Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.)
Abstract
:1. Introduction
2. Results
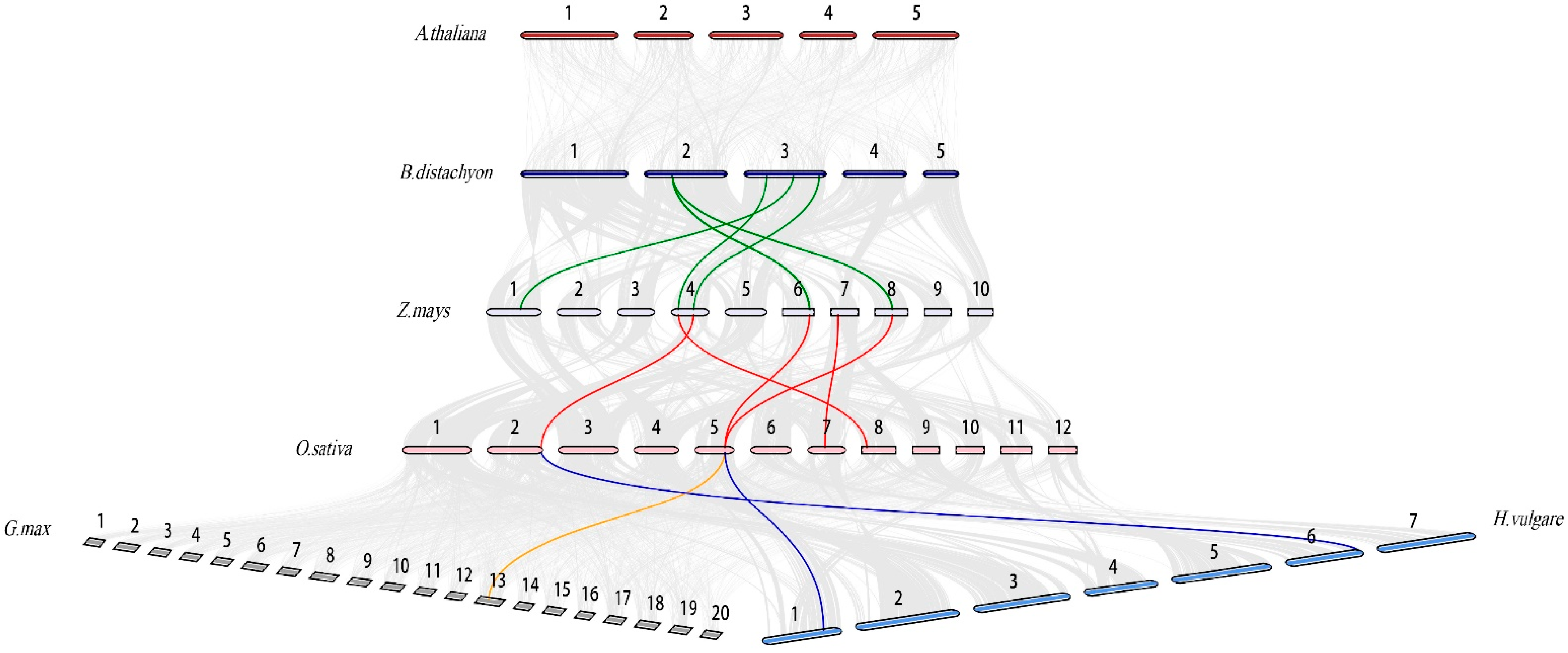
2.1. Physicochemical Properties, Chromosomal Distribution and Synteny Analysis of OsCOMTs
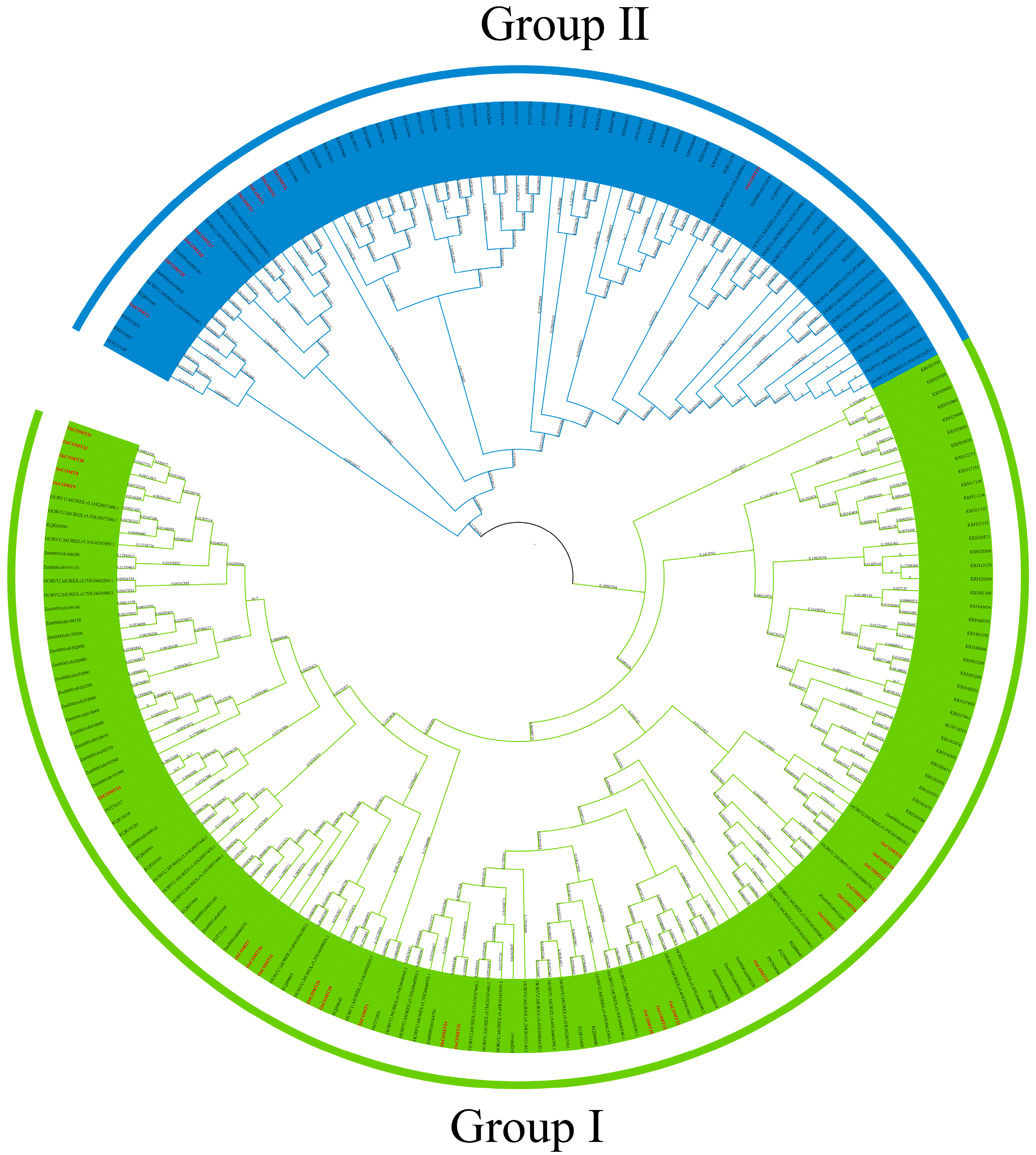
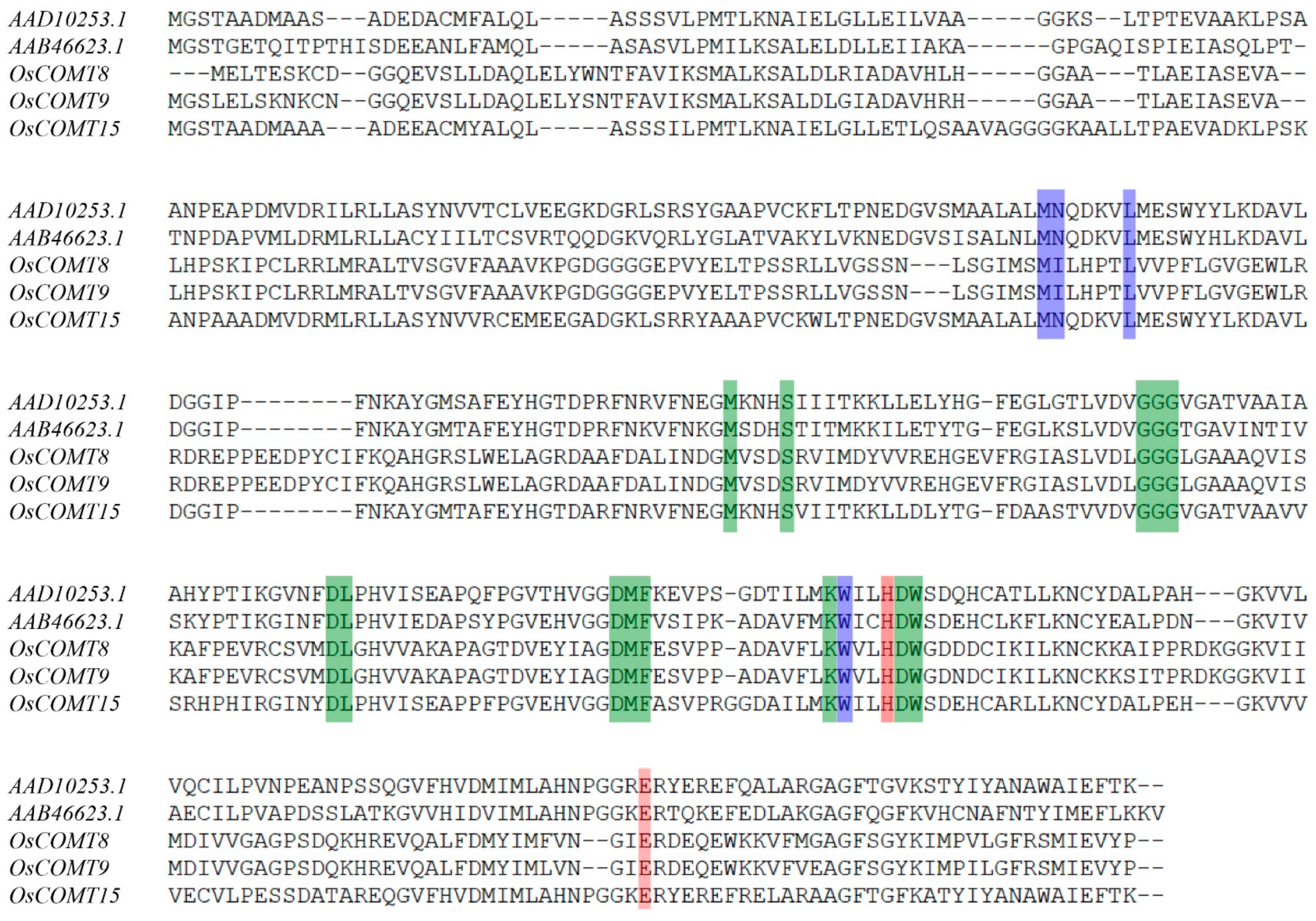
2.2. Phylogenetic Analysis, Motif, Conserved Domain and Gene Structure of COMT Genes in O. sativa
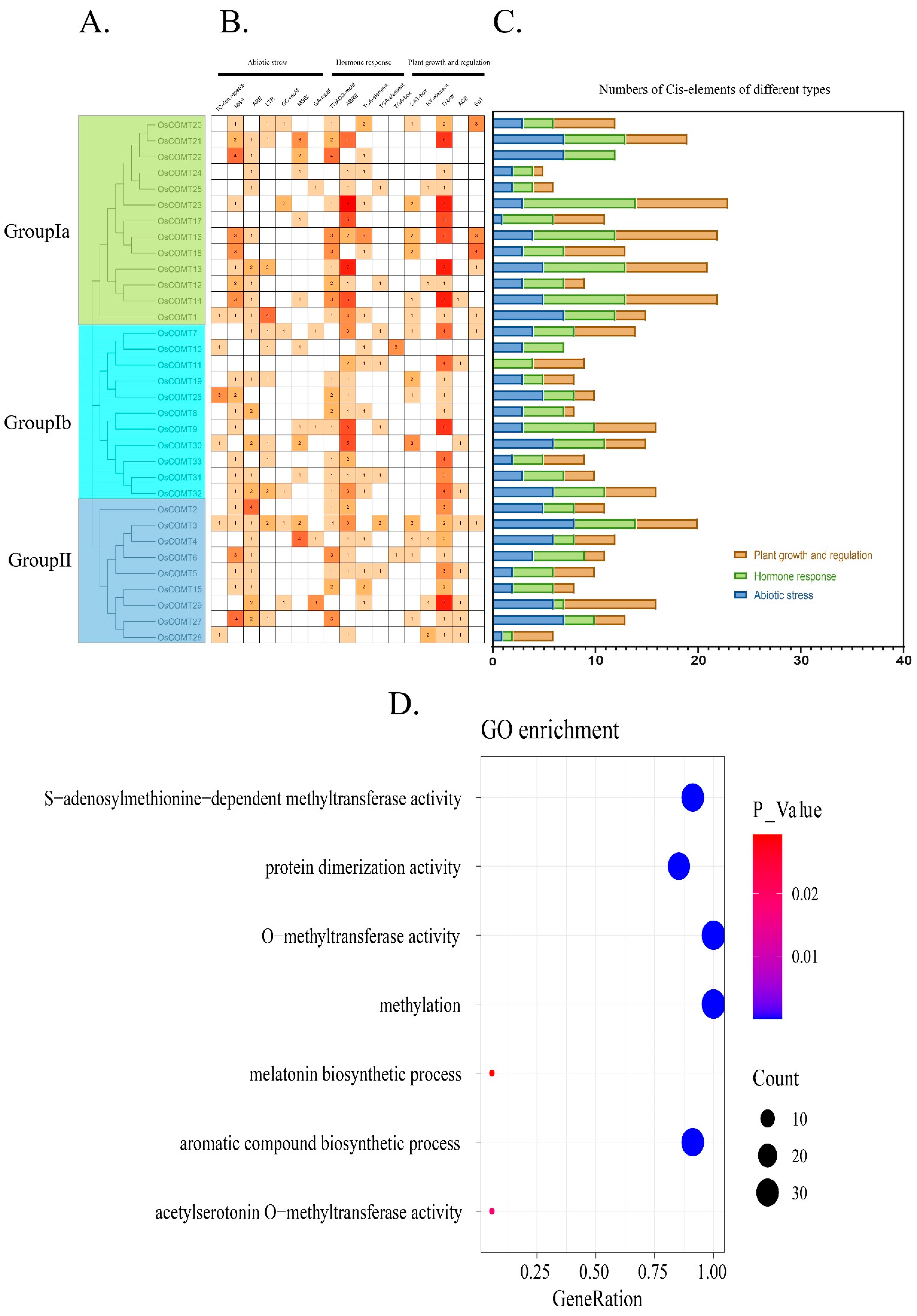
2.3. Cis-Acting Elements, Sequence Variation and Gene Ontology Annotation of OsCOMTs
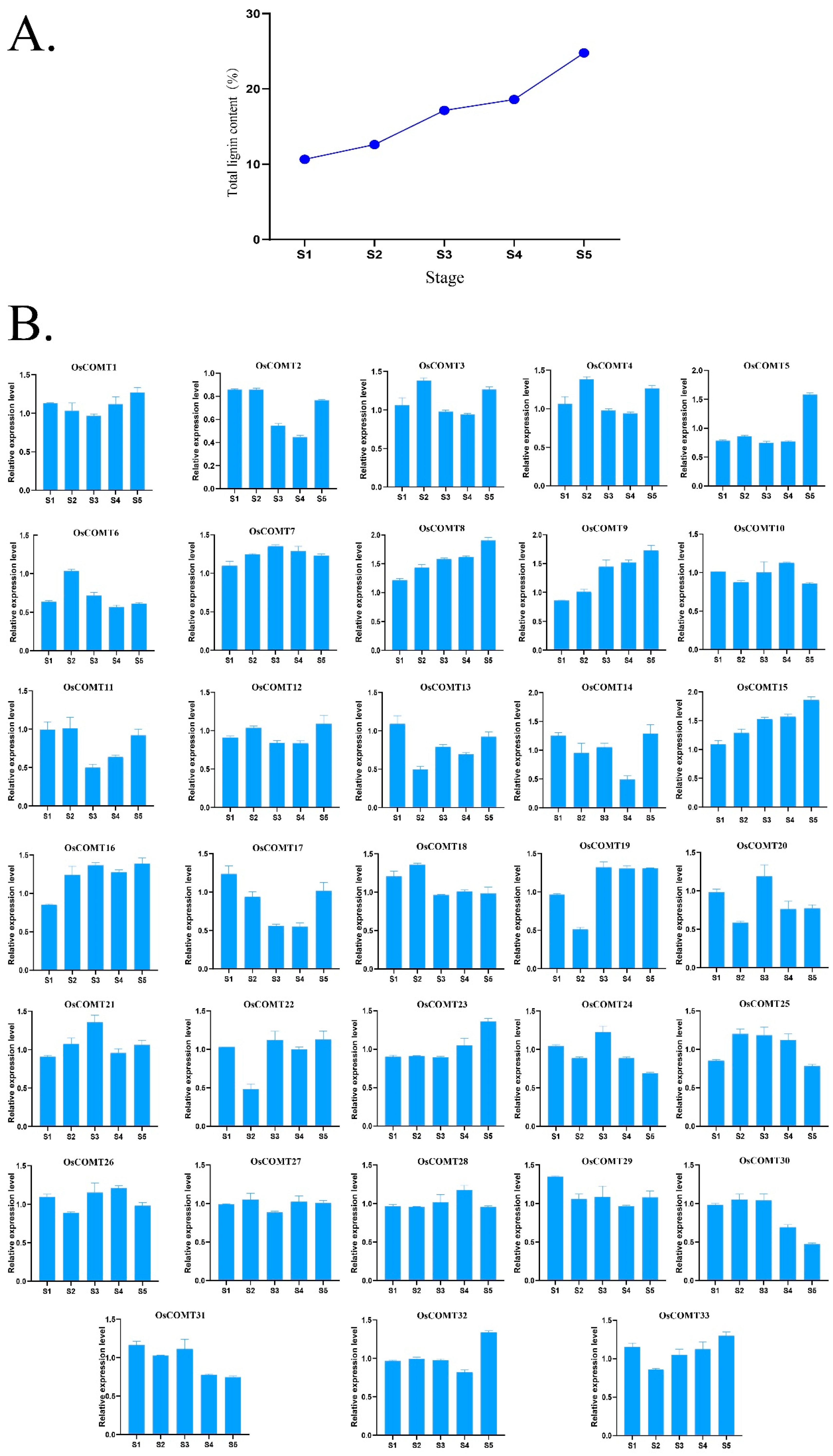
2.4. Expression Profiling of OsCOMTs
2.5. Spatial Expression of OsCOMTs in Stem
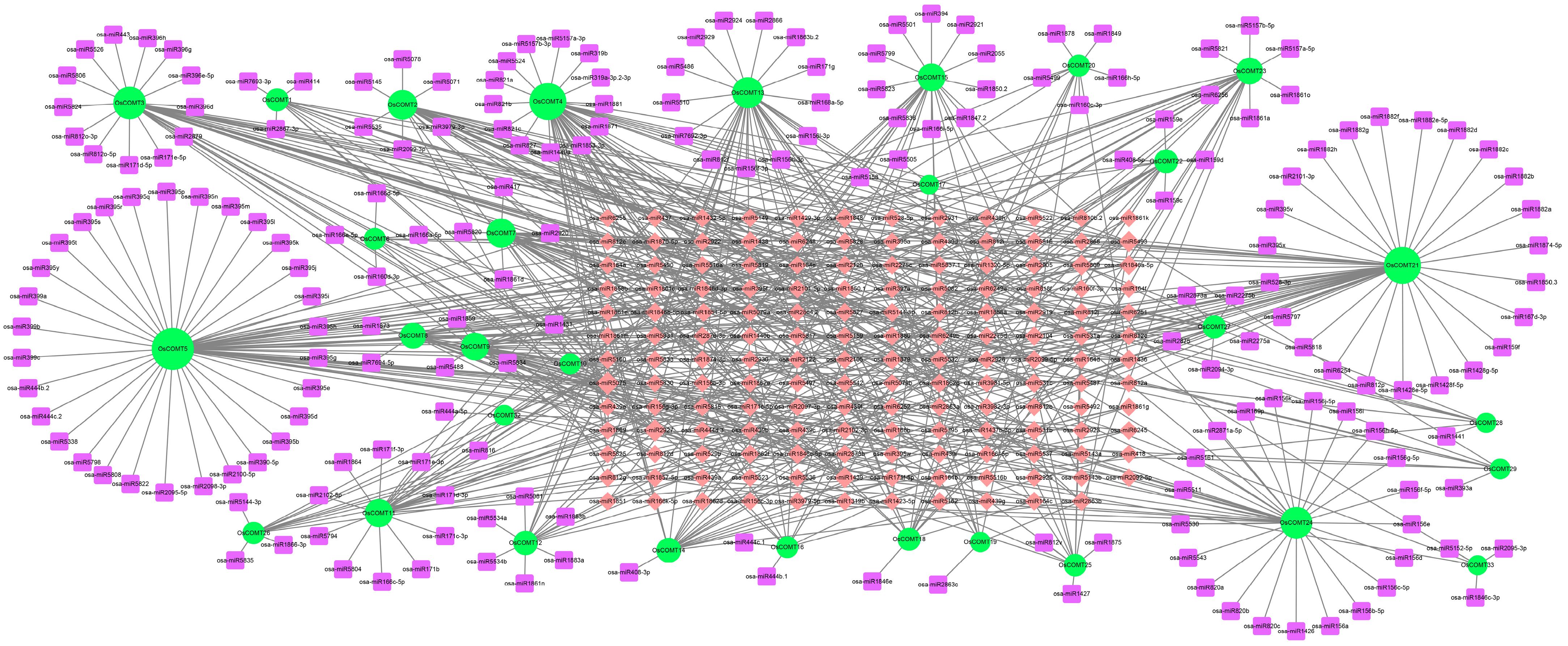
2.6. Network of Protein Interaction and miRNAs
3. Discussion
4. Materials and Methods
4.1. Identification of COMT Family Members in O. sativa and in Other Plants
4.2. Gene Structure, Conserved Motifs, and Phylogenetic Analysis
4.3. Gene Duplication Analysis
4.4. Cis-Acting Elements Analysis and miRNAs
4.5. Expression Analysis of COMT Genes in O. sativa
4.6. Plant Growth Conditions and Treatments
4.7. Expression Analysis of OsCOMTs in Rice by qRT-PCR
4.8. Lignin Content Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Recent advances in the dissection of drought-stress regulatory networks and strategies for development of drought-tolerant transgenic rice plants. Front. Plant Sci. 2015, 6, 84. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, H.; Wu, W.; Liu, H.; Wang, J.; Jia, Y.; Li, J.; Yang, L.; Lei, L.; Zou, D.; et al. QTL Mapping and Candidate Gene Analysis for Alkali Tolerance in Japonica Rice at the bud Stage Based on Linkage Mapping and Genome-Wide Association Study. Rice 2020, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Luis Espinoza-Acosta, J.; Torres-Chavez, P.I.; Olmedo-Martinez, J.L.; Vega-Rios, A.; Flores-Gallardo, S.; Armando Zaragoza-Contreras, E. Lignin in storage and renewable energy applications: A review. J. Energy Chem. 2018, 27, 1422–1438. [Google Scholar] [CrossRef]
- Cabane, M.; Afif, D.; Hawkins, S. Lignins and Abiotic Stresses. In Lignins: Biosynthesis, Biodegradation and Bioengineering; Jouann, L., Lapierre, C., Eds.; Academic Press: Cambridge, MA, USA, 2012; Volume 61, pp. 219–262. [Google Scholar]
- Ralph, J.; Lapierre, C.; Boerjan, W. Lignin structure and its engineering. Curr. Opin. Biotechnol. 2019, 56, 240–249. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, K.; Kim, H.; Hou, G.; Zhang, X.; Yang, H.; Feng, H.; Miller, L.; Ralph, J.; Liu, C.-J. Enhancing digestibility and ethanol yield of Populus wood via expression of an engineered monolignol 4-O-methyltransferase. Nat. Commun. 2016, 7, 11989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, Y.; Xu, H.; Zhao, Y.; Wu, H.; Lin, J. Plant lignification and its regulation. Sci. Sin. Vitae 2020, 50, 111–122. [Google Scholar]
- Guo, G.; Bai, F.; Liu, W.; Bi, C. Advances in Research of the Regulation of Transcription Factors of Lignin Biosynthesis. Sci. Agric. Sin. 2015, 48, 1277–1287. [Google Scholar]
- Dauwe, R.; Morreel, K.; Goeminne, G.; Gielen, B.; Rohde, A.; Van Beeumen, J.; Ralph, J.; Boudet, A.-M.; Kopka, J.; Rochange, S.F.; et al. Molecular phenotyping of lignin-modified tobacco reveals associated changes in cell-wall metabolism, primary metabolism, stress metabolism and photorespiration. Plant J. 2007, 52, 263–285. [Google Scholar] [CrossRef]
- Chun, H.J.; Baek, D.; Cho, H.M.; Lee, S.H.; Jin, B.J.; Yun, D.-J.; Hong, Y.-S.; Kim, M.C. Lignin biosynthesis genes play critical roles in the adaptation of Arabidopsis plants to high-salt stress. Plant Signal. Behav. 2019, 14, 1625697. [Google Scholar] [CrossRef] [PubMed]
- Zhao, D.; Luan, Y.; Shi, W.; Zhang, X.; Meng, J.; Tao, J. A Paeonia ostii caffeoyl-CoA O-methyltransferase confers drought stress tolerance by promoting lignin synthesis and ROS scavenging. Plant Sci. 2021, 303, 110765. [Google Scholar] [CrossRef] [PubMed]
- Whetten, R.; Sederoff, R. Lignin Biosynthesis. Plant Cell 1995, 7, 1001–1013. [Google Scholar] [CrossRef]
- Schubert, H.L.; Blumenthal, R.M.; Cheng, X.D. Many paths to methyltransfer: A chronicle of convergence. Trends Biochem. Sci. 2003, 28, 329–335. [Google Scholar] [CrossRef] [Green Version]
- Kozbial, P.Z.; Mushegian, A.R. Natural history of S-adenosylmethionine-binding proteins. BMC Struct. Biol. 2005, 5, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Vanholme, R.; De Meester, B.; Ralph, J.; Boerjan, W. Lignin biosynthesis and its integration into metabolism. Curr. Opin. Biotechnol. 2019, 56, 230–239. [Google Scholar] [CrossRef]
- Byeon, Y.; Lee, H.Y.; Lee, K.; Back, K. Caffeic acid O-methyltransferase is involved in the synthesis of melatonin by methylating N-acetylserotonin in Arabidopsis. J. Pineal Res. 2014, 57, 219–227. [Google Scholar] [CrossRef]
- Li, R.; Yang, R.; Zheng, W.; Wu, L.; Zhang, C.; Zhang, H. Melatonin Promotes SGT1-Involved Signals to Ameliorate Drought Stress Adaption in Rice. Int. J. Mol. Sci. 2022, 23, 599. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.Y.; Byeon, Y.; Lee, K.; Lee, H.-J.; Back, K. Cloning of Arabidopsis serotonin N-acetyltransferase and its role with caffeic acid O-methyltransferase in the biosynthesis of melatonin in vitro despite their different subcellular localizations. J. Pineal Res. 2014, 57, 418–426. [Google Scholar] [CrossRef]
- Li, J.-J.; Zhang, G.; Yu, J.-H.; Li, Y.-Y.; Huang, X.-H.; Wang, W.-J.; Tan, R.; Zhou, J.-Y.; Liao, H. Molecular cloning and characterization of caffeic acid 3-O-methyltransferase from the rhizome of Ligusticum chuanxiong. Biotechnol. Lett. 2015, 37, 2295–2302. [Google Scholar] [CrossRef]
- Fornale, S.; Rencoret, J.; Garcia-Calvo, L.; Encina, A.; Rigau, J.; Gutierrez, A.; Carlos del Rio, J.; Caparros-Ruiz, D. Changes in Cell Wall Polymers and Degradability in Maize Mutants Lacking 3′-and 5′-O-Methyltransferases Involved in Lignin Biosynthesis. Plant Cell Physiol. 2017, 58, 240–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, W.-J.; Du, Y.-T.; Zhou, Y.-B.; Chen, J.; Xu, Z.-S.; Ma, Y.-Z.; Chen, M.; Min, D.-H. Overexpression of TaCOMT Improves Melatonin Production and Enhances Drought Tolerance in Transgenic Arabidopsis. Int. J. Mol. Sci. 2019, 20, 652. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, Y.; Pei, J.; Li, Y.; Sun, H. Genome-wide identification and characterization of COMT gene family during the development of blueberry fruit. BMC Plant Biol. 2021, 21, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Lu, J.; Lu, K.; Yuan, J.; Huang, J.; Du, H.; Li, J. Cloning and Phylogenetic Analysis of Brassica napus L. Caffeic Acid O-Methyltransferase 1 Gene Family and Its Expression Pattern under Drought Stress. PLoS ONE 2016, 11, e0165975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Chen, B.; Wang, L.; Ali, S.; Guo, Y.; Liu, J.; Wang, J.; Xie, L.; Zhang, Q. Genome-Wide Identification and Characterization of Caffeic Acid O-Methyltransferase Gene Family in Soybean. Plants 2021, 10, 2816. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Han, Y.; Yan, Y.; Gong, B.; Shi, Q. Overexpression of caffeic acid-O-methyltransferase gene (COMT1) regulates physiological response of tomato seedlings to drought stress. Plant Physiol. J. 2019, 55, 1109–1122. [Google Scholar]
- Koshiba, T.; Hirose, N.; Mukai, M.; Yamamura, M.; Hattori, T.; Suzuki, S.; Sakamoto, M.; Umezawa, T. Characterization of 5-Hydroxyconiferaldehyde O-Methyltransferase in Oryza sativa. Plant Biotechnol. 2013, 30, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Hirano, K.; Aya, K.; Kondo, M.; Okuno, A.; Morinaka, Y.; Matsuoka, M. OsCAD2 is the major CAD gene responsible for monolignol biosynthesis in rice culm. Plant Cell Rep. 2012, 31, 91–101. [Google Scholar] [CrossRef] [PubMed]
- Petitot, A.-S.; Kyndt, T.; Haidar, R.; Dereeper, A.; Collin, M.; Engler, J.D.A.; Gheysen, G.; Fernandez, D. Transcriptomic and histological responses of African rice (Oryza glaberrima) to Meloidogyne graminicola provide new insights into root-knot nematode resistance in monocots. Ann. Bot. 2017, 119, 885–899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zubieta, C.; Kota, P.; Ferrer, J.-L.; Dixon, R.A.; Noel, J.P. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell 2002, 14, 1265–1277. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Yu, C.; Chen, C.; He, C.; Zhu, Y.; Huang, W. Identification of rice Di19 family reveals OsDi19-4 involved in drought resistance. Plant Cell Rep. 2014, 33, 2047–2062. [Google Scholar] [CrossRef]
- Radjendirane, V.; Jaiswal, A.K. Antioxidant response element-mediated 2,3,7,8-tetrachlorodibenzo-p-dioxin (TCDD) induction of human NAD(P)H:quinone oxidoreductase 1 gene expression. Biochem. Pharmacol. 1999, 58, 1649–1655. [Google Scholar] [CrossRef]
- Kumar, M.; Gho, Y.-S.; Jung, K.-H.; Kim, S.-R. Genome-Wide Identification and Analysis of Genes, Conserved between japonica and indica Rice Cultivars, that Respond to Low-Temperature Stress at the Vegetative Growth Stage. Front. Plant Sci. 2017, 8, 1120. [Google Scholar] [CrossRef] [Green Version]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Wang, Y.; Salasini, B.C.; Khan, M.; Devi, B.; Bush, M.; Subramaniam, R.; Hepworth, S.R. Clade I TGACG-Motif Binding Basic Leucine Zipper Transcription Factors Mediate BLADE-ON-PETIOLE-Dependent Regulation of Development. Plant Physiol. 2019, 180, 937–951. [Google Scholar] [CrossRef] [Green Version]
- Ezer, D.; Shepherd, S.J.K.; Brestovitsky, A.; Dickinson, P.; Cortijo, S.; Charoensawan, V.; Box, M.S.; Biswas, S.; Jaeger, K.E.; Wigge, P.A. The G-Box Transcriptional Regulatory Code in Arabidopsis. Plant Physiol. 2017, 175, 628–640. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, N.; Zheng, H.; Cui, J.; Wang, J.; Liu, H.; Sun, J.; Liu, T.; Zhao, H.; Lai, Y.; Zou, D. Genome-wide association study and candidate gene analysis of alkalinity tolerance in japonica rice germplasm at the seedling stage. Rice 2019, 12, 24. [Google Scholar] [CrossRef]
- Louie, G.V.; Bowman, M.E.; Tu, Y.; Mouradov, A.; Spangenberg, G.; Noel, J.P. Structure-Function Analyses of a Caffeic Acid O-Methyltransferase from Perennial Ryegrass Reveal the Molecular Basis for Substrate Preference. Plant Cell 2010, 22, 4114–4127. [Google Scholar] [CrossRef] [Green Version]
- Robin, A.Y.; Giustini, C.; Graindorge, M.; Matringe, M.; Dumas, R. Crystal structure of norcoclaurine-6-O-methyltransferase, a key rate-limiting step in the synthesis of benzylisoquinoline alkaloids. Plant J. 2016, 87, 641–653. [Google Scholar] [CrossRef] [Green Version]
- Bugos, R.C.; Chiang, V.L.; Campbell, W.H. cDNA cloning, sequence analysis and seasonal expression of lignin-bispecific caffeic acid/5-hydroxyferulic acid O-methyltransferase of aspen. Plant Mol. Biol. 1991, 17, 1203–1215. [Google Scholar] [CrossRef]
- He, M.-W.; Wang, Y.; Wu, J.-Q.; Shu, S.; Sun, J.; Guo, S.-R. Isolation and characterization of S-Adenosylmethionine synthase gene from cucumber and responsive to abiotic stress. Plant Physiol. Biochem. 2019, 141, 431–445. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Oh, M.W.; Komatsu, S. Characterization of S-adenosylmethionine synthetases in soybean under flooding and drought stresses. Biol. Plant. 2016, 60, 269–278. [Google Scholar] [CrossRef]
- Barros, J.; Serk, H.; Granlund, I.; Pesquet, E. The cell biology of lignification in higher plants. Ann. Bot. 2015, 115, 1053–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Curaba, J.; Spriggs, A.; Taylor, J.; Li, Z.; Helliwell, C. miRNA regulation in the early development of barley seed. BMC Plant Biol. 2012, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Wei, K.; Chen, H. Global identification, structural analysis and expression characterization of cytochrome P450 monooxygenase superfamily in rice. BMC Genomics 2018, 19, 35. [Google Scholar] [CrossRef] [Green Version]
- Kansal, S.; Mutum, R.D.; Balyan, S.C.; Arora, M.K.; Singh, A.K.; Mathur, S.; Raghuvanshi, S. Unique miRNome during anthesis in drought-tolerant indica rice var. Nagina 22. Planta 2015, 242, 365. [Google Scholar] [CrossRef] [Green Version]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-wide changes in microRNA expression during short and prolonged heat stress and recovery in contrasting rice cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive tree of life (iTOL) v3: An online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Otasek, D.; Morris, J.H.; Boucas, J.; Pico, A.R.; Demchak, B. Cytoscape Automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, J.; Gao, F.; Wu, X.; Lu, X.; Zeng, L.; Lv, J.; Su, X.; Luo, H.; Ren, G. Bph32, a novel gene encoding an unknown SCR domain-containing protein, confers resistance against the brown planthopper in rice. Sci. Rep. 2016, 6, 37645. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liang, S.; Xu, S.; Qu, D.; Yang, L.; Wang, J.; Liu, H.; Xin, W.; Zou, D.; Zheng, H. Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.). Int. J. Mol. Sci. 2022, 23, 8491. https://doi.org/10.3390/ijms23158491
Liang S, Xu S, Qu D, Yang L, Wang J, Liu H, Xin W, Zou D, Zheng H. Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.). International Journal of Molecular Sciences. 2022; 23(15):8491. https://doi.org/10.3390/ijms23158491
Chicago/Turabian StyleLiang, Shaoming, Shanbin Xu, Di Qu, Luomiao Yang, Jingguo Wang, Hualong Liu, Wei Xin, Detang Zou, and Hongliang Zheng. 2022. "Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.)" International Journal of Molecular Sciences 23, no. 15: 8491. https://doi.org/10.3390/ijms23158491
APA StyleLiang, S., Xu, S., Qu, D., Yang, L., Wang, J., Liu, H., Xin, W., Zou, D., & Zheng, H. (2022). Identification and Functional Analysis of the Caffeic Acid O-Methyltransferase (COMT) Gene Family in Rice (Oryza sativa L.). International Journal of Molecular Sciences, 23(15), 8491. https://doi.org/10.3390/ijms23158491





