LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis
Abstract
1. Introduction
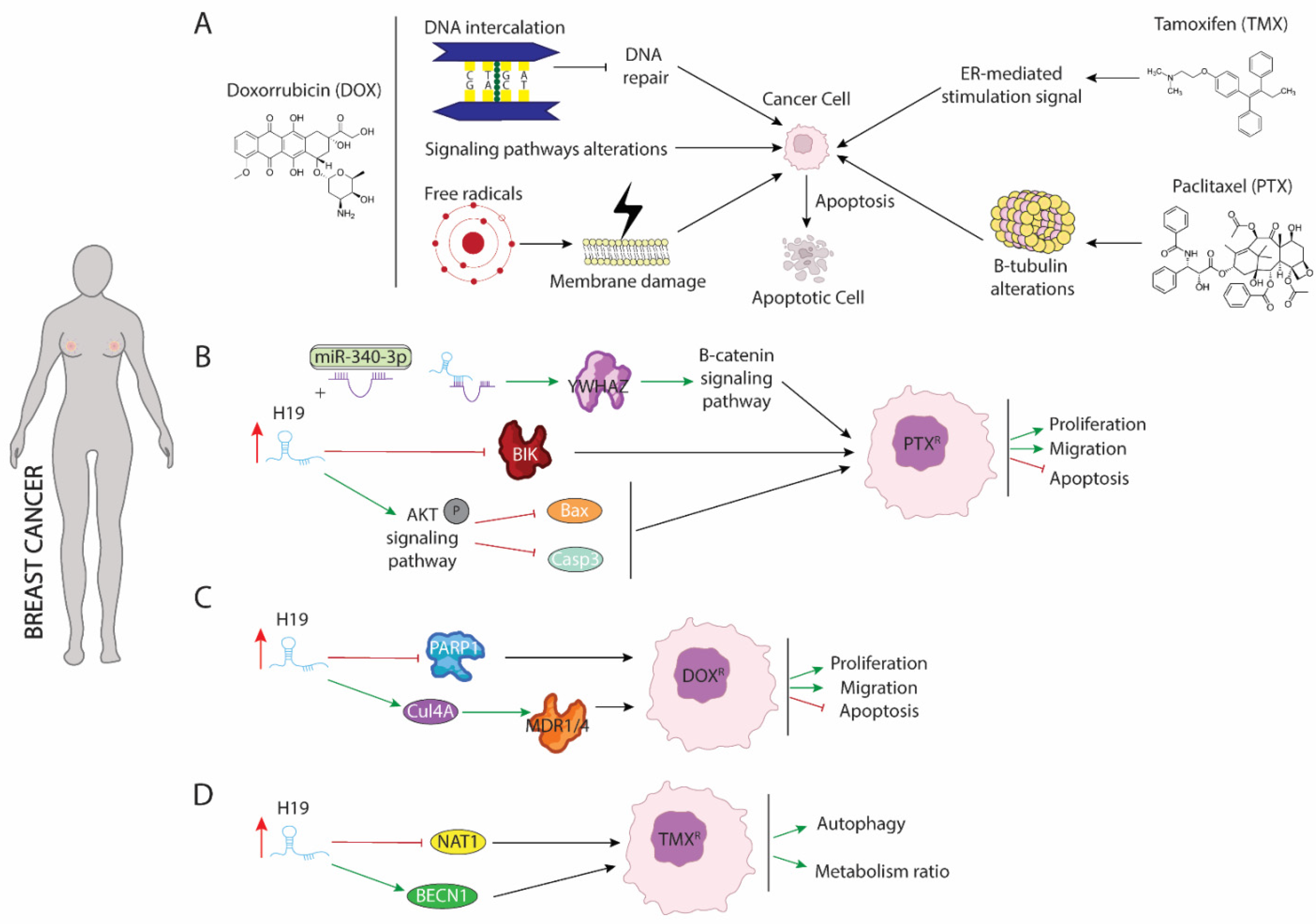
2. LncRNA H19 Impairs Chemo and Radiotherapy in Breast Cancer
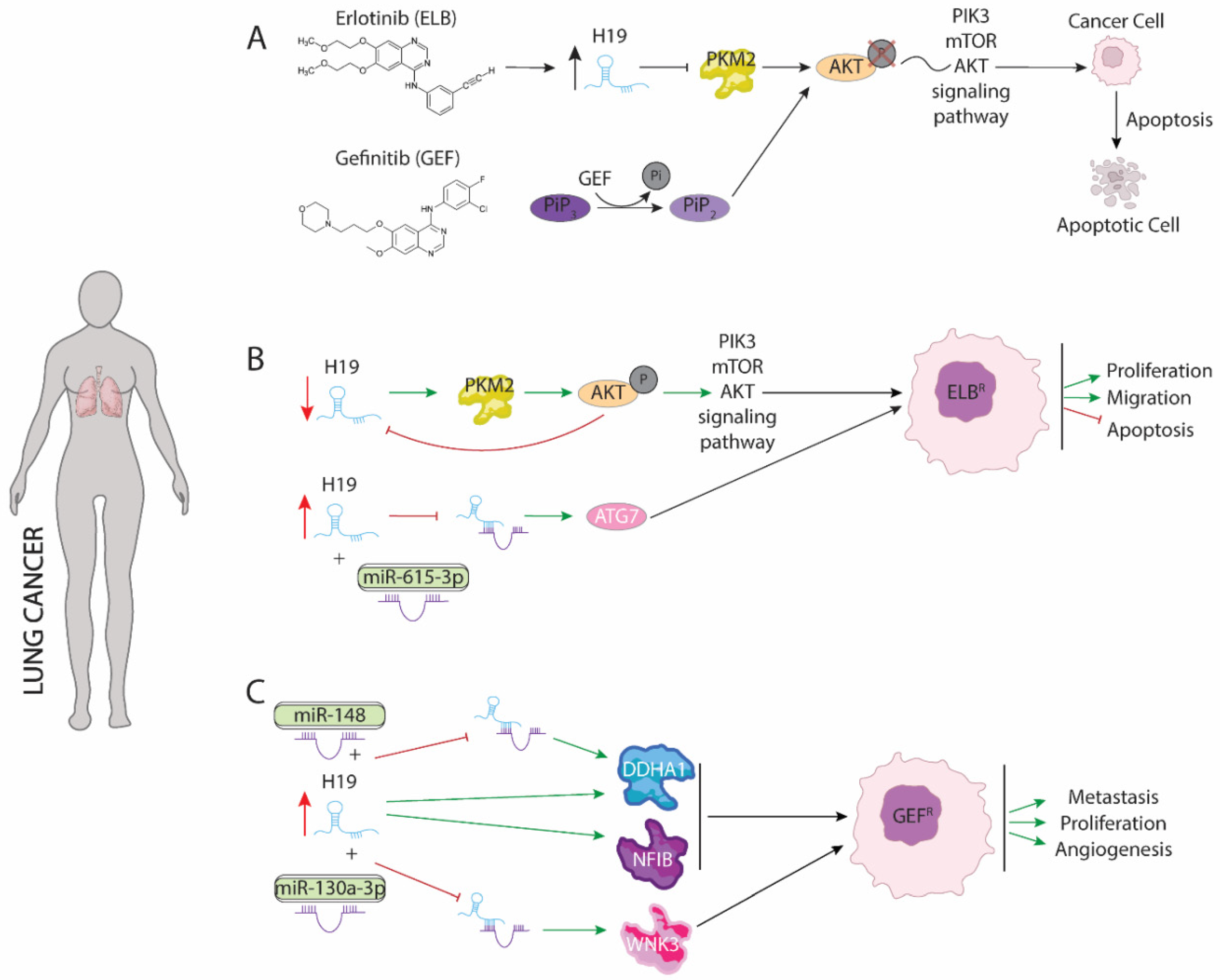
3. LncRNA H19 Impairs Chemo and Radiotherapy in Non-Small Cell Lung Cancer (NSCLC)
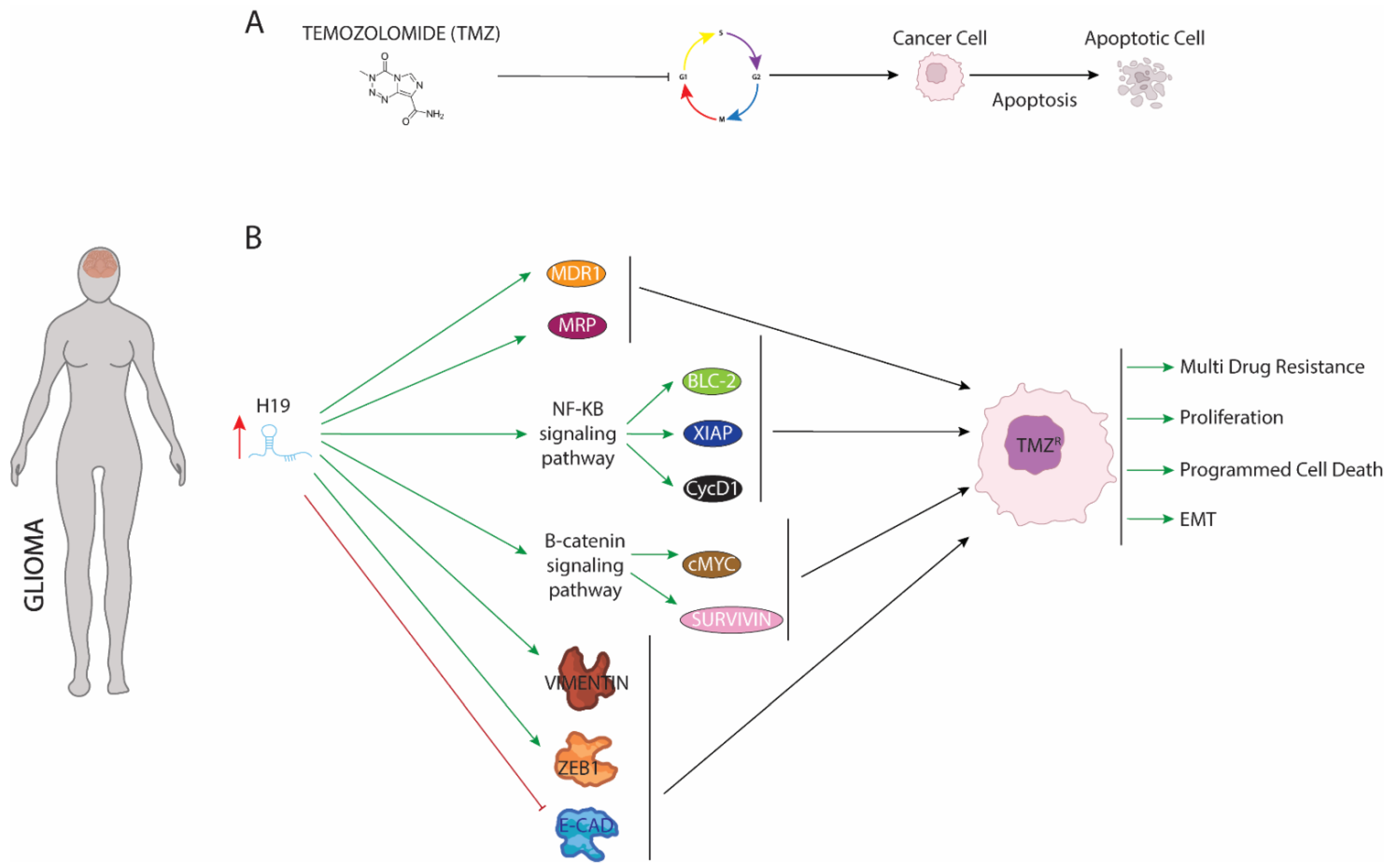
4. LncRNA H19 Impairs Chemo and Radiotherapy in Glioma
5. LncRNA H19 Impairs Chemo and Radiotherapy in Colorectal Cancer
6. Discussion and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Carninci, P.; Kasukawa, T.; Katayama, S.; Gough, J.; Frith, M.C.; Maeda, N.; Oyama, R.; Ravasi, T.; Lenhard, B.; Wells, C.; et al. The transcriptional landscape of the mammalian genome. Science 2005, 309, 1559–1563. [Google Scholar] [CrossRef]
- Harrow, J.; Frankish, A.; Gonzalez, J.M.; Tapanari, E.; Diekhans, M.; Kokocinski, F.; Aken, B.L.; Barrell, D.; Zadissa, A.; Searle, S.; et al. GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 2012, 22, 1760–1774. [Google Scholar] [CrossRef]
- García-Padilla, C.; Aránega, A.; Franco, D. The role of long non-coding RNAs in cardiac development and disease. AIMS Genet. 2018, 5, 124–140. [Google Scholar] [CrossRef]
- Expósito-Villén, A.; Aránega, A.E.; Franco, D. Functional Role of Non-Coding RNAs during Epithelial-To-Mesenchymal Transition. Non-Coding RNA 2018, 4, 14. [Google Scholar] [CrossRef]
- Shi, X.; Sun, M.; Liu, H.; Yao, Y.; Song, Y. Long non-coding RNAs: A new frontier in the study of human diseases. Cancer Lett. 2013, 339, 159–166. [Google Scholar] [CrossRef]
- Statello, L.; Guo, C.-J.; Chen, L.-L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Scott, M.S.; Ono, M. From snoRNA to miRNA: Dual function regulatory non-coding RNAs. Biochimie 2011, 93, 1987–1992. [Google Scholar] [CrossRef]
- Ma, L.; Bajic, V.B.; Zhang, Z. On the classification of long non-coding RNAs. RNA Biol. 2013, 10, 924–933. [Google Scholar] [CrossRef]
- Singh, S.R.; Rameshwar, P. MicroRNA in Development and in the Progression of Cancer; Springer: New York, NY, USA, 2014. [Google Scholar]
- Lu, M.; Zhang, Q.; Deng, M.; Miao, J.; Guo, Y.; Gao, W.; Cui, Q. An Analysis of Human MicroRNA and Disease Associations. PLoS ONE 2008, 3, e3420. [Google Scholar] [CrossRef]
- Garcia-Padilla, C.; Garcia-Lopez, V.; Aranega, A.; Franco, D.; Garcia-Martinez, V.; Lopez-Sanchez, C. Inhibition of RhoA and Cdc42 by miR-133a Modulates Retinoic Acid Signalling during Early Development of Posterior Cardiac Tube Segment. Int. J. Mol. Sci. 2022, 23, 4179. [Google Scholar] [CrossRef]
- Garcia-Padilla, C.; Dueñas, A.; Franco, D.; Garcia-Lopez, V.; Aranega, A.; Garcia-Martinez, V.; Lopez-Sanchez, C. Dynamic MicroRNA Expression Profiles During Embryonic Development Provide Novel Insights into Cardiac Sinus Venosus/Inflow Tract Differentiation. Front. Cell Dev. Biol. 2022, 9, 767954. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nat. Cell Biol. 2016, 539, 452–455. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, E.L.; Belhocine, M.; Dao, L.T.; Puthier, D.; Spicuglia, S. Functions of lncRNA in development and diseases. Med. Sci. 2014, 30, 790–796. (In French) [Google Scholar]
- García-Padilla, C.; Domínguez, J.N.; Lodde, V.; Munk, R.; Abdelmohsen, K.; Gorospe, M.; Jiménez-Sábado, V.; Ginel, A.; Hove-Madsen, L.; Aránega, A.E.; et al. Identification of atrial-enriched lncRNA Walras linked to cardiomyocyte cytoarchitecture and atrial fibrillation. FASEB J. 2022, 36, e22051. [Google Scholar] [CrossRef]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yuyang, L.J.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Milligan, L.; Forné, T.; Antoine, E.; Weber, M.; Hémonnot, B.; Dandolo, L.; Brunel, C.; Cathala, G. Turnover of primary transcripts is a major step in the regulation of mouse H19 gene expression. EMBO Rep. 2002, 3, 774–779. [Google Scholar] [CrossRef][Green Version]
- Schoenfelder, S.; Smits, G.; Fraser, P.; Reik, W.; Paro, R. Non-coding transcripts in the H19 imprinting control region mediate gene silencing in transgenic Drosophila. EMBO Rep. 2007, 8, 1068–1073. [Google Scholar] [CrossRef]
- Zemel, S.; Bartolomei, M.S.; Tilghman, S.M. Physical linkage of two mammalian imprinted genes, H19 and insulin-like growth factor 2. Nat. Genet. 1992, 2, 61–65. [Google Scholar] [CrossRef]
- García-Padilla, C.; Domínguez, J.N.; Aránega, A.E.; Franco, D. Differential chamber-specific expression and regulation of long non-coding RNAs during cardiac development. Biochim. Biophys. Acta Gene Regul. Mech. 2019, 1862, 194435. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, Z.; Fang, J. H19 functions as a competing endogenous RNA to regulate human epidermal growth factor receptor expression by sequestering let-7c in gastric cancer. Mol. Med. Rep. 2018, 17, 2600–2606. [Google Scholar] [CrossRef]
- Viereck, J.; Bührke, A.; Foinquinos, A.; Chatterjee, S.; Kleeberger, J.A.; Xiao, K.; Janssen-Peters, H.; Batkai, S.; Ramanujam, D.; Kraft, T.; et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur. Heart J. 2020, 41, 3462–3474. [Google Scholar] [CrossRef] [PubMed]
- Charalambous, M.; Menheniott, T.R.; Bennett, W.R.; Kelly, S.M.; Dell, G.; Dandolo, L.; Ward, A. An enhancer element at the Igf2/H19 locus drives gene expression in both imprinted and non-imprinted tissues. Dev. Biol. 2004, 271, 488–497. [Google Scholar] [CrossRef]
- Gabory, A.; Jammes, H.; Dandolo, L. The H19 locus: Role of an imprinted non-coding RNA in growth and development. Bioessays 2010, 32, 473–480. [Google Scholar] [CrossRef]
- Lv, J.; Wang, L.; Zhang, J.; Lin, R.; Wang, L.; Sun, W.; Wu, H.; Xin, S. Long noncoding RNA H19-derived miR-675 aggravates restenosis by targeting PTEN. Biochem. Biophys. Res. Commun. 2018, 497, 1154–1161. [Google Scholar] [CrossRef]
- Luo, H.; Wang, J.; Liu, D.; Zang, S.; Ma, N.; Zhao, L.; Zhang, L.; Zhang, X.; Qiao, C. The lncRNA H19/miR-675 axis regulates myocardial ischemic and reperfusion injury by targeting PPARα. Mol. Immunol. 2019, 105, 46–54. [Google Scholar] [CrossRef]
- Zhu, L.; Zhu, Y.; Han, S.; Chen, M.; Song, P.; Dai, D.; Xu, W.; Jiang, T.; Feng, L.; Shin, V.Y.; et al. Impaired autophagic degradation of lncRNA ARHGAP5-AS1 promotes chemoresistance in gastric cancer. Cell Death Dis. 2019, 10, 383. [Google Scholar] [CrossRef]
- Dong, H.; Wang, W.; Chen, R.; Zhang, Y.; Zou, K.; Ye, M.; He, X.; Zhang, F.; Han, J. Exosome-mediated transfer of lncRNA-SNHG14 promotes trastuzumab chemoresistance in breast cancer. Int. J. Oncol. 2018, 53, 1013–1026. [Google Scholar] [CrossRef]
- Luo, Y.; Zheng, S.; Wu, Q.; Wu, J.; Zhou, R.; Wang, C.; Wu, Z.; Rong, X.; Huang, N.; Sun, L.; et al. Long noncoding RNA (lncRNA) EIF3J-DT induces chemoresistance of gastric cancer via autophagy activation. Autophagy 2021, 17, 4083–4101. [Google Scholar] [CrossRef]
- Han, P.; Li, J.W.; Zhang, B.M.; Lv, J.C.; Li, Y.M.; Gu, X.Y.; Yu, Z.W.; Jia, Y.H.; Bai, X.F.; Li, L.; et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol. Cancer 2017, 16, 9. [Google Scholar] [CrossRef]
- He, W.; Liang, B.; Wang, C.; Li, S.; Zhao, Y.; Huang, Q.; Liu, Z.; Yao, Z.; Wu, Q.; Liao, W.; et al. MSC-regulated lncRNA MACC1-AS1 promotes stemness and chemoresistance through fatty acid oxidation in gastric cancer. Oncogene 2019, 38, 4637–4654. [Google Scholar] [CrossRef] [PubMed]
- Mansoori, B.; Mohammadi, A.; Davudian, S.; Shirjang, S.; Baradaran, B. The Different Mechanisms of Cancer Drug Resistance: A Brief Review. Adv. Pharm. Bull. 2017, 7, 339–348. [Google Scholar] [CrossRef]
- Wu, P.; Gao, W.; Su, M.; Nice, E.C.; Zhang, W.; Lin, J.; Xie, N. Adaptive Mechanisms of Tumor Therapy Resistance Driven by Tumor Microenvironment. Front. Cell Dev. Biol. 2021, 9, 641469. [Google Scholar] [CrossRef]
- Zheng, H.C. The molecular mechanisms of chemoresistance in cancers. Oncotarget 2017, 8, 59950–59964. [Google Scholar] [CrossRef]
- Xiu, M.; Wang, Y.; Li, B.; Wang, X.; Xiao, F.; Chen, S.; Zhang, L.; Zhou, B.; Hua, F. The Role of Notch3 Signaling in Cancer Stemness and Chemoresistance: Molecular Mechanisms and Targeting Strategies. Front. Mol. Biosci. 2021, 8, 694141. [Google Scholar] [CrossRef]
- Zhang, F.; Wang, H.; Yu, J.; Yao, X.; Yang, S.; Li, W.; Xu, L.; Zhao, L. LncRNA CRNDE attenuates chemoresistance in gastric cancer via SRSF6-regulated alternative splicing of PICALM. Mol. Cancer 2021, 20, 6. [Google Scholar] [CrossRef] [PubMed]
- Käsmann, L.; Dietrich, A.; Staab-Weijnitz, C.A.; Manapov, F.; Behr, J.; Rimner, A.; Jeremic, B.; Senan, S.; De Ruysscher, D.; Lauber, K.; et al. Radiation-induced lung toxicity—Cellular and molecular mechanisms of pathogenesis, management, and literature review. Radiat. Oncol. 2020, 15, 214. [Google Scholar] [CrossRef]
- Gan, L.; Lv, L.; Liao, S. Long non-coding RNA H19 regulates cell growth and metastasis via the miR-22-3p/Snail1 axis in gastric cancer. Int. J. Oncol. 2019, 54, 2157–2168. [Google Scholar] [CrossRef]
- Wang, J.; Sun, J.; Yang, F. The role of long non-coding RNA H19 in breast cancer. Oncol. Lett. 2020, 19, 7–16. [Google Scholar] [CrossRef]
- Yoshimura, H.; Matsuda, Y.; Yamamoto, M.; Michishita, M.; Takahashi, K.; Sasaki, N.; Ishikawa, N.; Aida, J.; Takubo, K.; Arai, T.; et al. Reduced expression of the H19 long non-coding RNA inhibits pancreatic cancer metastasis. Lab. Investig. 2018, 98, 814–824. [Google Scholar] [CrossRef]
- Huang, Z.; Chu, L.; Liang, J.; Tan, X.; Wang, Y.; Wen, J.; Chen, J.; Wu, Y.; Liu, S.; Liao, J.; et al. H19 Promotes HCC Bone Metastasis Through Reducing Osteoprotegerin Expression in a Protein Phosphatase 1 Catalytic Subunit Alpha/p38 Mitogen-Activated Protein Kinase-Dependent Manner and Sponging microRNA 200b-3p. Hepatology 2021, 74, 214–232. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Ding, Y.; Yang, X. Overexpression of Long Noncoding RNA H19 Downregulates miR-140-5p and Activates PI3K/AKT Signaling Pathway to Promote Invasion, Migration and Epithelial-Mesenchymal Transition of Ovarian Cancer Cells. Biomed. Res. Int. 2021, 2021, 6619730. [Google Scholar] [CrossRef]
- Liu, Z.Z.; Tian, Y.F.; Wu, H.; Ouyang, S.Y.; Kuang, W.L. LncRNA H19 promotes glioma angiogenesis through miR-138/HIF-1α/VEGF axis. Neoplasma 2020, 67, 111–118. [Google Scholar] [CrossRef]
- Liu, C.; Chen, Z.; Fang, J.; Xu, A.; Zhang, W.; Wang, Z. H19-derived miR-675 contributes to bladder cancer cell proliferation by regulating p53 activation. Tumour Biol. 2016, 37, 263–270. [Google Scholar] [CrossRef] [PubMed]
- Shermane Lim, Y.W.; Xiang, X.; Garg, M.; Le, M.T.; Li-Ann Wong, A.; Wang, L.; Goh, B.C. The double-edged sword of H19 lncRNA: Insights into cancer therapy. Cancer Lett. 2021, 500, 253–262. [Google Scholar] [CrossRef] [PubMed]
- Alipoor, B.; Parvar, S.N.; Sabati, Z.; Ghaedi, H.; Ghasemi, H. An updated review of the H19 lncRNA in human cancer: Molecular mechanism and diagnostic and therapeutic importance. Mol. Biol. Rep. 2020, 47, 6357–6374. [Google Scholar] [CrossRef]
- Katsura, C.; Ogunmwonyi, I.; Kankam, H.K.; Saha, S. Breast cancer: Presentation, investigation and management. Br. J. Hosp. Med. 2022, 83, 1–7. [Google Scholar] [CrossRef]
- Arciero, C.A.; Guo, Y.; Jiang, R.; Behera, M.; O’Regan, R.; Peng, L.; Li, X. ER+/HER2+ Breast Cancer Has Different Metastatic Patterns and Better Survival Than ER-/HER2+ Breast Cancer. Clin. Breast Cancer 2019, 19, 236–245. [Google Scholar] [CrossRef]
- Keenan, T.E.; Tolaney, S.M. Role of Immunotherapy in Triple-Negative Breast Cancer. J. Natl. Compr. Canc. Netw. 2020, 18, 479–489. [Google Scholar] [CrossRef] [PubMed]
- Belachew, E.B.; Sewasew, D.T. Molecular Mechanisms of Endocrine Resistance in Estrogen-Positive Breast Cancer. Front. Endocrinol. 2021, 12, 599586. [Google Scholar] [CrossRef]
- Fisusi, F.A.; Akala, E.O. Drug Combinations in Breast Cancer Therapy. Pharm. Nanotechnol. 2019, 7, 3–23. [Google Scholar] [CrossRef] [PubMed]
- Mills, J.N.; Rutkovsky, A.C.; Giordano, A. Mechanisms of resistance in estrogen receptor positive breast cancer: Overcoming resistance to tamoxifen/aromatase inhibitors. Curr. Opin. Pharmacol. 2018, 41, 59–65. [Google Scholar] [CrossRef]
- Shafei, A.; El-Bakly, W.; Sobhy, A.; Wagdy, O.; Reda, A.; Aboelenin, O.; Marzouk, A.; El Habak, K.; Mostafa, R.; Ali, M.A.; et al. A review on the efficacy and toxicity of different doxorubicin nanoparticles for targeted therapy in metastatic breast cancer. Biomed. Pharmacother. 2017, 95, 1209–1218. [Google Scholar] [CrossRef]
- Abu Samaan, T.M.; Samec, M.; Liskova, A.; Kubatka, P.; Büsselberg, D. Paclitaxel’s Mechanistic and Clinical Effects on Breast Cancer. Biomolecules 2019, 9, 789. [Google Scholar] [CrossRef]
- Hsu, M.Y.; Hsieh, C.H.; Huang, Y.T.; Chu, S.Y.; Chen, C.M.; Lee, W.J.; Liu, S.J. Enhanced Paclitaxel Efficacy to Suppress Triple-Negative Breast Cancer Progression Using Metronomic Chemotherapy with a Controlled Release System of Electrospun Poly-d-l-Lactide-Co-Glycolide (PLGA) Nanofibers. Cancers 2021, 13, 3350. [Google Scholar] [CrossRef]
- Kavallaris, M. Microtubules and resistance to tubulin-binding agents. Nat. Rev. Cancer. 2010, 10, 194–204, Erratum in Nat. Rev. Cancer 2010, 10, 309. [Google Scholar] [CrossRef] [PubMed]
- Jabbarzadeh Kaboli, P.; Salimian, F.; Aghapour, S.; Xiang, S.; Zhao, Q.; Li, M.; Wu, X.; Du, F.; Zhao, Y.; Shen, J.; et al. Akt-targeted therapy as a promising strategy to overcome drug resistance in breast cancer—A comprehensive review from chemotherapy to immunotherapy. Pharmacol. Res. 2020, 156, 104806. [Google Scholar] [CrossRef]
- Si, X.; Zang, R.; Zhang, E.; Liu, Y.; Shi, X.; Zhang, E.; Shao, L.; Li, A.; Yang, N.; Han, X.; et al. LncRNA H19 confers chemoresistance in ERα-positive breast cancer through epigenetic silencing of the pro-apoptotic gene BIK. Oncotarget 2016, 7, 81452–81462. [Google Scholar] [CrossRef] [PubMed]
- Pandya, V.; Glubrecht, D.; Vos, L.; Hanson, J.; Damaraju, S.; Mackey, J.; Hugh, J.; Goping, I.S. The pro-apoptotic paradox: The BH3-only protein Bcl-2 interacting killer (Bik) is prognostic for unfavorable outcomes in breast cancer. Oncotarget 2016, 7, 33272–33285. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Yang, S.; Yue, C.X.; Wei, X.Y.; Peng, W.; Dong, Z.Y.; Xu, H.N.; Chen, S.L.; Wang, W.R.; Chen, C.J.; et al. Long noncoding RNA H19 acts as a miR-340-3p sponge to promote epithelial-mesenchymal transition by regulating YWHAZ expression in paclitaxel-resistant breast cancer cells. Environ. Toxicol. 2020, 35, 1015–1028. [Google Scholar] [CrossRef]
- Han, J.; Han, B.; Wu, X.; Hao, J.; Dong, X.; Shen, Q.; Pang, H. Knockdown of lncRNA H19 restores chemo-sensitivity in paclitaxel-resistant triple-negative breast cancer through triggering apoptosis and regulating Akt signaling pathway. Toxicol. Appl. Pharmacol. 2018, 359, 55–61. [Google Scholar] [CrossRef]
- Thorn, C.F.; Oshiro, C.; Marsh, S.; Hernandez-Boussard, T.; McLeod, H.; Klein, T.E.; Altman, R.B. Doxorubicin pathways: Pharmacodynamics and adverse effects. Pharm. Genom. 2011, 21, 440–446. [Google Scholar] [CrossRef]
- Zhu, Q.N.; Wang, G.; Guo, Y.; Peng, Y.; Zhang, R.; Deng, J.L.; Li, Z.X.; Zhu, Y.S. LncRNA H19 is a major mediator of doxorubicin chemoresistance in breast cancer cells through a cullin4A-MDR1 pathway. Oncotarget 2017, 8, 91990–92003. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, P.; Li, P.; Yang, F.; Gao, X.Q. Long non-coding RNA H19 regulates proliferation and doxorubicin resistance in MCF-7 cells by targeting PARP1. Bioengineered 2020, 11, 536–546. [Google Scholar] [CrossRef] [PubMed]
- Saucedo-Cuevas, L.P.; Ruppen, I.; Ximénez-Embún, P.; Domingo, S.; Gayarre, J.; Muñoz, J.; Silva, J.M.; García, M.J.; Benítez, J. CUL4A contributes to the biology of basal-like breast tumors through modulation of cell growth and antitumor immune response. Oncotarget 2014, 5, 2330–2343. [Google Scholar] [CrossRef] [PubMed]
- Haque, A.; Sait, K.H.W.; Alam, Q.; Alam, M.Z.; Anfinan, N.; Wali, A.W.N.; Rasool, M. MDR1 Gene Polymorphisms and Its Association with Expression as a Clinical Relevance in Terms of Response to Chemotherapy and Prognosis in Ovarian Cancer. Front. Genet. 2020, 11, 516. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Li, H.; Li, Y.; Ruan, Y.; Quan, C. Identification of genes and pathways associated with MDR in MCF-7/MDR breast cancer cells by RNA-seq analysis. Mol. Med. Rep. 2018, 17, 6211–6226. [Google Scholar] [CrossRef]
- Martin, H.L.; Smith, L.; Tomlinson, D.C. Multidrug-resistant breast cancer: Current perspectives. Breast Cancer 2014, 6, 1–13. [Google Scholar] [CrossRef]
- Pignochino, Y.; Capozzi, F.; D’Ambrosio, L.; Dell’Aglio, C.; Basiricò, M.; Canta, M.; Lorenzato, A.; Vignolo Lutati, F.; Aliberti, S.; Palesandro, E.; et al. PARP1 expression drives the synergistic antitumor activity of trabectedin and PARP1 inhibitors in sarcoma preclinical models. Mol. Cancer 2017, 16, 86. [Google Scholar] [CrossRef]
- Weaver, A.N.; Yang, E.S. Beyond DNA Repair: Additional Functions of PARP-1 in Cancer. Front. Oncol. 2013, 3, 290. [Google Scholar] [CrossRef]
- Takagi, M.; Ogawa, C.; Aoki-Nogami, Y.; Iehara, T.; Ishibashi, E.; Imai, M.; Kihara, T.; Nobori, K.; Hasebe, K.; Mizutani, S.; et al. Phase I clinical study of oral olaparib in pediatric patients with refractory solid tumors: Study protocol. BMC Pediatrics 2019, 19, 31. [Google Scholar] [CrossRef]
- Wang, X.; Pei, X.; Guo, G.; Qian, X.; Dou, D.; Zhang, Z.; Xu, X.; Duan, X. Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J. Cell Physiol. 2020, 235, 6896–6904. [Google Scholar] [CrossRef] [PubMed]
- Elledge, R.M.; Lock-Lim, S.; Allred, D.C.; Hilsenbeck, S.G.; Cordner, L. p53 mutation and tamoxifen resistance in breast cancer. Clin. Cancer Res. 1995, 1, 1203–1208. [Google Scholar]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef] [PubMed]
- Riggins, R.B.; Schrecengost, R.S.; Guerrero, M.S.; Bouton, A.H. Pathways to tamoxifen resistance. Cancer Lett. 2007, 256, 1–24. [Google Scholar] [CrossRef]
- Zhu, Y.; Liu, Y.; Zhang, C.; Chu, J.; Wu, Y.; Li, Y.; Liu, J.; Li, Q.; Li, S.; Shi, Q.; et al. Tamoxifen-resistant breast cancer cells are resistant to DNA-damaging chemotherapy because of upregulated BARD1 and BRCA1. Nat. Commun. 2018, 9, 1595. [Google Scholar] [CrossRef]
- Wang, J.; Xie, S.; Yang, J.; Xiong, H.; Jia, Y.; Zhou, Y.; Chen, Y.; Ying, X.; Chen, C.; Ye, C.; et al. The long noncoding RNA H19 promotes tamoxifen resistance in breast cancer via autophagy. J. Hematol. Oncol. 2019, 12, 81. [Google Scholar] [CrossRef]
- Gao, H.; Hao, G.; Sun, Y.; Li, L.; Wang, Y. Long noncoding RNA H19 mediated the chemosensitivity of breast cancer cells via Wnt pathway and EMT process. Oncol. Targets Ther. 2018, 11, 8001–8012. [Google Scholar] [CrossRef]
- Matsuda, N.; Lim, B.; Wang, X.; Ueno, N.T. Early clinical development of epidermal growth factor receptor targeted therapy in breast cancer. Expert Opin. Investig. Drugs 2017, 26, 463–479. [Google Scholar] [CrossRef] [PubMed]
- Xu, B.; Huo, Z.; Huang, H.; Ji, W.; Bian, Z.; Jiao, J.; Sun, J.; Shao, J. The expression and prognostic value of the epidermal growth factor receptor family in glioma. BMC Cancer 2021, 21, 451. [Google Scholar] [CrossRef]
- Grapa, C.M.; Mocan, T.; Gonciar, D.; Zdrehus, C.; Mosteanu, O.; Pop, T.; Mocan, L. Epidermal Growth Factor Receptor and Its Role in Pancreatic Cancer Treatment Mediated by Nanoparticles. Int. J. Nanomed. 2019, 14, 9693–9706. [Google Scholar] [CrossRef]
- Du, Z.; Brown, B.P.; Kim, S.; Ferguson, D.; Pavlick, D.C.; Jayakumaran, G.; Benayed, R.; Gallant, J.N.; Zhang, Y.K.; Yan, Y.; et al. Structure-function analysis of oncogenic EGFR Kinase Domain Duplication reveals insights into activation and a potential approach for therapeutic targeting. Nat. Commun. 2021, 12, 1382. [Google Scholar] [CrossRef]
- Jänne, P.A.; Engelman, J.A.; Johnson, B.E. Epidermal growth factor receptor mutations in non-small-cell lung cancer: Implications for treatment and tumor biology. J. Clin. Oncol. 2005, 23, 3227–3234. [Google Scholar] [CrossRef]
- Chen, G.; Kronenberger, P.; Teugels, E.; Umelo, I.A.; De Grève, J. Targeting the epidermal growth factor receptor in non-small cell lung cancer cells: The effect of combining RNA interference with tyrosine kinase inhibitors or cetuximab. BMC Med. 2012, 10, 28. [Google Scholar] [CrossRef]
- Piperdi, B.; Perez-Soler, R. Role of erlotinib in the treatment of non-small cell lung cancer: Clinical outcomes in wild-type epidermal growth factor receptor patients. Drugs 2012, 72 (Suppl. S1), 11–19. [Google Scholar] [CrossRef]
- Nurwidya, F.; Takahashi, F.; Takahashi, K. Gefitinib in the treatment of nonsmall cell lung cancer with activating epidermal growth factor receptor mutation. J. Nat. Sci. Biol. Med. 2016, 7, 119–123. [Google Scholar] [CrossRef]
- Chen, C.; Liu, W.R.; Zhang, B.; Zhang, L.M.; Li, C.G.; Liu, C.; Zhang, H.; Huo, Y.S.; Ma, Y.C.; Tian, P.F.; et al. LncRNA H19 downregulation confers erlotinib resistance through upregulation of PKM2 and phosphorylation of AKT in EGFR-mutant lung cancers. Cancer Lett. 2020, 486, 58–70. [Google Scholar] [CrossRef]
- Pan, R.; Zhou, H. Exosomal Transfer of lncRNA H19 Promotes Erlotinib Resistance in Non-Small Cell Lung Cancer via miR-615-3p/ATG7 Axis. Cancer Manag. Res. 2020, 12, 4283–4297. [Google Scholar] [CrossRef]
- Zhang, P.; Ling, L.; Zheng, Z.; Zhang, Y.; Wang, R.; Wu, M.; Zhang, N.; Hu, M.; Yang, X. ATG7-dependent and independent autophagy determine the type of treatment in lung cancer. Pharmacol. Res. 2021, 163, 105324. [Google Scholar] [CrossRef] [PubMed]
- Cho, K.J.; Shin, S.Y.; Moon, H.; Kim, B.K.; Ro, S.W. Knockdown of Atg7 suppresses Tumorigenesis in a murine model of liver cancer. Transl. Oncol. 2021, 14, 101158. [Google Scholar] [CrossRef]
- Zhou, Y.; Zhang, Y. Inhibition of LncRNAH19 has the effect of anti-tumour and enhancing sensitivity to Gefitinib and Chemotherapy in Non-small-cell lung cancer in vivo. J. Cell Mol. Med. 2020, 24, 5811–5816. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Gregorc, V.; Rossi, E.; Cancellieri, A.; Magrini, E.; Paties, C.T.; Ceresoli, G.; Lombardo, L.; Bartolini, S.; Calandri, C.; et al. Gefitinib in pretreated non-small-cell lung cancer (NSCLC): Analysis of efficacy and correlation with HER2 and epidermal growth factor receptor expression in locally advanced or metastatic NSCLC. J. Clin. Oncol. 2003, 21, 2658–2663. [Google Scholar] [CrossRef]
- Huang, Z.; Ma, Y.; Zhang, P.; Si, J.; Xiong, Y.; Yang, Y. Long non-coding RNA H19 confers resistance to gefitinib via miR-148b-3p/DDAH1 axis in lung adenocarcinoma. Anticancer Drugs 2020, 31, 44–54. [Google Scholar] [CrossRef]
- Lei, Y.; Guo, W.; Chen, B.; Chen, L.; Gong, J.; Li, W. Tumor-released lncRNA H19 promotes gefitinib resistance via packaging into exosomes in non-small cell lung cancer. Oncol. Rep. 2018, 40, 3438–3446. [Google Scholar] [CrossRef]
- Zhao, X.; Jin, X.; Zhang, Q.; Liu, R.; Luo, H.; Yang, Z.; Geng, Y.; Feng, S.; Li, C.; Wang, L.; et al. Silencing of the lncRNA H19 enhances sensitivity to X-ray and carbon-ions through the miR-130a-3p /WNK3 signaling axis in NSCLC cells. Cancer Cell Int. 2021, 21, 644. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Friedman, H.S.; Kerby, T.; Calvert, H. Temozolomide and treatment of malignant glioma. Clin. Cancer Res. 2000, 6, 2585–2597. [Google Scholar]
- Duan, S.; Li, M.; Wang, Z.; Wang, L.; Liu, Y. H19 induced by oxidative stress confers temozolomide resistance in human glioma cells via activating NF-κB signaling. Oncol. Targets Ther. 2018, 11, 6395–6404. [Google Scholar] [CrossRef]
- Chien, C.H.; Hsueh, W.T.; Chuang, J.Y.; Chang, K.Y. Dissecting the mechanism of temozolomide resistance and its association with the regulatory roles of intracellular reactive oxygen species in glioblastoma. J. Biomed. Sci. 2021, 28, 18. [Google Scholar] [CrossRef]
- Campbell, K.J.; Tait, S. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Fang, W.; Che, X.; Li, G.; Wang, A.; Wang, Y.; Shi, X.; Hou, K.; Zhang, X.; Qu, X.; Liu, Y. Sur-X, a novel peptide, kills colorectal cancer cells by targeting survivin-XIAP complex. J. Exp. Clin. Cancer Res. 2020, 39, 82. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef]
- Fu, B.; Zhang, A.; Li, M.; Pan, L.; Tang, W.; An, M.; Liu, W.; Zhang, J. Circular RNA profile of breast cancer brain metastasis: Identification of potential biomarkers and therapeutic targets. Epigenomics 2018, 10, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Jiang, P.; Wang, P.; Sun, X.; Yuan, Z.; Zhan, R.; Ma, X.; Li, W. Knockdown of long noncoding RNA H19 sensitizes human glioma cells to temozolomide therapy. Oncol. Targets Ther. 2016, 9, 3501–3509. [Google Scholar] [CrossRef][Green Version]
- Kuang, Y.; Bing, Z.; Jin, X.; Li, Q. LncRNA H19 Upregulation Participates in the Response of Glioma Cells to Radiation. Biomed. Res. Int. 2021, 2021, 1728352. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.F.; Liang, W.C.; Feng, L.; Pang, J.X.; Waye, M.M.; Zhang, J.F.; Fu, W.M. H19 mediates methotrexate resistance in colorectal cancer through activating Wnt/β-catenin pathway. Exp. Cell Res. 2017, 350, 312–317. [Google Scholar] [CrossRef]
- Singh, R.; Fouladi-Nashta, A.A.; Li, D.; Halliday, N.; Barrett, D.A.; Sinclair, K.D. Methotrexate induced differentiation in colon cancer cells is primarily due to purine deprivation. J. Cell Biochem. 2006, 99, 146–155. [Google Scholar] [CrossRef]
- Raimondi, M.V.; Randazzo, O.; La Franca, M.; Barone, G.; Vignoni, E.; Rossi, D.; Collina, S. DHFR Inhibitors: Reading the Past for Discovering Novel Anticancer Agents. Molecules 2019, 24, 1140. [Google Scholar] [CrossRef]
- Arango, D.; Wilson, A.J.; Shi, Q.; Corner, G.A.; Arañes, M.J.; Nicholas, C.; Lesser, M.; Mariadason, J.M.; Augenlicht, L.H. Molecular mechanisms of action and prediction of response to oxaliplatin in colorectal cancer cells. Br. J. Cancer 2004, 91, 1931–1946. [Google Scholar] [CrossRef]
- Ren, J.; Ding, L.; Zhang, D.; Shi, G.; Xu, Q.; Shen, S.; Wang, Y.; Wang, T.; Hou, Y. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8, 3932–3948. [Google Scholar] [CrossRef]
- Vodenkova, S.; Buchler, T.; Cervena, K.; Veskrnova, V.; Vodicka, P.; Vymetalkova, V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: Past, present and future. Pharmacol. Ther. 2020, 206, 107447. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, Y.; Sakatani, T.; Wada, R.; Ishino, K.; Kudo, M.; Koizumi, M.; Yamada, T.; Yoshida, H.; Naito, Z. In vitro and in vivo studies on the association of long non-coding RNAs H19 and urothelial cancer associated 1 with the susceptibility to 5-fluorouracil in rectal cancer. Int. J. Oncol. 2019, 55, 1361–1371. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Padilla, C.; Lozano-Velasco, E.; Muñoz-Gallardo, M.d.M.; Castillo-Casas, J.M.; Caño-Carrillo, S.; Martínez-Amaro, F.J.; García-López, V.; Aránega, A.; Franco, D.; García-Martínez, V.; et al. LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis. Int. J. Mol. Sci. 2022, 23, 8309. https://doi.org/10.3390/ijms23158309
Garcia-Padilla C, Lozano-Velasco E, Muñoz-Gallardo MdM, Castillo-Casas JM, Caño-Carrillo S, Martínez-Amaro FJ, García-López V, Aránega A, Franco D, García-Martínez V, et al. LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis. International Journal of Molecular Sciences. 2022; 23(15):8309. https://doi.org/10.3390/ijms23158309
Chicago/Turabian StyleGarcia-Padilla, Carlos, Estefanía Lozano-Velasco, María del Mar Muñoz-Gallardo, Juan Manuel Castillo-Casas, Sheila Caño-Carrillo, Francisco José Martínez-Amaro, Virginio García-López, Amelia Aránega, Diego Franco, Virginio García-Martínez, and et al. 2022. "LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis" International Journal of Molecular Sciences 23, no. 15: 8309. https://doi.org/10.3390/ijms23158309
APA StyleGarcia-Padilla, C., Lozano-Velasco, E., Muñoz-Gallardo, M. d. M., Castillo-Casas, J. M., Caño-Carrillo, S., Martínez-Amaro, F. J., García-López, V., Aránega, A., Franco, D., García-Martínez, V., & López-Sánchez, C. (2022). LncRNA H19 Impairs Chemo and Radiotherapy in Tumorigenesis. International Journal of Molecular Sciences, 23(15), 8309. https://doi.org/10.3390/ijms23158309







