Five Rice Seed-Specific NF-YC Genes Redundantly Regulate Grain Quality and Seed Germination via Interfering Gibberellin Pathway
Abstract
:1. Introduction
2. Results and Discussion
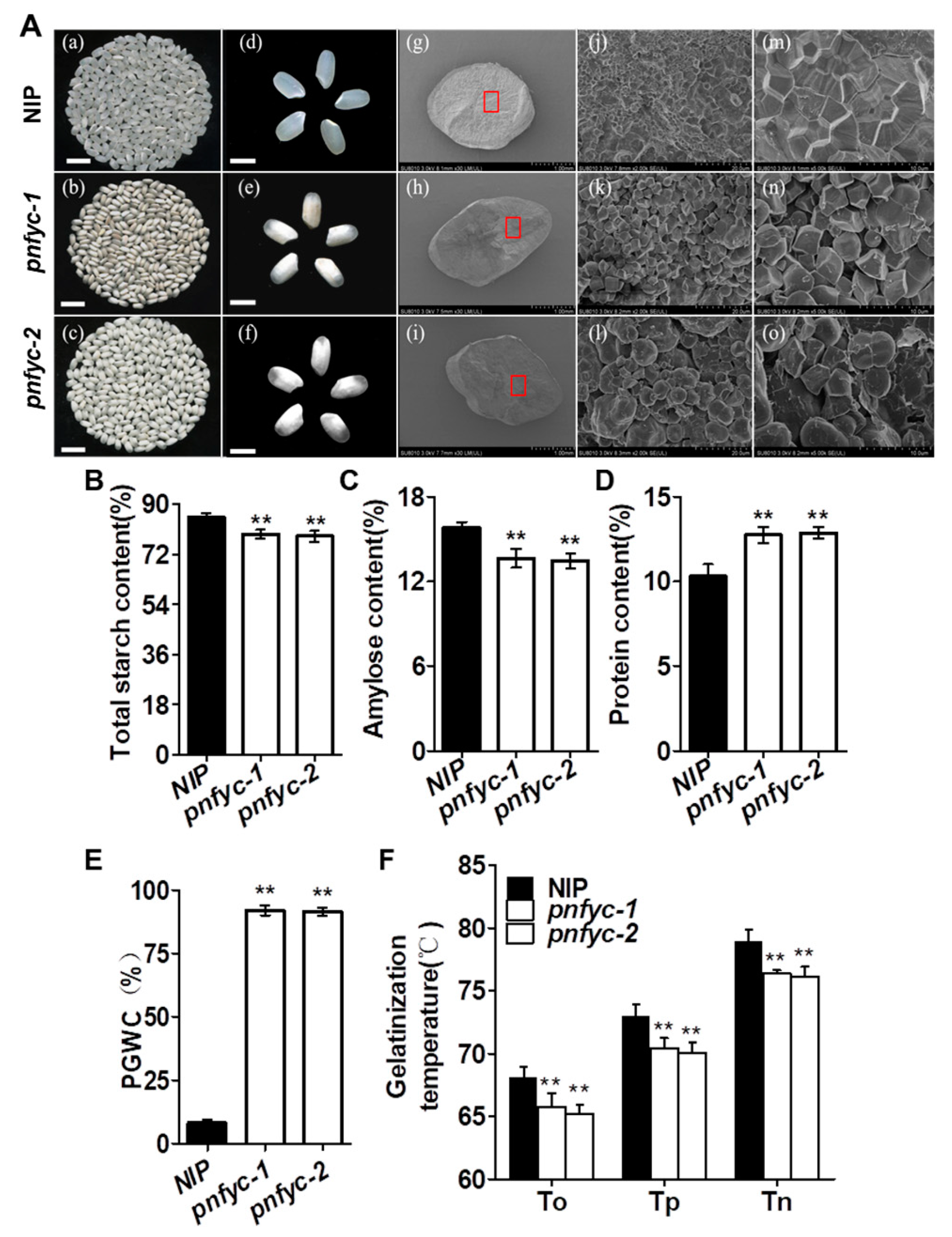
2.1. Five Seed-Specific NY-YCs Works Redundantly to Regulate Grain Quality
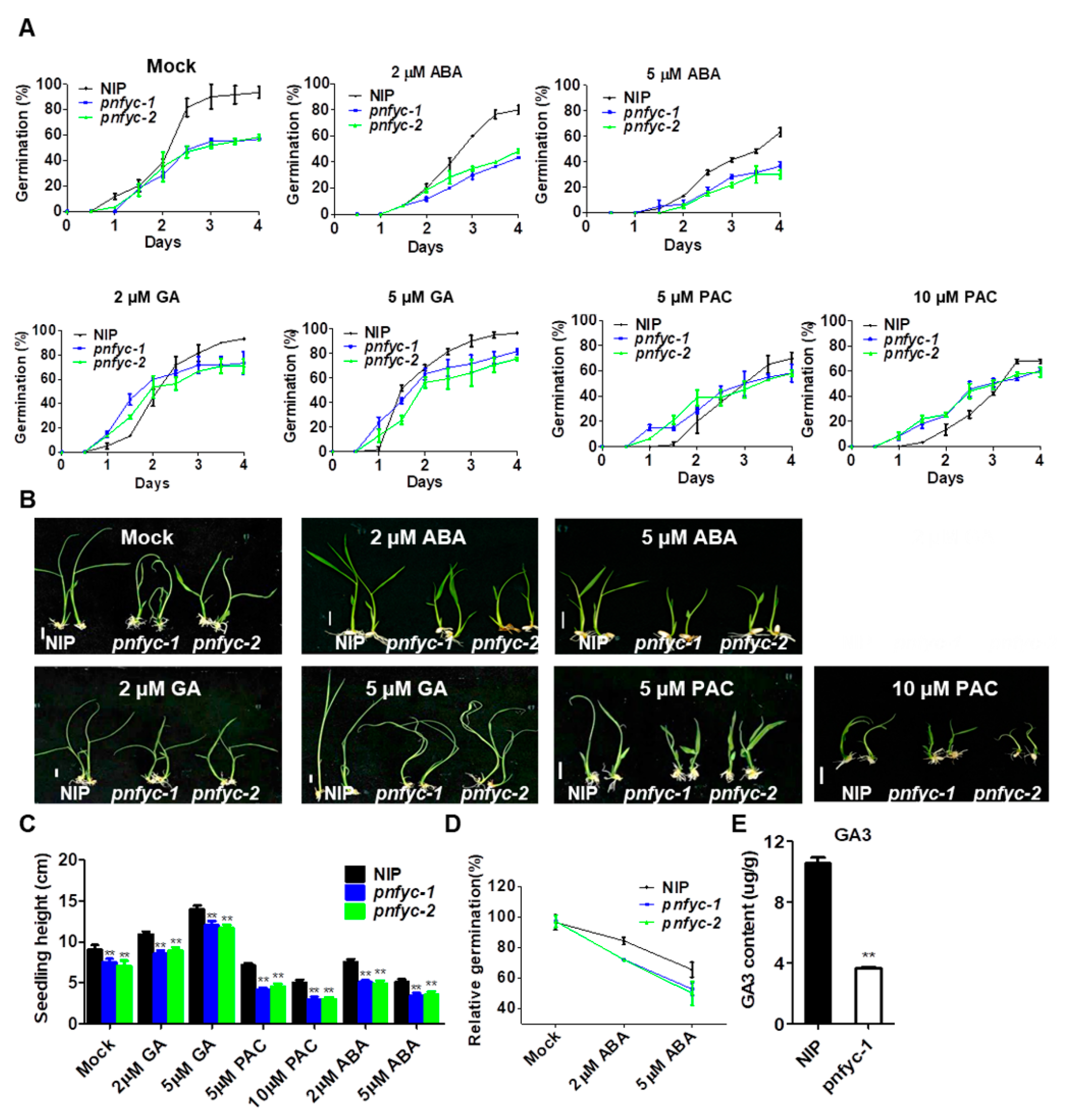
2.2. pnfyc Are GA-Deficient with Retarded Seed Germination
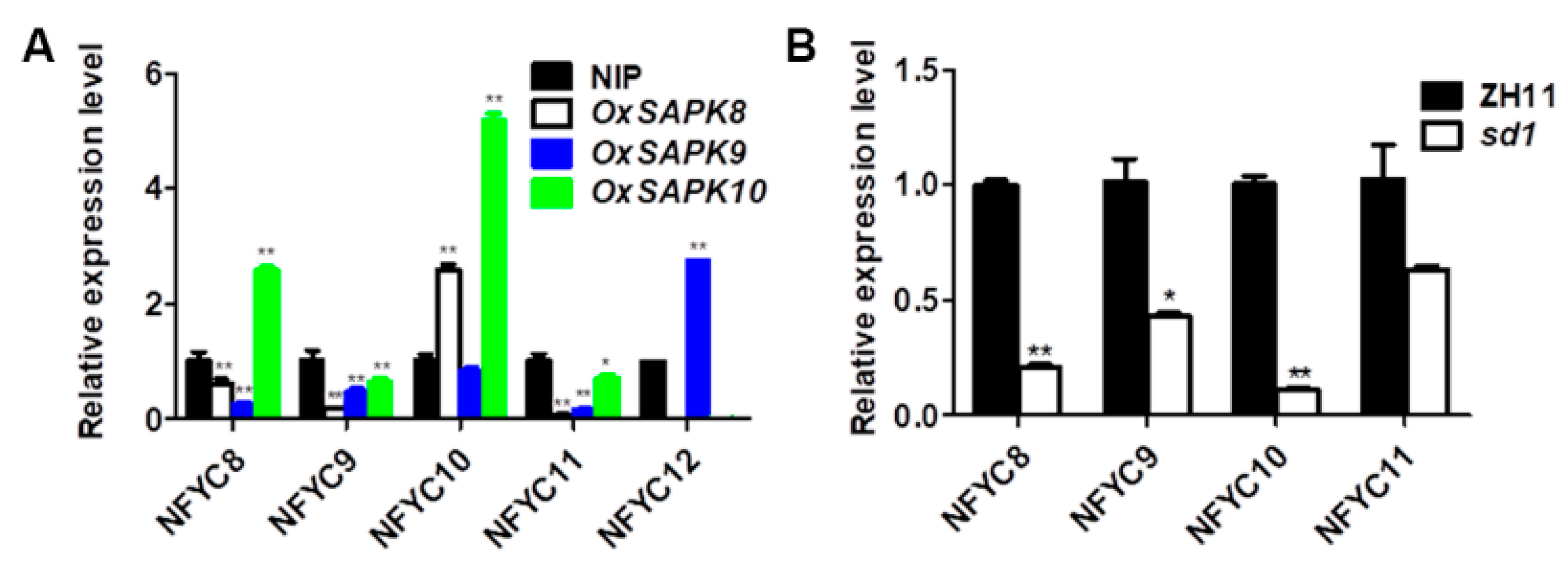
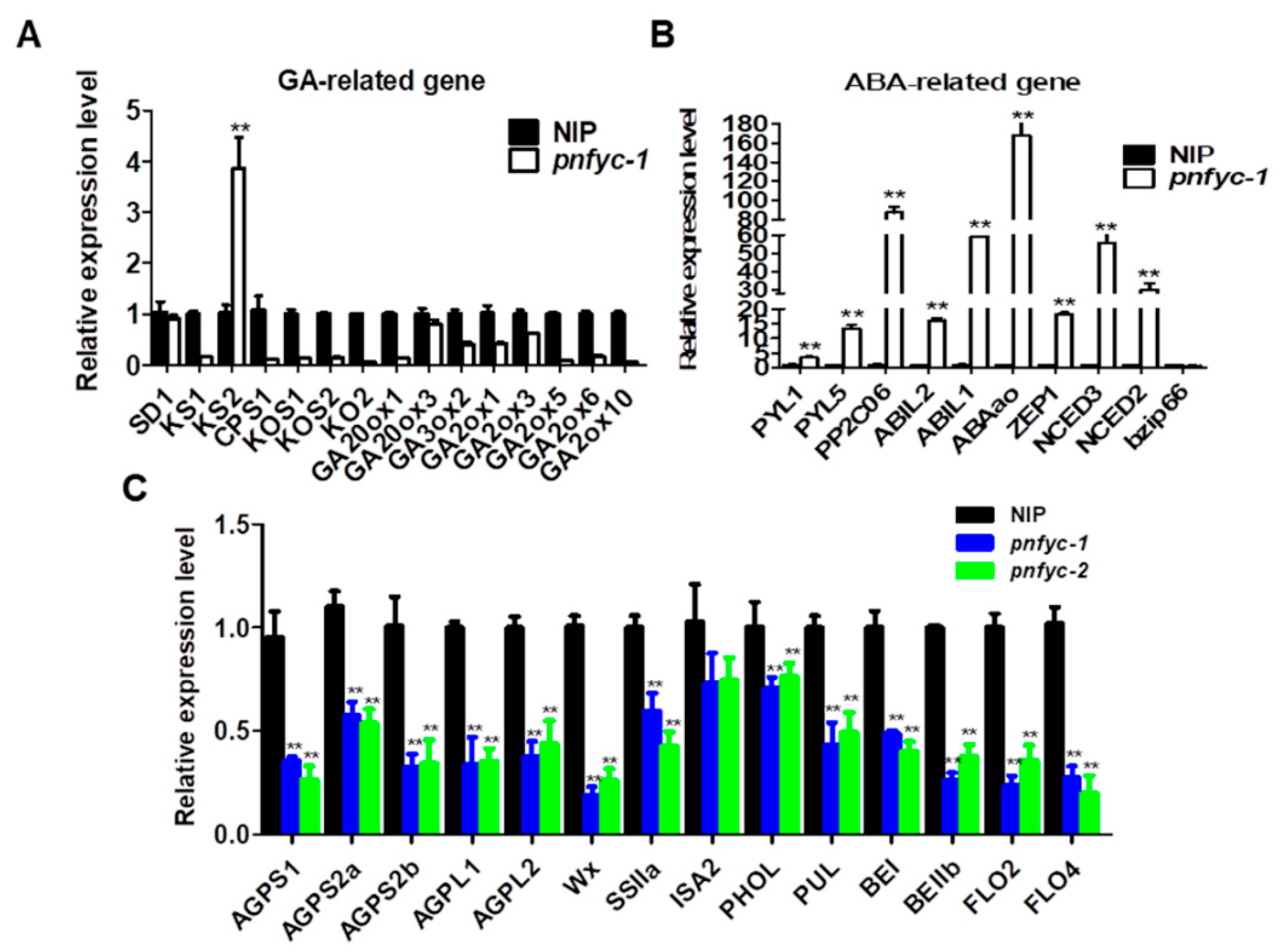
2.3. NF-YCs Regulates the Transcription of ABA and GA Pathway Genes
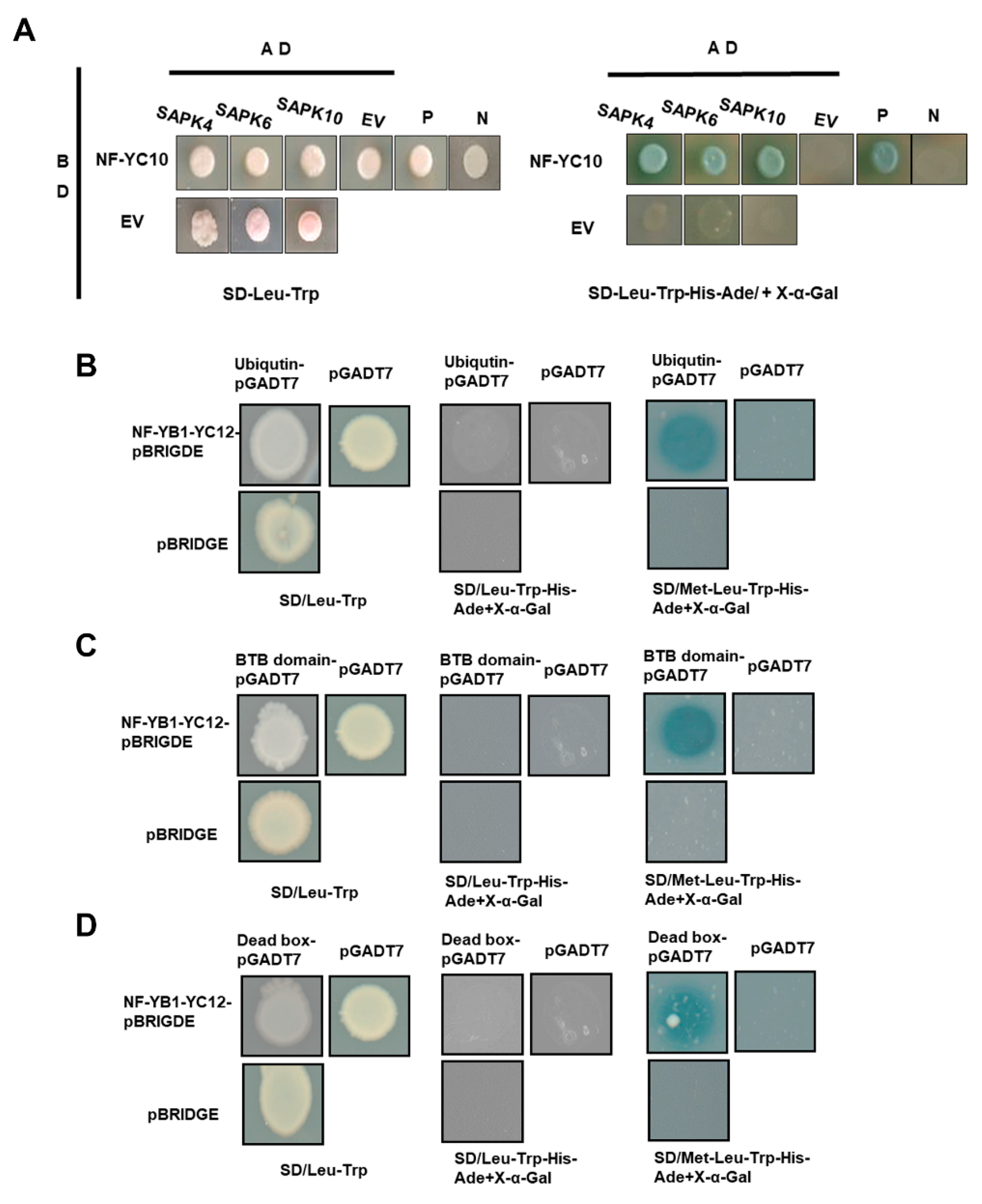
2.4. Potential Interactive Proteins of the Five NF-YCs
3. Materials and Methods
3.1. Plant Growth Conditions and Phenotype Measurement
3.2. Physicochemical Properties of Seed Grain
3.3. Scanning Electron Microscopy (SEM) Assay
3.4. Seed Germination and Phenotypic Assay
3.5. RNA-Seq Analysis and RT-PCR Analysis
3.6. Yeast-Two-Hybrid Assay
3.7. Yeast-Three-Hybrid Assay
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, N.L.; Long, T.; Yao, W.; Xiong, L.Z.; Zhang, Q.F.; Wu, C.Y. Mutant resources for the functional analysis of the rice genome. Mol. Plant 2013, 6, 596–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Locascio, A.; Roig-Villanova, I.; Bernardi, J.; Varotto, S. Current perspectives on the hormonal control of seed development in Arabidopsis and maize: A focus on auxin. Front Plant Sci. 2014, 5, 412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rijavec, T.; Dermastia, M. Cytokinins and their function in developing seeds. Acta Chim. Slov. 2010, 57, 617–629. [Google Scholar]
- Abbas, M.; Alabadi, D.; Blazquez, M.A. Differential growth at the apical hook: All roads lead to auxin. Front. Plant Sci. 2013, 4, 441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hedden, P.; Thomas, S.G. Plant Hormone Signaling. Annu. Plant Rev. 2006, 24, 229–255. [Google Scholar]
- Yang, J.; Zhang, J.; Wang, Z.; Zhu, Q.; Wang, W. Hormonal changes in the grains of rice subjected to water stress during grain filling. Plant Physiol. 2001, 127, 315–323. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; Lu, Z.; Xiong, Y.; Yao, J. Genome-wide identification and co-expression network analysis of the OsNF-Y gene family in rice. Crop J. 2017, 5, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Bello, B.K.; Hou, Y.X.; Zhao, J.; Jiao, G.A.; Wu, Y.W.; Li, Z.Y.; Wang, Y.F.; Tong, X.H.; Wang, W.; Yuan, W.Y.; et al. NF-YB1-YC12-bHLH144 complex directly activates Wx to regulate grain quality in rice (Oryza sativa L.). Plant Biotechnol. J. 2019, 17, 1222–1235. [Google Scholar] [CrossRef] [Green Version]
- Yamakawa, H.; Hirose, T.; Kuroda, M.; Yamaguchi, T. Comprehensive expression profiling of rice grain filling-related genes under high temperature using DNA microarray. Plant Physiol. 2007, 144, 258–277. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.F.; Ren, Y.; Li, W.; Wu, F.S.; Yang, W.J.; Huang, X.L.; Yao, J.L. NF-YC12 is a key multi-functional regulator of accumulation of seed storage substances in rice. J. Exp. Bot. 2019, 70, 3765–3780. [Google Scholar] [CrossRef] [Green Version]
- Manimaran, P.; Venkata Reddy, S.; Moin, M.; Raghurami Reddy, M.; Yugandhar, P.; Mohanraj, S.S.; Balachandran, S.M.; Kirti, P.B. Activation-tagging in indica rice identifies a novel transcription factor subunit, NF-YC13 associated with salt tolerance. Sci. Rep. 2017, 7, 9341. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.K.; Park, H.Y.; Jang, Y.H.; Lee, K.C.; Chung, Y.S.; Lee, J.H.; Kim, J.K. OsNF-YC2 and OsNF-YC4 proteins inhibit flowering under long-day conditions in rice. Planta 2016, 243, 563–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laloum, T.; De Mita, S.; Gamas, P.; Baudin, M.; Niebel, A. CCAAT-box binding transcription factors in plants: Y so many? Trends Plant Sci. 2013, 18, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Wu, D.; Kong, F.; Lin, K.; Zhang, H.; Li, G. The Arabidopsis thaliana nuclear factor Y transcription factors. Front. Plant Sci. 2017, 7, 2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swain, S.; Myers, Z.A.; Siriwardana, C.L.; Holt, B.F., 3rd. The multifaceted roles of NUCLEAR FACTOR-Y in Arabidopsis thaliana development and stress responses. Biochim. Biophys. Acta Gene Regul. Mech. 2017, 1860, 636–644. [Google Scholar] [CrossRef] [PubMed]
- Petroni, K.; Kumimoto, R.W.; Gnesutta, N.; Calvenzani, V.; Fornari, M.; Tonelli, C.; Holt, B.F.; Mantovani, R. The promiscuous life of plant NUCLEAR FACTOR Y transcription factors. Plant Cell 2012, 24, 4777–4792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thirumurugan, T.; Ito, Y.; Kubo, T.; Serizawa, A.; Kurata, N. Identification, characterization and interaction of HAP family genes in rice. Mol. Genet. Genom. 2008, 279, 279–289. [Google Scholar] [CrossRef]
- Lee, D.K.; Kim, H.I.; Jang, G.; Chung, P.J.; Jeong, J.S.; Kim, Y.S.; Bang, S.W.; Jung, H.; Do Choi, Y.; Kim, J.K. The NF-YA transcription factor OsNF-YA7 confers drought stress tolerance of rice in an abscisic acid independent manner. Plant Sci. 2015, 241, 199–210. [Google Scholar] [CrossRef]
- Li, L.; Zheng, W.; Zhu, Y.; Ye, H.; Tang, B.; Arendsee, Z.W.; Jones, D.; Li, R.; Ortiz, D.; Zhao, X. QQS orphan gene regulates carbon and nitrogen partitioning across species via NF-YC interactions. Proc. Natl. Acad. Sci. USA 2015, 112, 14734–14739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adachi, S.; Yoshikawa, K.; Yamanouchi, U.; Tanabata, T.; Sun, J.; Ookawa, T.; Yamamoto, T.; Sage, R.F.; Hirasawa, T.; Yonemaru, J. Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front. Plant Sci. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.H.; Wang, P.; Chen, H.X.; Zhou, H.J.; Li, Q.P.; Wang, C.R.; Ding, Z.H.; Zhang, Y.S.; Yu, S.B.; Xing, Y.Z. A major QTL, Ghd8, plays pleiotropic roles in regulating grain productivity, plant height, and heading date in rice. Mol. Plant 2011, 4, 319–330. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.J.; Xue, H.W. OsLEC1/OsHAP3E participates in the determination of meristem identity in both vegetative and reproductive developments of rice F. J. Integr. Plant Biol. 2013, 55, 232–249. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Thirumurugan, T.; Serizawa, A.; Hiratsu, K.; Ohme-Takagi, M.; Kurata, N. Aberrant vegetative and reproductive development by overexpression and lethality by silencing of OsHAP3E in rice. Plant Sci. 2011, 181, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Miyoshi, K.; Ito, Y.; Serizawa, A.; Kurata, N. OsHAP3 genes regulate chloroplast biogenesis in rice. Plant J. 2003, 36, 532–540. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.C.; Ling, S.; Lu, Z.H.; Ouyang, Y.D.; Liu, S.S.; Yao, J.L. OsNF-YB1, a rice endosperm-specific gene, is essential for cell proliferation in endosperm development. Gene 2014, 551, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Bai, A.N.; Lu, X.D.; Li, D.Q.; Liu, J.X.; Liu, C.M. NF-YB1-regulated expression of sucrose transporters in aleurone facilitates sugar loading to rice endosperm. Cell Res. 2016, 26, 384. [Google Scholar] [CrossRef] [Green Version]
- Nie, D.M.; Ouyang, Y.D.; Wang, X.; Zhou, W.; Hu, C.-G.; Yao, J. Genome-wide analysis of endosperm-specific genes in rice. Gene 2013, 530, 236–247. [Google Scholar] [CrossRef]
- Xu, J.J.; Zhang, X.F.; Xue, H.W. Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 2016, 67, 6399–6411. [Google Scholar] [CrossRef]
- Yamamoto, A.; Kagaya, Y.; Toyoshima, R.; Kagaya, M.; Takeda, S.; Hattori, T. Arabidopsis NF-YB subunits LEC1 and LEC1-LIKE activate transcription by interacting with seed-specific ABRE-binding factors. Plant J. 2009, 58, 843–856. [Google Scholar] [CrossRef] [PubMed]
- E, Z.G.; Li, T.T.; Zhang, H.Y.; Liu, Z.H.; Deng, H.; Sharma, S.; Wei, X.F.; Wang, L.; Niu, B.X.; Chen, C. A group of nuclear factor Y transcription factors are sub-functionalized during endosperm development in monocots. J. Exp. Bot. 2018, 69, 2495–2510. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.Z.; Xiong, Y.F.; Xiao, P.P.; Wang, X.; Yao, J.L. OsNF-YC10, a seed preferentially expressed gene regulates grain width by affecting cell proliferation in rice. Plant Sci. 2019, 280, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Murata, M.; Minami, H.; Yamamoto, S.; Kagaya, Y.; Hobo, T.; Yamamoto, A.; Hattori, T. Abscisic acid-activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 2005, 44, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Ayele, B.T.; Magome, H.; Lee, S.; Shin, K.; Kamiya, Y.; Soh, M.S.; Yamaguchi, S. GA-sensitive dwarf1-1D (gsd1-1D) Defines a New Mutation that Controls Endogenous GA Levels in Arabidopsis. J. Plant Growth Regul. 2014, 33, 340–354. [Google Scholar] [CrossRef]
- Cui, W.; Song, Q.; Zuo, B.; Han, Q.; Jia, Z. Effects of Gibberellin (GA4+7) in Grain Filling, Hormonal Behavior, and Antioxidants in High-Density Maize (Zea mays L.). Plants 2020, 9, 978. [Google Scholar] [CrossRef]
- Liu, X.; Hu, P.; Huang, M.; Tang, Y.; Li, Y.; Li, L.; Hou, X. The NF-YC-RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat. Commun. 2016, 7, 12768. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkelstein, R.R.; Lynch, T.J. The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 2000, 12, 599–609. [Google Scholar] [CrossRef] [Green Version]
- Carles, C.; Bies-Etheve, N.; Aspart, L.; Léon-Kloosterziel, K.M.; Koornneef, M.; Echeverria, M.; Delseny, M. Regulation of Arabidopsis thaliana Em genes: Role of ABI5. Plant J. 2002, 30, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Yang, Y.N.; Xue, L.J.; Zou, M.J.; Liu, J.Y.; Chen, F.; Xue, H.W. Rice ABI5-Like1 regulates abscisic acid and auxin responses by affecting the expression of ABRE-containing genes. Plant Physiol. 2011, 156, 1397–1409. [Google Scholar] [CrossRef] [Green Version]
- Tang, N.; Zhang, H.; Li, X.; Xiao, J.; Xiong, L. Constitutive activation of transcription factor OsbZIP46 improves drought tolerance in rice. Plant Physiol. 2012, 158, 1755–1768. [Google Scholar] [CrossRef] [Green Version]
- Joo, J.; Lee, Y.H.; Song, S.I. Overexpression of the rice basic leucine zipper transcription factor OsbZIP12 confers drought tolerance to rice and makes seedlings hypersensitive to ABA. Plant Biotechnol. Rep. 2014, 8, 431–441. [Google Scholar] [CrossRef]
- Hossain, M.A.; Lee, Y.; Cho, J.-I.; Ahn, C.-H.; Lee, S.-K.; Jeon, J.-S.; Kang, H.; Lee, C.-H.; An, G.; Park, P.B. The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 2010, 72, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Gao, F.; Xiong, A.; Peng, R.; Jin, X.; Xu, J.; Zhu, B.; Chen, J.; Yao, Q. OsNAC52, a rice NAC transcription factor, potentially responds to ABA and confers drought tolerance in transgenic plants. Plant Cell Tissue Organ Cult. (PCTOC) 2010, 100, 255–262. [Google Scholar] [CrossRef]
- Zou, J.; Liu, A.; Chen, X.; Zhou, X.; Gao, G.; Wang, W.; Zhang, X. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J. Plant Physiol. 2009, 166, 851–861. [Google Scholar] [CrossRef]
- Lee, S.-K.; Hwang, S.-K.; Han, M.; Eom, J.-S.; Kang, H.-G.; Han, Y.; Choi, S.-B.; Cho, M.-H.; Bhoo, S.H.; An, G. Identification of the ADP-glucose pyrophosphorylase isoforms essential for starch synthesis in the leaf and seed endosperm of rice (Oryza sativa L.). Plant Mol. Biol. 2007, 65, 531–546. [Google Scholar] [CrossRef]
- Tang, X.J.; Peng, C.; Zhang, J.; Cai, Y.; You, X.-M.; Kong, F.; Yan, H.-G.; Wang, G.-X.; Wang, L.; Jin, J. ADP-glucose pyrophosphorylase large subunit 2 is essential for storage substance accumulation and subunit interactions in rice endosperm. Plant Sci. 2016, 249, 70–83. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Jiao, G.; Lin, H.; Sheng, Z.; Shao, G.; Xie, L.; Tang, S.; Xu, Q.; Hu, P. GRAIN INCOMPLETE FILLING 2 regulates grain filling and starch synthesis during rice caryopsis development. J. Integr. Plant Biol. 2017, 59, 134–153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ryoo, N.; Yu, C.; Park, C.-S.; Baik, M.-Y.; Park, I.M.; Cho, M.-H.; Bhoo, S.H.; An, G.; Hahn, T.-R.; Jeon, J.-S. Knockout of a starch synthase gene OsSSIIIa/Flo5 causes white-core floury endosperm in rice (Oryza sativa L.). Plant Cell Rep. 2007, 26, 1083–1095. [Google Scholar] [CrossRef]
- Ma, X.; Zhang, Q.; Zhu, Q.; Liu, W.; Chen, Y.; Qiu, R.; Wang, B.; Yang, Z.; Li, H.; Lin, Y. A robust CRISPR/Cas9 system for convenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol. Plant 2015, 8, 1274–1284. [Google Scholar] [CrossRef]
- Kang, H.G.; Park, S.; Matsuoka, M.; An, G. White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J. 2005, 42, 901–911. [Google Scholar] [CrossRef] [Green Version]
- Nishi, A.; Nakamura, Y.; Tanaka, N.; Satoh, H. Biochemical and genetic analysis of the effects ofamylose-extender mutation in rice endosperm. Plant Physiol. 2001, 127, 459–472. [Google Scholar] [CrossRef] [PubMed]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, H.; Li, S.; Kazeem, B.B.; Ajadi, A.A.; Luo, J.; Yin, M.; Liu, X.; Chen, L.; Ying, J.; Tong, X.; et al. Five Rice Seed-Specific NF-YC Genes Redundantly Regulate Grain Quality and Seed Germination via Interfering Gibberellin Pathway. Int. J. Mol. Sci. 2022, 23, 8382. https://doi.org/10.3390/ijms23158382
Xu H, Li S, Kazeem BB, Ajadi AA, Luo J, Yin M, Liu X, Chen L, Ying J, Tong X, et al. Five Rice Seed-Specific NF-YC Genes Redundantly Regulate Grain Quality and Seed Germination via Interfering Gibberellin Pathway. International Journal of Molecular Sciences. 2022; 23(15):8382. https://doi.org/10.3390/ijms23158382
Chicago/Turabian StyleXu, Huayu, Shufan Li, Bello Babatunde Kazeem, Abolore Adijat Ajadi, Jinjin Luo, Man Yin, Xinyong Liu, Lijuan Chen, Jiezheng Ying, Xiaohong Tong, and et al. 2022. "Five Rice Seed-Specific NF-YC Genes Redundantly Regulate Grain Quality and Seed Germination via Interfering Gibberellin Pathway" International Journal of Molecular Sciences 23, no. 15: 8382. https://doi.org/10.3390/ijms23158382
APA StyleXu, H., Li, S., Kazeem, B. B., Ajadi, A. A., Luo, J., Yin, M., Liu, X., Chen, L., Ying, J., Tong, X., Wang, Y., Niu, B., Chen, C., Zeng, X., & Zhang, J. (2022). Five Rice Seed-Specific NF-YC Genes Redundantly Regulate Grain Quality and Seed Germination via Interfering Gibberellin Pathway. International Journal of Molecular Sciences, 23(15), 8382. https://doi.org/10.3390/ijms23158382









