Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L.
Abstract
:1. Introduction
2. Results
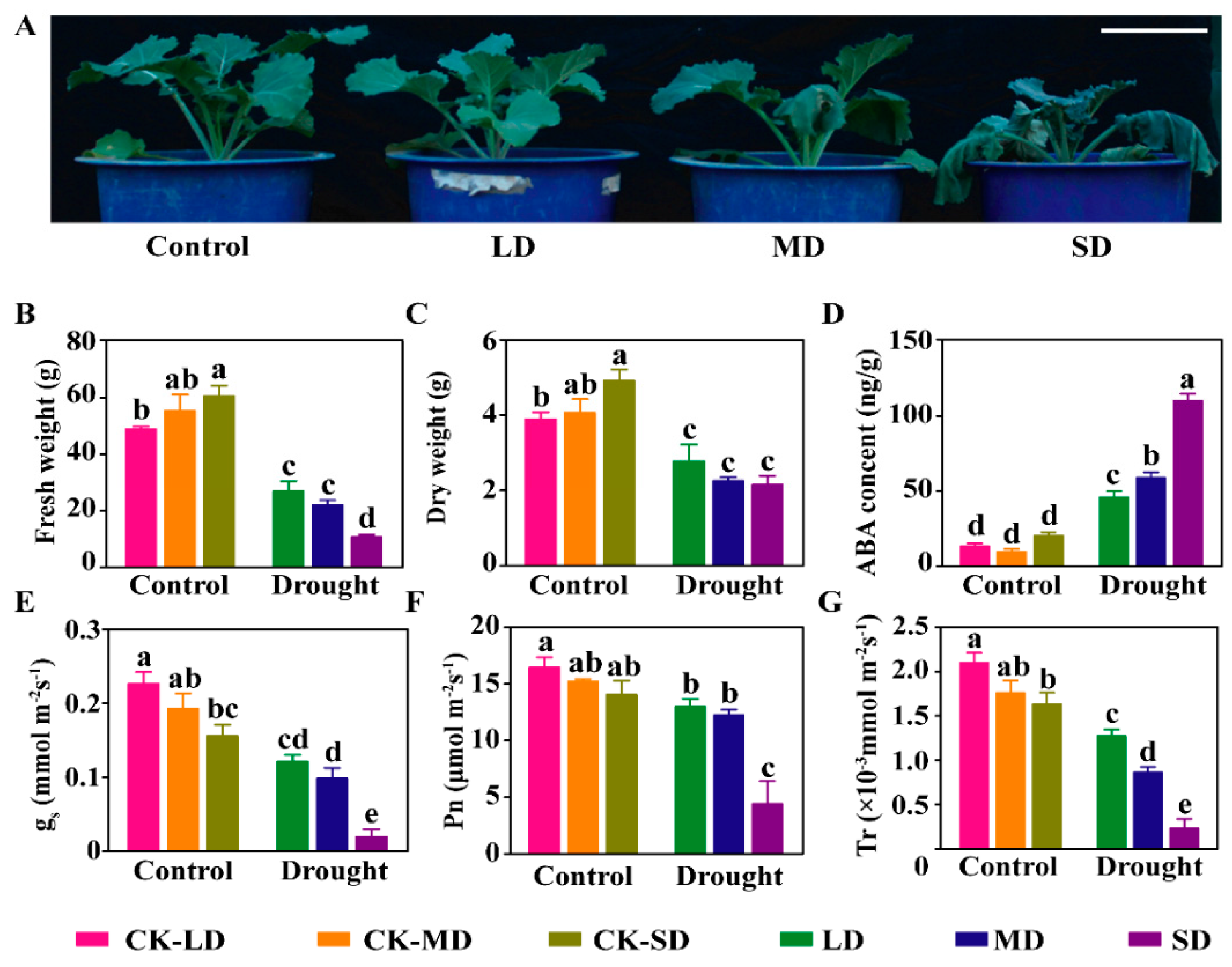
2.1. Characterization of Physio-Biochemical Traits at Seedling Stage under Varying Degrees of Drought Stress in B. napus
2.2. Changes in Crystal Morphology, Content and Composition of Epidermal Wax under Varying Degrees of Drought Stress in B. napus
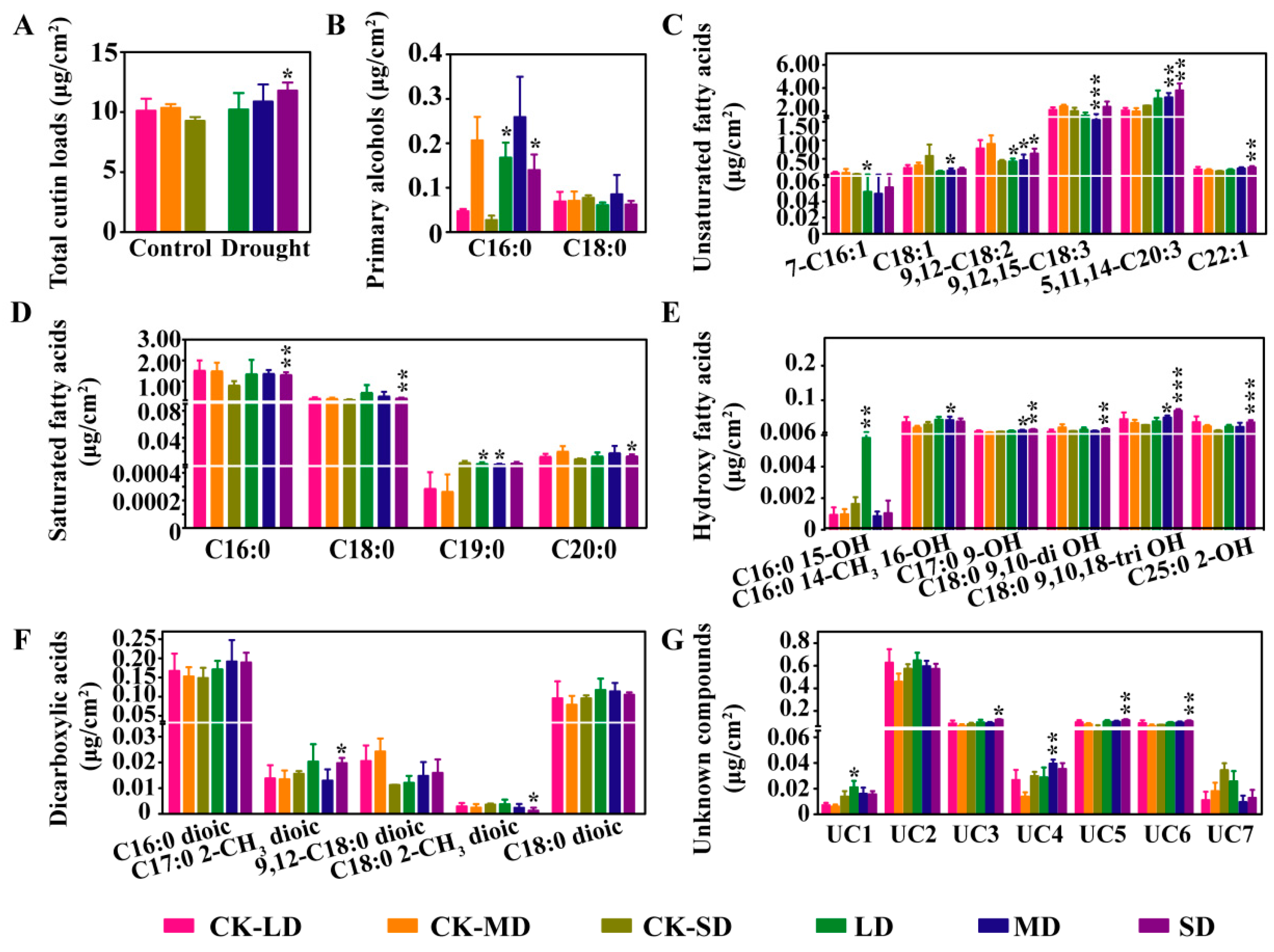
2.3. Changes in Cutin Content and Composition under Varying Degrees of Drought Stress in B. napus
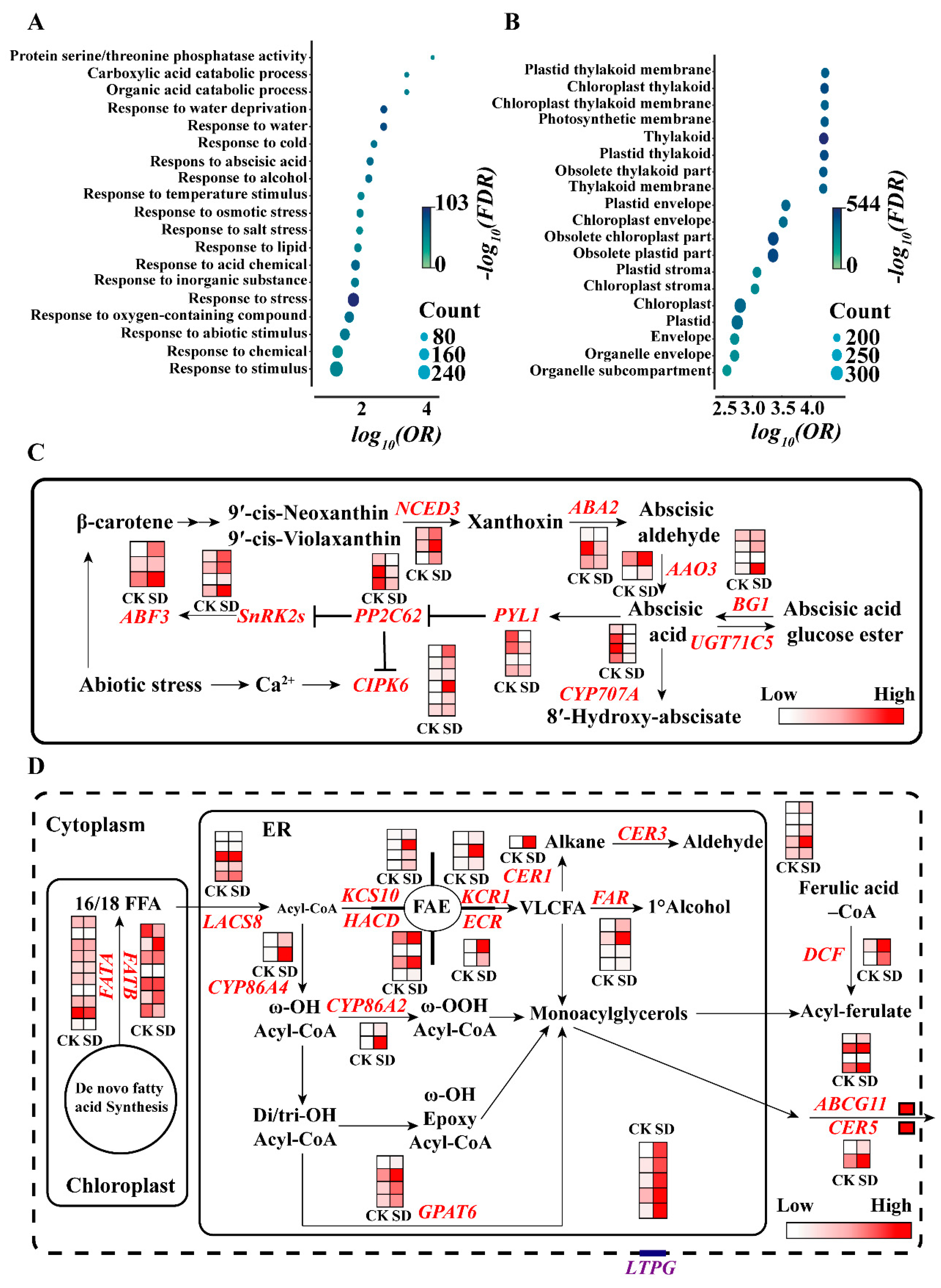
2.4. Transcriptome Analysis Revealed Gene Expression Alternation under Drought Stress
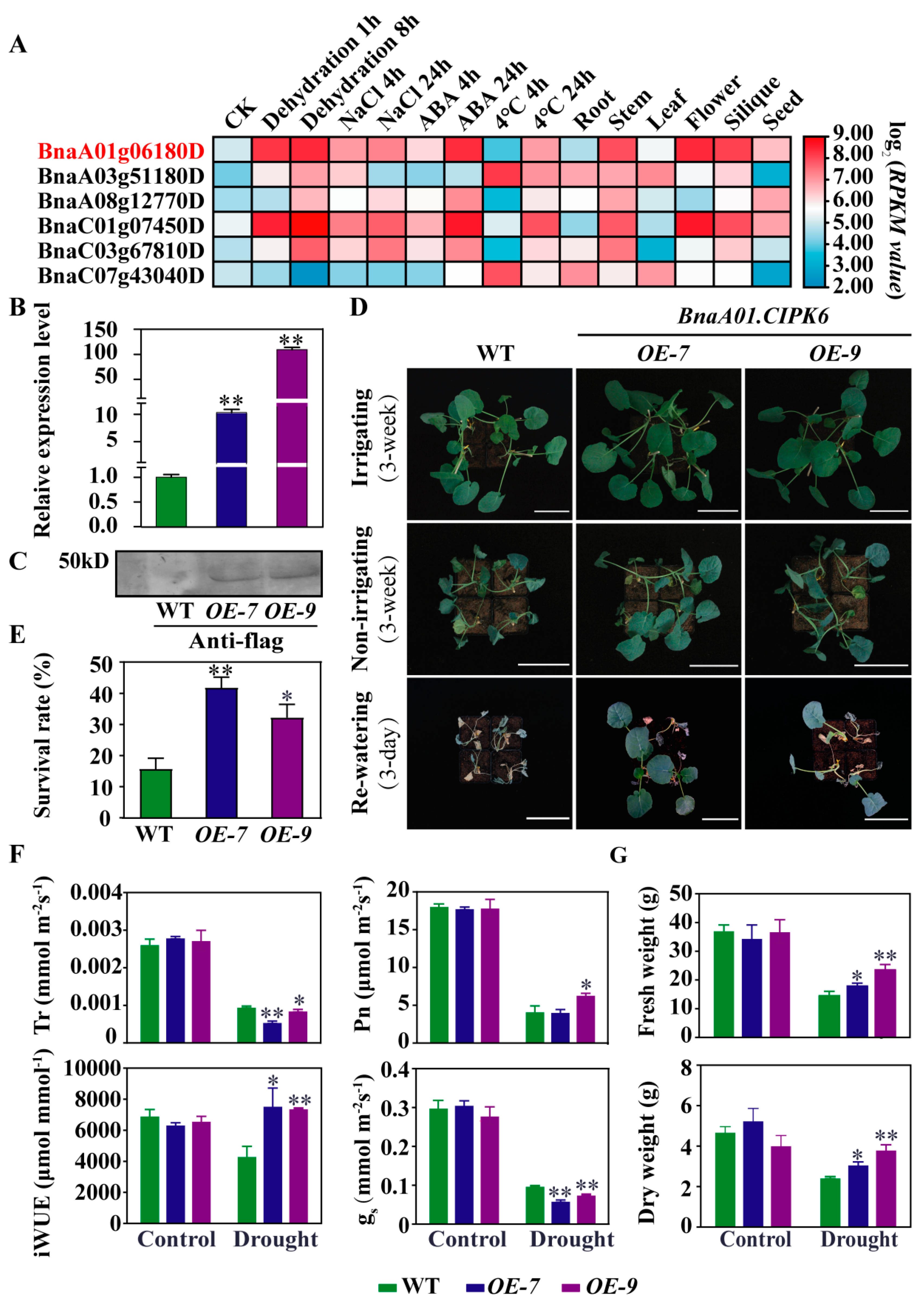
2.5. Over-expression of BnaA01.CIPK6 Confers Drought Tolerance in B. napus
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Drought Stress Treatments
4.2. Determination of ABA Content
4.3. RNA-Seq Alignment and Differential Expression Analysis
4.4. Quantitative Real-Time PCR Analysis
4.5. Scanning Electron Microscopy
4.6. Photosynthetic Parameter Measurements
4.7. Determination of Leaf Epicuticular Wax
4.8. Determination of Leaf Cutin
4.9. Vector Construction and Genetic Transformation
4.10. Western Blot Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gupta, A.; Rico-Medina, A.; Cano-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Karbulkova, J.; Schreiber, L.; Macek, P.; Santrucek, J. Differences between water permeability of astomatous and stomatous cuticular membranes: Effects of air humidity in two species of contrasting drought-resistance strategy. J. Exp. Bot. 2008, 59, 3987–3995. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, H.; Sun, H.; Bao, L.; Han, S.; Hui, T.; Zhang, R.; Zhang, M.; Su, C.; Qian, Y.; Jiao, F. Secondary Metabolism and Hormone Response Reveal the Molecular Mechanism of Triploid Mulberry (Morus alba L.) Trees Against Drought. Front. Plant Sci. 2021, 12, 720452. [Google Scholar] [CrossRef] [PubMed]
- Holmgren, P.; Jarvis, P.G.; Jarvis, M.S. Resistances to Carbon Dioxide and Water Vapour Transfer in Leaves of Different Plant Species. Physiol. Plant. 1965, 18, 557–573. [Google Scholar] [CrossRef]
- Jensen, A.B.; Busk, P.K.; Figueras, M.; Alba, M.M.; Peracchia, G.; Messeguer, R.; Goday, A.; Pages, M. Drought signal transduction in plants. Plant Growth Regul. 1996, 20, 105–110. [Google Scholar] [CrossRef]
- Schreiber, L. Transport barriers made of cutin, suberin and associated waxes. Trends Plant Sci. 2010, 15, 546–553. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.K.; Dubeaux, G.; Takahashi, Y.; Schroeder, J.I. Signaling mechanisms in abscisic acid-mediated stomatal closure. Plant J. 2021, 105, 307–321. [Google Scholar] [CrossRef]
- Santiago, J.; Rodrigues, A.; Saez, A.; Rubio, S.; Antoni, R.; Dupeux, F.; Park, S.Y.; Marquez, J.A.; Cutler, S.R.; Rodriguez, P.L. Modulation of drought resistance by the abscisic acid receptor PYL5 through inhibition of clade A PP2Cs. Plant J. 2009, 60, 575–588. [Google Scholar] [CrossRef]
- Ramachandra Reddy, A.; Chaitanya, K.V.; Vivekanandan, M. Drought-induced responses of photosynthesis and antioxidant metabolism in higher plants. J. Plant Physiol. 2004, 161, 1189–1202. [Google Scholar] [CrossRef] [PubMed]
- Sato, H.; Takasaki, H.; Takahashi, F.; Suzuki, T.; Iuchi, S.; Mitsuda, N.; Ohme-Takagi, M.; Ikeda, M.; Seo, M.; Yamaguchi-Shinozaki, K.; et al. Arabidopsis thaliana NGATHA1 transcription factor induces ABA biosynthesis by activating NCED3 gene during dehydration stress. Proc. Natl. Acad. Sci. USA 2018, 115, E11178–E11187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Zhang, J.; Sun, M.; He, C.; Yu, K.; Zhao, B.; Li, R.; Li, J.; Yang, Z.; Wang, X.; et al. Multi-Omics Analyses Reveal Systemic Insights into Maize Vivipary. Plants 2021, 10, 2437. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [Green Version]
- Verma, R.K.; Santosh Kumar, V.V.; Yadav, S.K.; Pushkar, S.; Rao, M.V.; Chinnusamy, V. Overexpression of ABA Receptor PYL10 Gene Confers Drought and Cold Tolerance to Indica Rice. Front. Plant Sci. 2019, 10, 1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- He, Z.; Wu, J.; Sun, X.; Dai, M. The Maize Clade A PP2C Phosphatases Play Critical Roles in Multiple Abiotic Stress Responses. Int. J. Mol. Sci. 2019, 20, 3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, S.; Sharma, P.; Singh, R.; Tiwari, R.; Singh, G.P. Genome-wide identification and expression analysis of sucrose nonfermenting-1-related protein kinase (SnRK) genes in Triticum aestivum in response to abiotic stress. Sci. Rep. 2021, 11, 22477. [Google Scholar] [CrossRef]
- Park, S.Y.; Fung, P.; Nishimura, N.; Jensen, D.R.; Fujii, H.; Zhao, Y.; Lumba, S.; Santiago, J.; Rodrigues, A.; Chow, T.F.; et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 2009, 324, 1068–1071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Chen, M.; Zhang, Y.; Kong, X.; Du, Z.; Zhou, H.; Yu, Z.; Qin, J.; Chen, C. Leaf Cuticular Transpiration Barrier Organization in Tea Tree Under Normal Growth Conditions. Front. Plant Sci. 2021, 12, 655799. [Google Scholar] [CrossRef]
- Kunst, L.; Samuels, L. Plant cuticles shine: Advances in wax biosynthesis and export. Curr. Opin. Plant Biol. 2009, 12, 721–727. [Google Scholar] [CrossRef]
- Yeats, T.H.; Rose, J.K. The formation and function of plant cuticles. Plant Physiol 2013, 163, 5–20. [Google Scholar] [CrossRef] [Green Version]
- Schnurr, J.; Shockey, J.; Browse, J. The acyl-CoA synthetase encoded by LACS2 is essential for normal cuticle development in Arabidopsis. Plant Cell 2004, 16, 629–642. [Google Scholar] [CrossRef] [Green Version]
- Li-Beisson, Y.; Shorrosh, B.; Beisson, F.; Andersson, M.X.; Arondel, V.; Bates, P.D.; Baud, S.; Bird, D.; Debono, A.; Durrett, T.P.; et al. Acyl-lipid metabolism. Arab. Book 2013, 11, e0161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beisson, F.; Li-Beisson, Y.; Pollard, M. Solving the puzzles of cutin and suberin polymer biosynthesis. Curr. Opin. Plant Biol. 2012, 15, 329–337. [Google Scholar] [CrossRef]
- Petit, J.; Bres, C.; Mauxion, J.P.; Tai, F.W.; Martin, L.B.; Fich, E.A.; Joubes, J.; Rose, J.K.; Domergue, F.; Rothan, C. The Glycerol-3-Phosphate Acyltransferase GPAT6 from Tomato Plays a Central Role in Fruit Cutin Biosynthesis. Plant Physiol. 2016, 171, 894–913. [Google Scholar] [CrossRef]
- Fich, E.A.; Segerson, N.A.; Rose, J.K. The Plant Polyester Cutin: Biosynthesis, Structure, and Biological Roles. Annu. Rev. Plant Biol. 2016, 67, 207–233. [Google Scholar] [CrossRef]
- Zhang, X.; Ni, Y.; Xu, D.; Busta, L.; Xiao, Y.; Jetter, R.; Guo, Y. Integrative analysis of the cuticular lipidome and transcriptome of Sorghum bicolor reveals cultivar differences in drought tolerance. Plant Physiol. Biochem. 2021, 163, 285–295. [Google Scholar] [CrossRef]
- Hosseini, S.A.; Rethore, E.; Pluchon, S.; Ali, N.; Billiot, B.; Yvin, J.C. Calcium Application Enhances Drought Stress Tolerance in Sugar Beet and Promotes Plant Biomass and Beetroot Sucrose Concentration. Int. J. Mol. Sci. 2019, 20, 3777. [Google Scholar] [CrossRef] [Green Version]
- Farber, M.; Attia, Z.; Weiss, D. Cytokinin activity increases stomatal density and transpiration rate in tomato. J. Exp. Bot. 2016, 67, 6351–6362. [Google Scholar] [CrossRef]
- Liu, R.; Xu, Y.H.; Jiang, S.C.; Lu, K.; Lu, Y.F.; Feng, X.J.; Wu, Z.; Liang, S.; Yu, Y.T.; Wang, X.F.; et al. Light-harvesting chlorophyll a/b-binding proteins, positively involved in abscisic acid signalling, require a transcription repressor, WRKY40, to balance their function. J. Exp. Bot. 2013, 64, 5443–5456. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Z.; Ma, J.; Lei, P.; Wang, Q.; Feng, X.; Xu, H. Poly-gamma-glutamic acid induces system tolerance to drought stress by promoting abscisic acid accumulation in Brassica napus L. Sci. Rep. 2020, 10, 252. [Google Scholar] [CrossRef] [Green Version]
- Niu, J.; Zhang, S.; Liu, S.; Ma, H.; Chen, J.; Shen, Q.; Ge, C.; Zhang, X.; Pang, C.; Zhao, X. The compensation effects of physiology and yield in cotton after drought stress. J. Plant Physiol. 2018, 224–225, 30–48. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Wang, P.; Jia, X.; Huo, L.; Che, R.; Ma, F. Improvement of drought tolerance by overexpressing MdATG18a is mediated by modified antioxidant system and activated autophagy in transgenic apple. Plant Biotechnol. J. 2018, 16, 545–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, J.; Yin, X.; Struik, P.C.; Stomph, T.J.; Wang, H. Using chromosome introgression lines to map quantitative trait loci for photosynthesis parameters in rice (Oryza sativa L.) leaves under drought and well-watered field conditions. J. Exp. Bot. 2012, 63, 455–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almeida, D.M.; Oliveira, M.M.; Saibo, N.J.M. Regulation of Na+ and K+ homeostasis in plants: Towards improved salt stress tolerance in crop plants. Genet. Mol. Biol. 2017, 40 (Suppl. 1), 326–345. [Google Scholar] [CrossRef] [Green Version]
- Chaimala, A.; Jogloy, S.; Vorasoot, N.; Toomsan, B.; Jongrungklang, N.; Kesmala, T.; Holbrook, C.C.; Kvien, C.K. Responses of Total Biomass, Shoot Dry Weight, Yield and Yield Components of Jerusalem Artichoke (Helianthus tuberosus L.) Varieties under Different Terminal Drought Duration. Agriculture 2020, 10, 198. [Google Scholar] [CrossRef]
- Rollins, J.A.; Habte, E.; Templer, S.E.; Colby, T.; Schmidt, J.; von Korff, M. Leaf proteome alterations in the context of physiological and morphological responses to drought and heat stress in barley (Hordeum vulgare L.). J. Exp. Bot. 2013, 64, 3201–3212. [Google Scholar] [CrossRef] [Green Version]
- Nambara, E.; Marion-Poll, A. Abscisic acid biosynthesis and catabolism. Annu. Rev. Plant Biol. 2005, 56, 165–185. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.C.; Song, R.F.; Zheng, S.Q.; Li, T.T.; Zhang, B.L.; Gao, X.; Lu, Y.T. Coordination of plant growth and abiotic stress responses by tryptophan synthase beta subunit 1 through modulation of tryptophan and ABA homeostasis in Arabidopsis. Mol. Plant 2022, 15, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Su, G.; Li, M.; Ke, Q.; Kim, S.Y.; Li, H.; Huang, J.; Xu, B.; Deng, X.P.; Kwak, S.S. Overexpressing Arabidopsis ABF3 increases tolerance to multiple abiotic stresses and reduces leaf size in alfalfa. Plant Physiol. Biochem. 2016, 109, 199–208. [Google Scholar] [CrossRef] [PubMed]
- Miyazono, K.; Miyakawa, T.; Sawano, Y.; Kubota, K.; Kang, H.J.; Asano, A.; Miyauchi, Y.; Takahashi, M.; Zhi, Y.; Fujita, Y.; et al. Structural basis of abscisic acid signalling. Nature 2009, 462, 609–614. [Google Scholar] [CrossRef]
- Wang, D.; Gao, Z.; Du, P.; Xiao, W.; Tan, Q.; Chen, X.; Li, L.; Gao, D. Expression of ABA Metabolism-Related Genes Suggests Similarities and Differences Between Seed Dormancy and Bud Dormancy of Peach (Prunus persica). Front. Plant Sci. 2015, 6, 1248. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Yang, B.; Liu, W.Z.; Li, H.; Wang, L.; Wang, B.; Deng, M.; Liang, W.; Deyholos, M.K.; Jiang, Y.Q. Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 2014, 14, 8. [Google Scholar] [CrossRef] [Green Version]
- Tripathi, V.; Parasuraman, B.; Laxmi, A.; Chattopadhyay, D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009, 58, 778–790. [Google Scholar] [CrossRef]
- Tsou, P.L.; Lee, S.Y.; Allen, N.S.; Winter-Sederoff, H.; Robertson, D. An ER-targeted calcium-binding peptide confers salt and drought tolerance mediated by CIPK6 in Arabidopsis. Planta 2012, 235, 539–552. [Google Scholar] [CrossRef]
- Sardar, A.; Nandi, A.K.; Chattopadhyay, D. CBL-interacting protein kinase 6 negatively regulates immune response to Pseudomonas syringae in Arabidopsis. J. Exp. Bot. 2017, 68, 3573–3584. [Google Scholar] [CrossRef] [Green Version]
- Willick, I.R.; Lahlali, R.; Vijayan, P.; Muir, D.; Karunakaran, C.; Tanino, K.K. Wheat flag leaf epicuticular wax morphology and composition in response to moderate drought stress are revealed by SEM, FTIR-ATR and synchrotron X-ray spectroscopy. Physiol. Plant 2018, 162, 316–332. [Google Scholar] [CrossRef] [Green Version]
- Tapia, G.; Morales-Quintana, L.; Parra, C.; Berbel, A.; Alcorta, M. Study of nsLTPs in Lotus japonicus genome reveal a specific epidermal cell member (LjLTP10) regulated by drought stress in aerial organs with a putative role in cutin formation. Plant Mol. Biol. 2013, 82, 485–501. [Google Scholar] [CrossRef]
- Guo, J.; Xu, W.; Yu, X.; Shen, H.; Li, H.; Cheng, D.; Liu, A.; Liu, J.; Liu, C.; Zhao, S.; et al. Cuticular Wax Accumulation Is Associated with Drought Tolerance in Wheat Near-Isogenic Lines. Front. Plant Sci. 2016, 7, 1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barthlott, W.; Neinhuis, C.; Cutler, D.; Ditsch, F.; Meusel, I.; Theisen, I.; Wilhelmi, H. Classification and terminology of plant epicuticular waxes. Bot. J. Linn. Soc. 1998, 126, 237–260. [Google Scholar] [CrossRef]
- Kosma, D.K.; Bourdenx, B.; Bernard, A.; Parsons, E.P.; Lu, S.; Joubes, J.; Jenks, M.A. The impact of water deficiency on leaf cuticle lipids of Arabidopsis. Plant Physiol. 2009, 151, 1918–1929. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.; Xiong, L. Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc. Natl. Acad. Sci. USA 2013, 110, 17790–17795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Wei, W.; Zhu, H.; Challa, G.S.; Bi, C.; Trick, H.N.; Li, W. W3 Is a New Wax Locus That Is Essential for Biosynthesis of beta-Diketone, Development of Glaucousness, and Reduction of Cuticle Permeability in Common Wheat. PLoS ONE 2015, 10, e0140524. [Google Scholar] [CrossRef] [Green Version]
- Li, L.; Du, Y.; He, C.; Dietrich, C.R.; Li, J.; Ma, X.; Wang, R.; Liu, Q.; Liu, S.; Wang, G.; et al. Maize glossy6 is involved in cuticular wax deposition and drought tolerance. J. Exp. Bot. 2019, 70, 3089–3099. [Google Scholar] [CrossRef] [Green Version]
- Xiao, F.; Goodwin, S.M.; Xiao, Y.; Sun, Z.; Baker, D.; Tang, X.; Jenks, M.A.; Zhou, J.M. Arabidopsis CYP86A2 represses Pseudomonas syringae type III genes and is required for cuticle development. EMBO J. 2004, 23, 2903–2913. [Google Scholar] [CrossRef]
- Zhang, Y.; Ali, U.; Zhang, G.; Yu, L.; Fang, S.; Iqbal, S.; Li, H.; Lu, S.; Guo, L. Transcriptome analysis reveals genes commonly responding to multiple abiotic stresses in rapeseed. Mol. Breed. 2019, 39, 158. [Google Scholar] [CrossRef]
- Jin, S.; Zhang, S.; Liu, Y.; Jiang, Y.; Wang, Y.; Li, J.; Ni, Y. A combination of genome-wide association study and transcriptome analysis in leaf epidermis identifies candidate genes involved in cuticular wax biosynthesis in Brassica napus. BMC Plant Biol. 2020, 20, 458. [Google Scholar] [CrossRef]
- Liu, C.; Zhang, X.; Zhang, K.; An, H.; Hu, K.; Wen, J.; Shen, J.; Ma, C.; Yi, B.; Tu, J.; et al. Comparative Analysis of the Brassica napus Root and Leaf Transcript Profiling in Response to Drought Stress. Int. J. Mol. Sci. 2015, 16, 18752–18777. [Google Scholar] [CrossRef] [Green Version]
- Dai, C.; Li, Y.; Li, L.; Du, Z.; Lin, S.; Tian, X.; Li, S.; Yang, B.; Yao, W.; Wang, J.; et al. An efficient Agrobacterium-mediated transformation method using hypocotyl as explants for Brassica napus. Mol. Breed. 2020, 40, 96. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, S.; Zhao, P.; Tan, Z.; Peng, Y.; Xu, L.; Jin, Y.; Wei, F.; Guo, L.; Yao, X. Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L. Int. J. Mol. Sci. 2022, 23, 8555. https://doi.org/10.3390/ijms23158555
Fang S, Zhao P, Tan Z, Peng Y, Xu L, Jin Y, Wei F, Guo L, Yao X. Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L. International Journal of Molecular Sciences. 2022; 23(15):8555. https://doi.org/10.3390/ijms23158555
Chicago/Turabian StyleFang, Shuai, Peimin Zhao, Zengdong Tan, Yan Peng, Lintang Xu, Yutong Jin, Fang Wei, Liang Guo, and Xuan Yao. 2022. "Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L." International Journal of Molecular Sciences 23, no. 15: 8555. https://doi.org/10.3390/ijms23158555
APA StyleFang, S., Zhao, P., Tan, Z., Peng, Y., Xu, L., Jin, Y., Wei, F., Guo, L., & Yao, X. (2022). Combining Physio-Biochemical Characterization and Transcriptome Analysis Reveal the Responses to Varying Degrees of Drought Stress in Brassica napus L. International Journal of Molecular Sciences, 23(15), 8555. https://doi.org/10.3390/ijms23158555





