microsc. Res. Tech. 2009, 72, 323–332. [Google Scholar] [CrossRef]
- Robert, T.Y.; Haibing, T. Measuring protein dynamics in live cells: Protocols and practical considerations for fluorescence fluctuation microscopy. J. Biomed. Opt. 2014, 19, 090801. [Google Scholar]
- Yang, Y.-P.; Ma, H.; Starchenko, A.; Huh, W.J.; Li, W.; Hickman, F.E.; Zhang, Q.; Franklin, J.L.; Mortlock, D.P.; Fuhrmann, S.; et al. A Chimeric Egfr Protein Reporter Mouse Reveals Egfr Localization and Trafficking In Vivo. Cell Rep. 2017, 19, 1257–1267. [Google Scholar] [CrossRef]
- Carter, R.E.; Sorkin, A. Endocytosis of Functional Epidermal Growth Factor Receptor-Green Fluorescent Protein Chimera. J. Biol. Chem. 1998, 273, 35000–35007. [Google Scholar] [CrossRef]
- Sigismund, S.; Argenzio, E.; Tosoni, D.; Cavallaro, E.; Polo, S.; Di Fiore, P.P. Clathrin-Mediated Internalization Is Essential for Sustained EGFR Signaling but Dispensable for Degradation. Dev. Cell 2008, 15, 209–219. [Google Scholar] [CrossRef]
- Dobie, C.; Skropeta, D. Insights into the role of sialylation in cancer progression and metastasis. Br. J. Cancer 2021, 124, 76–90. [Google Scholar] [CrossRef]
- Du, J.; Meledeo, M.A.; Wang, Z.; Khanna, H.S.; Paruchuri, V.D.P.; Yarema, K.J. Metabolic glycoengineering: Sialic acid and beyond. Glycobiology 2009, 19, 1382–1401. [Google Scholar] [CrossRef]
- Agard, N.J.; Prescher, J.A.; Bertozzi, C.R. A Strain-Promoted [3 + 2] Azide−Alkyne Cycloaddition for Covalent Modification of Biomolecules in Living Systems. J. Am. Chem. Soc. 2004, 126, 15046–15047. [Google Scholar] [CrossRef] [PubMed]
- Needham, S.R.; Roberts, S.K.; Arkhipov, A.; Mysore, V.P.; Tynan, C.J.; Zanetti-Domingues, L.C.; Kim, E.T.; Losasso, V.; Korovesis, D.; Hirsch, M.; et al. EGFR oligomerization organizes kinase-active dimers into competent signalling platforms. Nat. Commun. 2016, 7, 13307. [Google Scholar] [CrossRef] [PubMed]
- Digman, M.A.; Dalal, R.; Horwitz, A.F.; Gratton, E. Mapping the Number of Molecules and Brightness in the Laser Scanning Microscope. Biophys. J. 2008, 94, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Adu-Gyamfi, E.; Soni, S.P.; Xue, Y.; Digman, M.A.; Gratton, E.; Stahelin, R.V. The Ebola Virus Matrix Protein Penetrates into the Plasma Membrane. J. Biol. Chem. 2013, 288, 5779–5789. [Google Scholar] [CrossRef] [PubMed]
- Casaletto, J.B.; McClatchey, A.I. Spatial regulation of receptor tyrosine kinases in development and cancer. Nat. Rev. Cancer 2012, 12, 387–400. [Google Scholar] [CrossRef] [PubMed]
- Overeem, N.J.; van der Vries, E.; Huskens, J. A Dynamic, Supramolecular View on the Multivalent Interaction between Influenza Virus and Host Cell. Small 2021, 17, 2007214. [Google Scholar] [CrossRef]
- Fricke, F.; Dietz, M.S.; Heilemann, M. Single-Molecule Methods to Study Membrane Receptor Oligomerization. ChemPhysChem 2015, 16, 713–721. [Google Scholar] [CrossRef]
- Fernández-Suárez, M.; Ting, A.Y. Fluorescent probes for super-resolution imaging in living cells. Nat. Rev. Mol. Cell Biol. 2008, 9, 929–943. [Google Scholar] [CrossRef]
- Tanaka, T.; Zhou, Y.; Ozawa, T.; Okizono, R.; Banba, A.; Yamamura, T.; Oga, E.; Muraguchi, A.; Sakurai, H. Ligand-activated epidermal growth factor receptor (EGFR) signaling governs endocytic trafficking of unliganded receptor monomers by non-canonical phosphorylation. J. Biol. Chem. 2018, 293, 2288–2301. [Google Scholar] [CrossRef]
- Nolan, R.; Iliopoulou, M.; Alvarez, L.; Padilla-Parra, S. Detecting protein aggregation and interaction in live cells: A guide to number and brightness. Methods 2018, 140–141, 172–177. [Google Scholar] [CrossRef]
- Guillot, A.; Dauchez, M.; Belloy, N.; Jonquet, J.; Duca, L.; Romier, B.; Maurice, P.; Debelle, L.; Martiny, L.; Durlach, V.; et al. Impact of sialic acids on the molecular dynamic of bi-antennary and tri-antennary glycans. Sci. Rep. 2016, 6, 35666. [Google Scholar] [CrossRef] [PubMed]
- Mozzi, A.; Forcella, M.; Riva, A.; Difrancesco, C.; Molinari, F.; Martin, V.; Papini, N.; Bernasconi, B.; Nonnis, S.; Tedeschi, G.; et al. NEU3 activity enhances EGFR activation without affecting EGFR expression and acts on its sialylation levels. Glycobiology 2015, 25, 855–868. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kohnz, R.A.; Roberts, L.S.; DeTomaso, D.; Bideyan, L.; Yan, P.; Bandyopadhyay, S.; Goga, A.; Yosef, N.; Nomura, D.K. Protein Sialylation Regulates a Gene Expression Signature that Promotes Breast Cancer Cell Pathogenicity. ACS Chem. Biol. 2016, 11, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Swanson, C.D.; Akama-Garren, E.H.; Stein, E.A.; Petralia, J.D.; Ruiz, P.J.; Edalati, A.; Lindstrom, T.M.; Robinson, W.H. Inhibition of Epidermal Growth Factor Receptor Tyrosine Kinase Ameliorates Collagen-Induced Arthritis. J. Immunol. 2012, 188, 3513–3521. [Google Scholar] [CrossRef] [PubMed]
- Romano, R.; Bucci, C. Role of EGFR in the Nervous System. Cells 2020, 9, 1887. [Google Scholar] [CrossRef]
- Makki, N.; Thiel, K.W.; Miller, J.F.J. The Epidermal Growth Factor Receptor and Its Ligands in Cardiovascular Disease. Int. J. Mol. Sci. 2013, 14, 20597–20613. [Google Scholar] [CrossRef]
- Rossow, M.J.; Sasaki, J.M.; Digman, M.A.; Gratton, E. Raster image correlation spectroscopy in live cells. Nat. Protoc. 2010, 5, 1761–1774. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Leong, S.K.; Hsiao, J.-C.; Shie, J.-J. A Multiscale Molecular Dynamic Analysis Reveals the Effect of Sialylation on EGFR Clustering in a CRISPR/Cas9-Derived Model. Int. J. Mol. Sci. 2022, 23, 8754. https://doi.org/10.3390/ijms23158754
Leong SK, Hsiao J-C, Shie J-J. A Multiscale Molecular Dynamic Analysis Reveals the Effect of Sialylation on EGFR Clustering in a CRISPR/Cas9-Derived Model. International Journal of Molecular Sciences. 2022; 23(15):8754. https://doi.org/10.3390/ijms23158754
Chicago/Turabian StyleLeong, Shwee Khuan, Jye-Chian Hsiao, and Jiun-Jie Shie. 2022. "A Multiscale Molecular Dynamic Analysis Reveals the Effect of Sialylation on EGFR Clustering in a CRISPR/Cas9-Derived Model" International Journal of Molecular Sciences 23, no. 15: 8754. https://doi.org/10.3390/ijms23158754
APA StyleLeong, S. K., Hsiao, J.-C., & Shie, J.-J. (2022). A Multiscale Molecular Dynamic Analysis Reveals the Effect of Sialylation on EGFR Clustering in a CRISPR/Cas9-Derived Model. International Journal of Molecular Sciences, 23(15), 8754. https://doi.org/10.3390/ijms23158754





