Overexpression of CpADC from Chinese Cherry (Cerasus pseudocerasus Lindl. ‘Manaohong’) Promotes the Ability of Response to Drought in Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
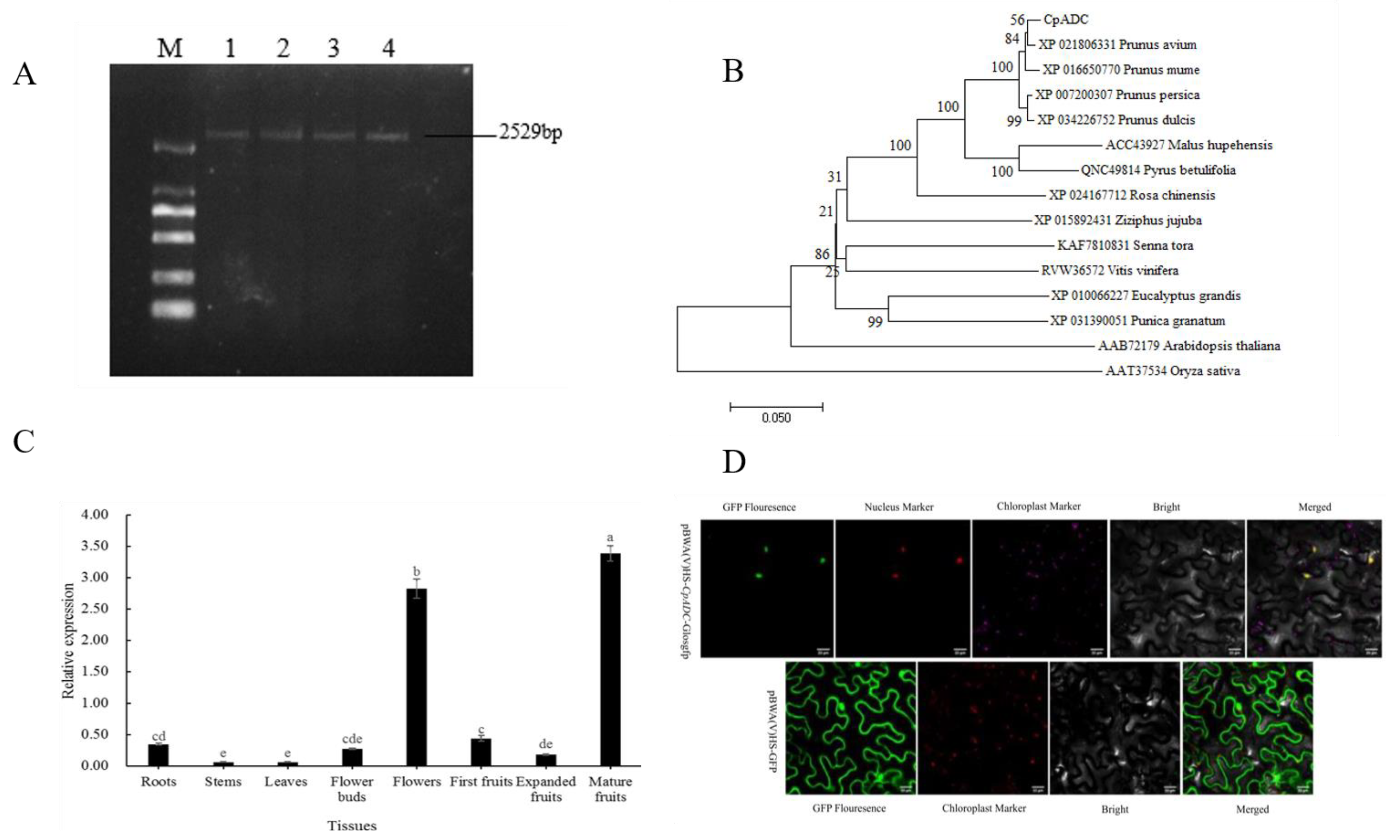
2.1. Obtaining of CpADC
2.2. Physiological Analysis and Sequence Alignment
2.3. Tissue-Specific Expression of CpADC Gene
2.4. CpADC Is Localized in the Nucleus
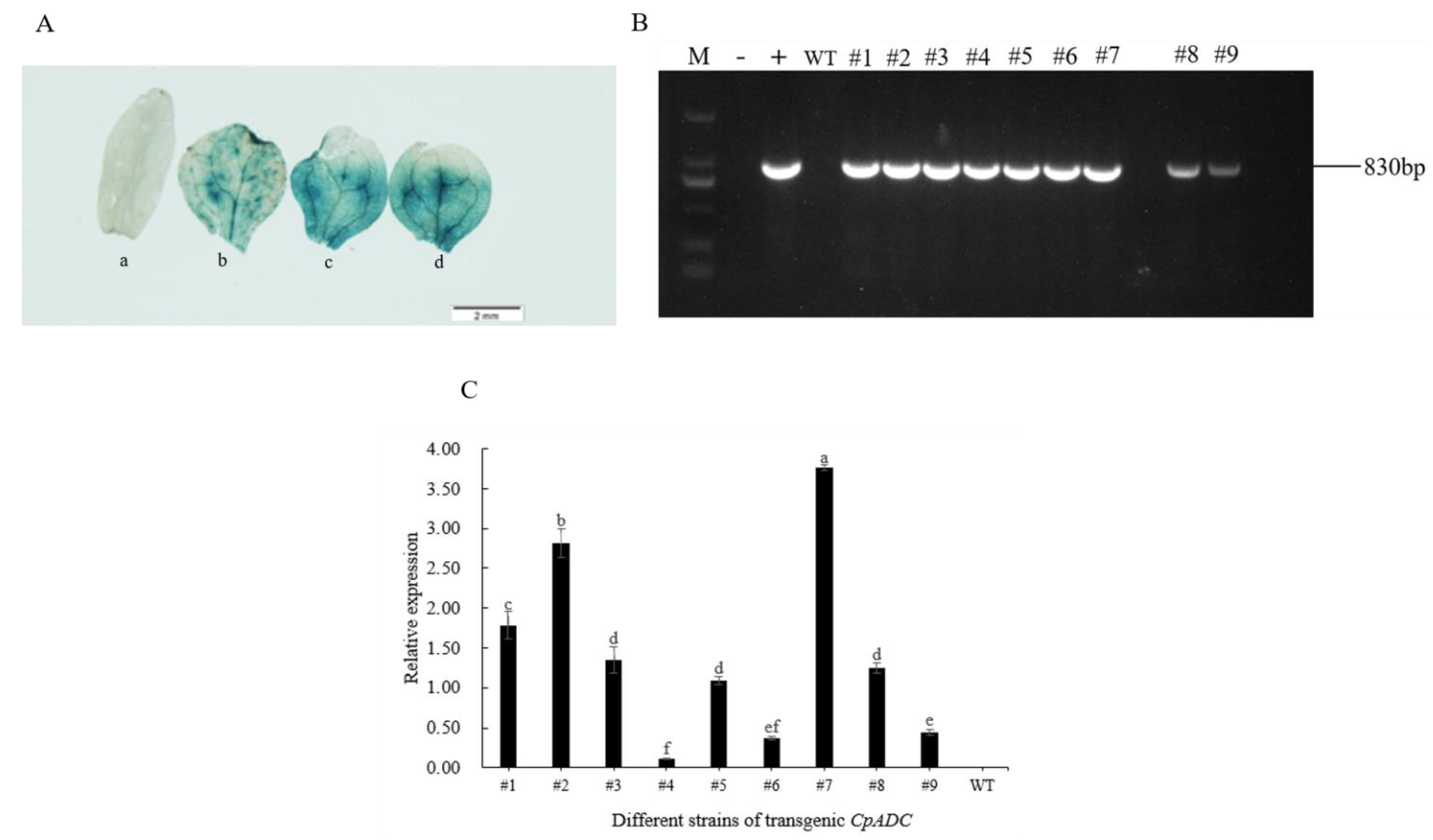
2.5. Transformation and Characterization of Transgenic Arabidopsis thaliana
2.6. Quantification of Free Polyamine Contene
2.7. Exposure to Drought Environment Led to Enhanced ADC Activity in AtADC Overexpression Line
2.8. MDA and Pro Levels Were Increased in Transgenic Arabidopsis Plant under Drought Tress
3. Discussion
4. Materials and Methods
4.1. Plant Materials Growth Conditions and Drought Stress Treatment
4.2. Cloning of CpADC Cording Sequence
4.3. Sequence Analysis of CpADC
4.4. Subcellular Localization of CpADC Protein
4.5. Analysis of CpADC Gene Expression by Quantitative Real-Time PCR (qRT-PCR)
4.6. Generation of Transgenic Plants Arabidopsis thaliana by Agrobacterium Tumefaciens Mediated Transformation
4.7. Quantification of Free Polyamine Content
4.8. Arginine Decarboxylase Activity Determination, MDA and Pro Content Measurement
4.9. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rastogi, R.; Dulson, J.; Rothstein, S.J. Cloning of tomato (Lycopersicon esculentum Mill.) arginine decarboxylase gene and its expression during fruit ripening. Plant Physiol. 1993, 103, 829–834. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Berberich, T.; Takahashi, Y.; Kusano, T.; Yamaguchi, K. Advances in polyamine research in 2007. J. Plant Res. 2007, 120, 345–350. [Google Scholar] [CrossRef]
- Groppa, M.D.; Benavides, M.P. Polyamines and abiotic stress: Recent advances Review Article. Amino Acids 2008, 34, 35–45. [Google Scholar] [CrossRef]
- Sarfraz, S.; Ali, M.; Ahmad, M.; Siddique, K.H.M. Polyamines: Natural and engineered abiotic and biotic stress tolerance in plants. Biotechnol. Adv. 2011, 29, 300–311. [Google Scholar] [CrossRef]
- Chen, D.; Shao, Q.; Yin, L.; Younis, A.; Zheng, B. Polyamine function in plants: Metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 2019, 9, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Walden, K.; Robertson, H.M. Letter to the Editor: Ancient DNA from Amber Fossil Bees? Mol. Biol. Evol. 1997, 14, 1075–1077. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Nada, K.; Pang, X.; Honda, C.; Kitashiba, H.; Moriguchi, T. Role of polyamines in peach fruit development and storage. Tree Physiol. 2006, 26, 791–798. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Guo, C.; Wu, H.; Chen, C. Arginine decarboxylase ADC2 enhances salt tolerance through increasing ROS scavenging enzyme activity in Arabidopsis thaliana. Plant Growth Regul. 2017, 83, 253–263. [Google Scholar] [CrossRef]
- Lenis, Y.Y.; Elmetwally, M.A.; Maldonado-estrada, J.G.; Bazer, F.W. Physiological importance of polyamines 3. Zygote 2017, 25, 244–255. [Google Scholar] [CrossRef]
- Mo, H.; Pua, E.C. Up-regulation of arginine decarboxylase gene expression and accumulation of polyamines in mustard (Brassica juncea) in response to stress. Physiol. Plant. 2002, 114, 439–449. [Google Scholar] [CrossRef]
- Yin, X.; Yang, Y.; Lv, Y.; Li, Y.; Yang, D.; Yue, Y.; Yang, Y. BrrICE1.1 is associated with putrescine synthesis through regulation of the arginine decarboxylase gene in freezing tolerance of turnip (Brassica rapa var. rapa). BMC Plant Biol. 2020, 20, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.J.; Kitashiba, H.; Honda, C.; Nada, K.; Moriguchi, T. Expression of arginine decarboxylase and ornithine decarboxylase genes in apple cells and stressed shoots. J. Exp. Bot. 2005, 56, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alcázar, R.; Altabella, T.; Marco, F.; Bortolotti, C.; Reymond, M.; Koncz, C.; Carrasco, P.; Tiburcio, A.F. Polyamines: Molecules with regulatory functions in plant abiotic stress tolerance. Planta 2010, 231, 1237–1249. [Google Scholar] [CrossRef]
- Liu, J.H.; Nada, K.; Honda, C.; Kitashiba, H.; Wen, X.P.; Pang, X.M.; Moriguchi, T. Polyamine biosynthesis of apple callus under salt stress: Importance of the arginine decarboxylase pathway in stress response. J. Exp. Bot. 2006, 57, 2589–2599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Capell, T.; Bassie, L.; Christou, P. Modulation of the polyamine biosynthetic pathway in transgenic rice confers tolerance to drought stress. Proc. Natl. Acad. Sci. USA 2004, 101, 9909–9914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, M.; Wu, R. Arginine decarboxylase transgene expression and analysis of environmental stress tolerance in transgenic rice. Plant Sci. 2001, 160, 869–875. [Google Scholar] [CrossRef] [PubMed]
- Kou, S.; Chen, L.; Tu, W.; Scossa, F.; Wang, Y.; Liu, J.; Fernie, A.R.; Song, B.; Xie, C. The arginine decarboxylase gene ADC1, associated to the putrescine pathway, plays an important role in potato cold-acclimated freezing tolerance as revealed by transcriptome and metabolome analyses. Plant J. 2018, 96, 1283–1298. [Google Scholar] [CrossRef] [Green Version]
- Thomas, S.; Krishna, G.K.; Yadav, P.; Pal, M. Cloning and abiotic stress responsive expression analysis of arginine decarboxylase genes in contrasting rice genotypes. Indian J. Genet. Plant Breed 2019, 79, 411–419. [Google Scholar] [CrossRef]
- Song, C.M.; Wen, X.P. Using ISSR Marker to Explore the Origin of Guizhou Local Cherry (Prunus pseudocerasus Lindl. “Manaohong”). J. Beijing For. Univ. 2011, 33, 94–97. [Google Scholar]
- Chen, Z.Y.; Zheng, Y.H.; Xu, F.J. Breeding of a new early-maturing cherry (Prunus pseudocerasus Lindl) variety Manaohong. China Fruits 2013, 1, 8–10, 187. [Google Scholar]
- Tian, T.; Qiao, G.; Deng, B.; Wen, Z.; Hong, Y.; Wen, X. The effects of rain shelter coverings on the vegetative growth and fruit characteristics of Chinese cherry (Prunus pseudocerasus Lindl.). Sci. Hortic. 2019, 254, 228–235. [Google Scholar] [CrossRef]
- Wu, M.; Tian, T.; Liu, H.; Zhang, H.; Qiu, Z.; Qiao, G.; Wen, X. Shelter covering altered polyamines accumulation of cherry (Prunus pseudocerasus Lindl.) via elevating the expression of spermidine synthase (SPDS) gene. Sci. Hortic. 2020, 270, 109440. [Google Scholar] [CrossRef]
- Schmitz-Eiberger, M.A.; Blanke, M.M. Bioactive components in forced sweet cherry fruit (Prunus avium L.), antioxidative capacity and allergenic potential as dependent on cultivation under cover. LWT Food Sci. Technol. 2012, 46, 388–392. [Google Scholar] [CrossRef]
- Jin, L.F.; Guo, D.Y.; Ning, D.Y.; Hussain, S.B.; Liu, Y.Z. Covering the trees of Kinokuni tangerine with plastic film during fruit ripening improves sweetness and alters the metabolism of cell wall components. Acta Physiol. Plant. 2018, 40, 1–10. [Google Scholar] [CrossRef]
- Movahed, N.; Eshghi, S.; Jamali, B. Effects of polyamines on vegetative characteristics, growth, flowering and yield of strawberry (’Paros’ and’ Selva’). Acta Hortic. 2012, 926, 287–294. [Google Scholar] [CrossRef]
- Yariuchi, Y.; Okamoto, T.; Noutoshi, Y.; Takahashi, T. Responses of Polyamine-Metabolic Genes to Polyamines and Plant Stress Hormones in Arabidopsis Seedlings. Cells 2021, 10, 3283. [Google Scholar] [CrossRef]
- Hosseini, M.S.; Samsampour, D.; Zahedi, S.M.; Zamanian, K.; Rahman, M.M.; Mostofa, M.G.; Tran, L.P. Melatonin alleviates drought impact on growth and essential oil yield of lemon verbena by enhancing antioxidant responses, mineral balance, and abscisic acid content. Physiol Plant. 2021, 172, 1363–1375. [Google Scholar] [CrossRef]
- Zahedi, S.M.; Hosseini, M.S.; Naghmeh, D.H.M.; Abadia, J.; Germ, M.; Gholami, R.; Abdelrahman, M. Evaluation of drought tolerance in three commercial pomegranate cultivars using photosynthetic pigments, yield parameters and biochemical traits as biomarkers. Agric. Water Manag. 2022, 261, 107357. [Google Scholar] [CrossRef]
- Gholami, R.; Fahadi Hoveizeh, N.; Zahedi, S.M.; Gholami, H.; Carillo, P. Effect of three water-regimes on morpho-physiological, biochemical and yield responses of local and foreign olive cultivars under field conditions. BMC Plant Biol. 2022, 22, 477. [Google Scholar] [CrossRef]
- Takahashi, Y.; Uemura, T.; Teshima, Y. Polyamine oxidase 2 is involved in regulating excess spermidine contents during seed germination and early seedling development in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2019, 516, 1248–1251. [Google Scholar] [CrossRef]
- Zhang, Q.X.; Xu, J.G.; Bao, M.Y.; Liu, M.M.; Xie, A.Q.; Zhang, D.L.; Sun, X. Effect of waterlogging stress on root morphology and in vivo polyamine content of Paeonia lactiflora. J. Plant Physiol. 2020, 56, 1445–1457. [Google Scholar]
- Shi, H.T.; Chan, Z.L. Improvement of plant abiotic stress tolerance through modulation of the polyamine pathway. J. Integr. Plant Biol. 2014, 56, 114–121. [Google Scholar] [CrossRef] [PubMed]
- Shaddad, M.; EI-Samad, M.; Mohammed, H.T. Interactive Effects of Drought Stress and Phytohormones or Polyamines on Growth and Yield of Two M (Zea maize L.) Genotypes. Am. J. Plant Sci. 2011, 2, 790–807. [Google Scholar] [CrossRef]
- Mattoo, A.K.; Handa, A.K. Higher polyamines restore and enhance metabolic memory in ripening fruit. Plant Sci. 2008, 174, 386–393. [Google Scholar] [CrossRef]
- Vuosku, J.; Karppinen, K.; Muilu-Mäkelä, R.; Kusano, T.; Sagor, G.H.M.; Avia, K.; Alakärppä, E.; Kestilä, J.; Suokas, M.; Nickolov, K.; et al. Scots pine aminopropyltransferases shed new light on evolution of the polyamine biosynthesis pathway in seed plants. Ann. Bot. 2018, 121, 1243–1256. [Google Scholar] [CrossRef] [Green Version]
- Bortolotti, C.; Cordeiro, A.; Alcázar, R.; Borrell, A.; Culiañez-Macià, F.A.; Tiburcio, A.F.; Altabella, T. Localization of arginine decarboxylase in tobacco plants. Physiol. Plant. 2004, 120, 84–92. [Google Scholar] [CrossRef]
- Maruri-López, I.; Jiménez-Bremont, J.F. Hetero- and homodimerization of Arabidopsis thaliana arginine decarboxylase AtADC1 and AtADC2. Biochem. Biophys. Res. Commun. 2017, 484, 508–513. [Google Scholar] [CrossRef]
- Sánchez-Rangel, D.; Chávez-Martínez, A.I.; Rodríguez-Hernández, A.A.; Maruri-López, I.; Urano, K.; Shinozaki, K.; Jiménez-Bremont, J.F. Simultaneous silencing of two arginine decarboxylase genes alters development in arabidopsis. Front. Plant Sci. 2016, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Sun, P.P.; Chen, C.L.; Wang, Y.; Fu, X.Z.; Liu, J.H. An arginine decarboxylase gene PtADC from Poncirus trifoliata confers abiotic stress tolerance and promotes primary root growth in Arabidopsis. J. Exp. Bot. 2011, 62, 2899–2914. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.P.; Dong, B.H.; Zhang, Y.Y.; Liu, Z.P.; Liu, Y.L. Relationship between osmotic stress and the levels of free, conjugated and bound polyamines in leaves of wheat seedlings. Plant Sci. 2004, 166, 1261–1267. [Google Scholar] [CrossRef]
- Wang, J. Cloning and Characterization of Two Polyamine Biosynthesis Genes and Changes in Polyamines Under Abiotic Stresses in Citrus Plants. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2009. [Google Scholar]
- Wang, B.Q.; Zhang, X.N.; Li, G.H. Study of tomato dwarfism and late flowering due to overexpression of the PpADC gene in peach trees. Henan Agric. Sci. 2020, 49, 101–107. [Google Scholar]
- Del Rio, D.; Stewart, A.J.; Pellegrini, N. A review of recent studies on malondialdehyde as toxic molecule and biological marker of oxidative stress. Nutr. Metab. Cardiovasc. Dis. 2005, 15, 316–328. [Google Scholar] [CrossRef] [PubMed]
- Furlan, A.L.; Bianucci, E.; Giordano, W.; Castro, S.; Becker, D.F. Proline metabolic dynamics and implications in drought tolerance of peanut plants. Plant Physiol. Biochem. 2020, 151, 566–578. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.; Wen, X.; Wen, Z.; Qiu, Z.; Hou, Q.; Li, Z.; Mei, L.; Yu, H.; Qiao, G. Genome-wide identification and expression analysis of bHLH transcription factor family in response to cold stress in sweet cherry (Prunus avium L.). Sci. Hortic. 2021, 279, 109905. [Google Scholar] [CrossRef]
- Ghedira, R.; De Buck, S.; Nolf, J.; Depicker, A. The efficiency of Arabidopsis thaliana floral dip transformation is determined not only by the Agrobacterium strain used but also by the physiology and the ecotype of the dipped plant. Mol. Plant-Microbe Interact. 2013, 26, 823–832. [Google Scholar] [CrossRef]


| Primer Name | Primer Sequence | Purpose |
|---|---|---|
| CpADC-F1 | GCCAGCCATTACAAACTCACAA | Clone for CpADC |
| CpADC-R1 | TAGAAGAGGCGGGAATAGGG | |
| CpADC-F2 | CAGTCGTCTCACAACATGCCGGCCCTGGCTTGTTG | Subcellular localization of CpADC protein |
| CpADC-R2 | CAGTCGTCTCATACAAGCACAGCAGTAAGACCACT | |
| CpADC-qF | AGACGTTCCCAATAGTCCCGA | qRT-PCR for CpADC |
| CpADC-qR | TGATAAGCCCCGCCCAAG | |
| CpADC-cF | CAGTCGTCTCACAACATGCCGGCCCTGGCTTGTTG | Constructing plant expression vector |
| CpADC-cR | CAGTCGTCTCATACATCAAGCACAGCAGTAAGACC | |
| EF-F | TGAGAGGCTGACTGTGCTGTTC | Internal reference gene for qRT-PCR |
| EF-R | GGAGTAGTGGCATCCATCTTGTT | |
| CpADC-JF | CCCACCCACGAGGAGCAT | Characterization of transgenic Agrobacterium tumefaciens |
| CpADC-JR | GCGAGCAAACAGAGCCAGAG | |
| At-action7-F | AGCTAGAGACAGCCAAGAGC | Internal reference gene for transgenic line qRT-PCR |
| At-action7-R | GCTTCCATTCCGATGAGCGA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ran, J.; Shang, C.; Mei, L.; Li, S.; Tian, T.; Qiao, G. Overexpression of CpADC from Chinese Cherry (Cerasus pseudocerasus Lindl. ‘Manaohong’) Promotes the Ability of Response to Drought in Arabidopsis thaliana. Int. J. Mol. Sci. 2022, 23, 14943. https://doi.org/10.3390/ijms232314943
Ran J, Shang C, Mei L, Li S, Tian T, Qiao G. Overexpression of CpADC from Chinese Cherry (Cerasus pseudocerasus Lindl. ‘Manaohong’) Promotes the Ability of Response to Drought in Arabidopsis thaliana. International Journal of Molecular Sciences. 2022; 23(23):14943. https://doi.org/10.3390/ijms232314943
Chicago/Turabian StyleRan, Jiaxin, Chunqiong Shang, Lina Mei, Shuang Li, Tian Tian, and Guang Qiao. 2022. "Overexpression of CpADC from Chinese Cherry (Cerasus pseudocerasus Lindl. ‘Manaohong’) Promotes the Ability of Response to Drought in Arabidopsis thaliana" International Journal of Molecular Sciences 23, no. 23: 14943. https://doi.org/10.3390/ijms232314943






