AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in Arabidopsis by Regulating the Expression of Flowering-Related Genes
Abstract
:1. Introduction
2. Results
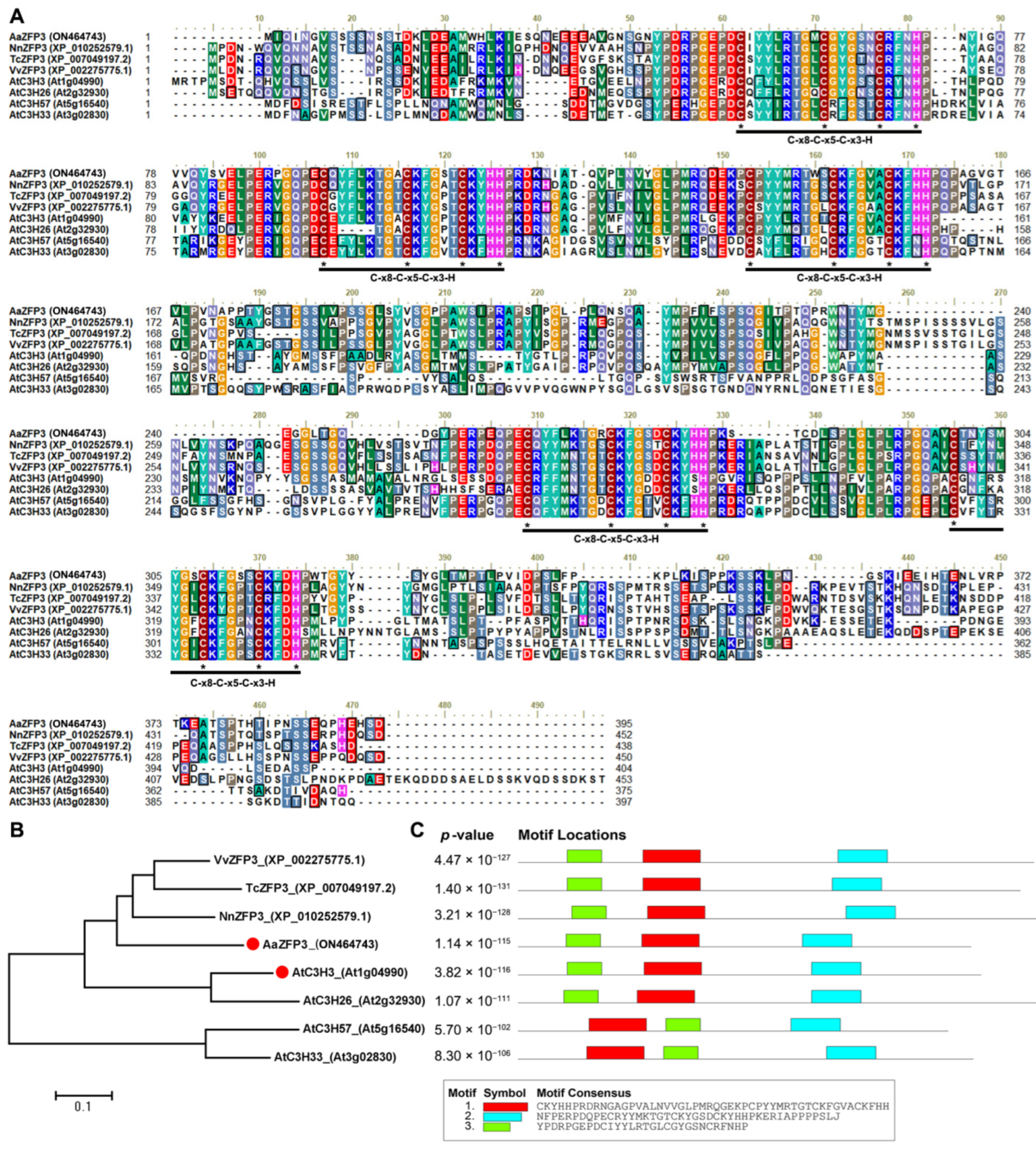
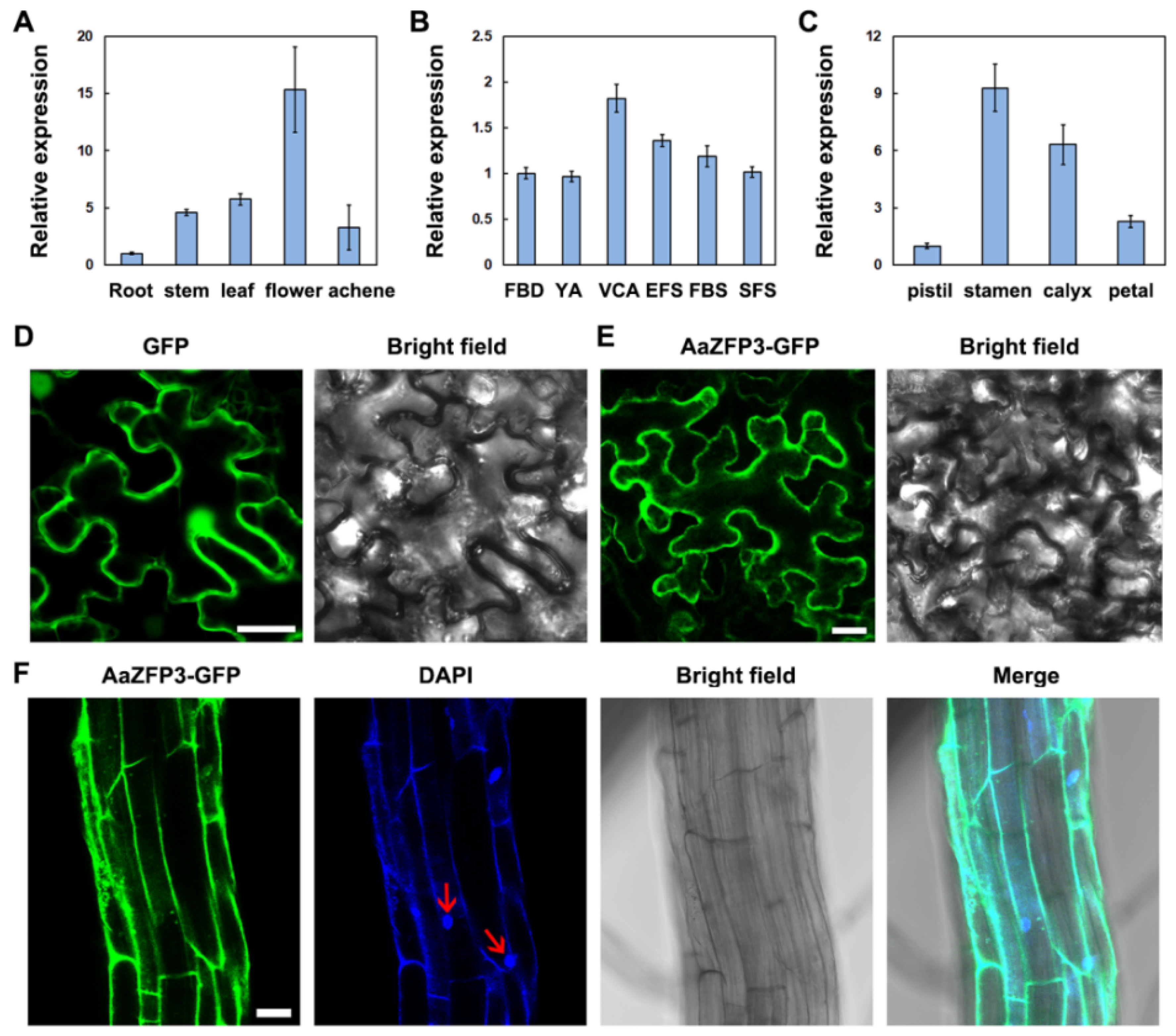
2.1. Sequence and Expression Analysis of AaZFP3
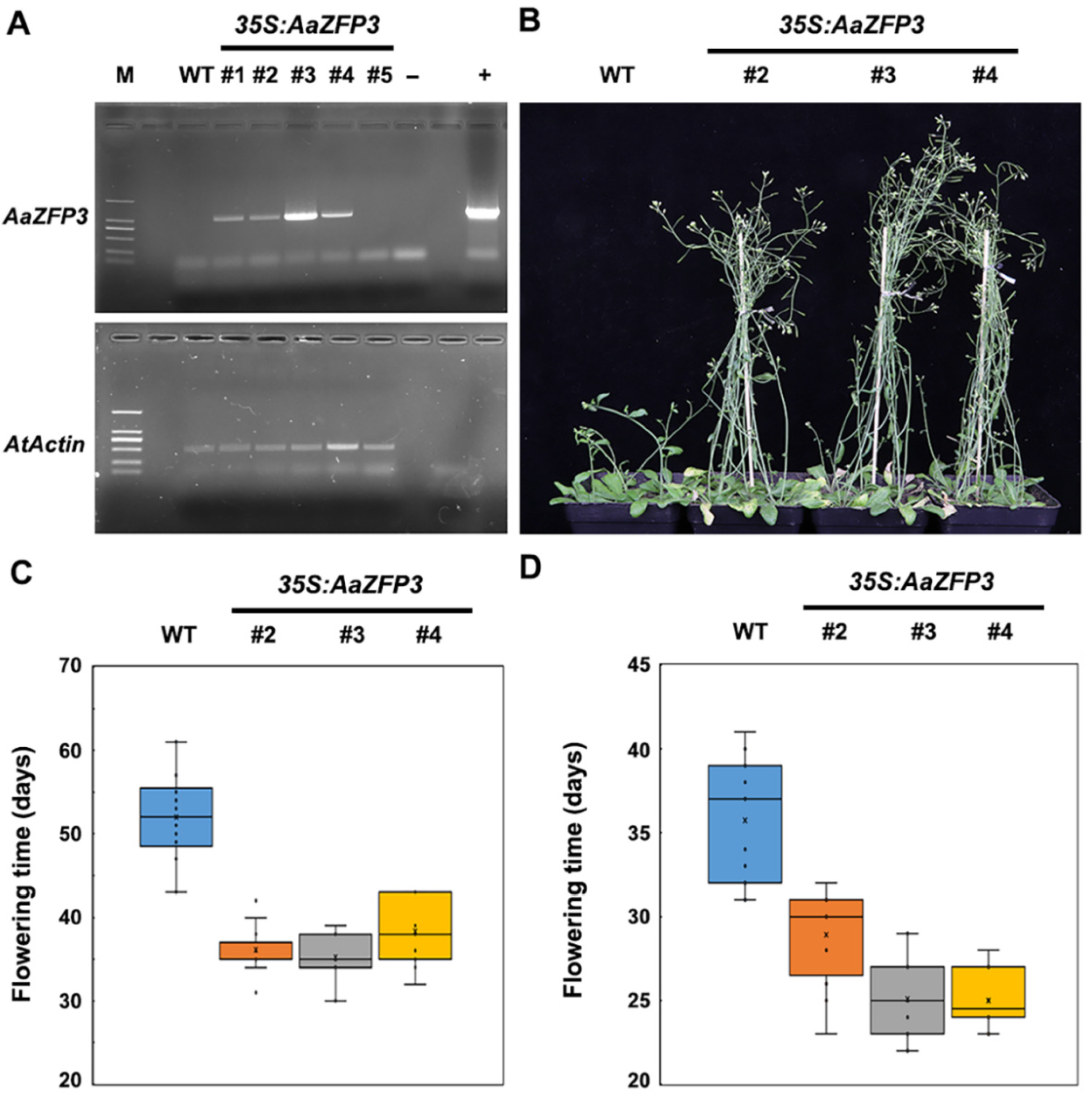
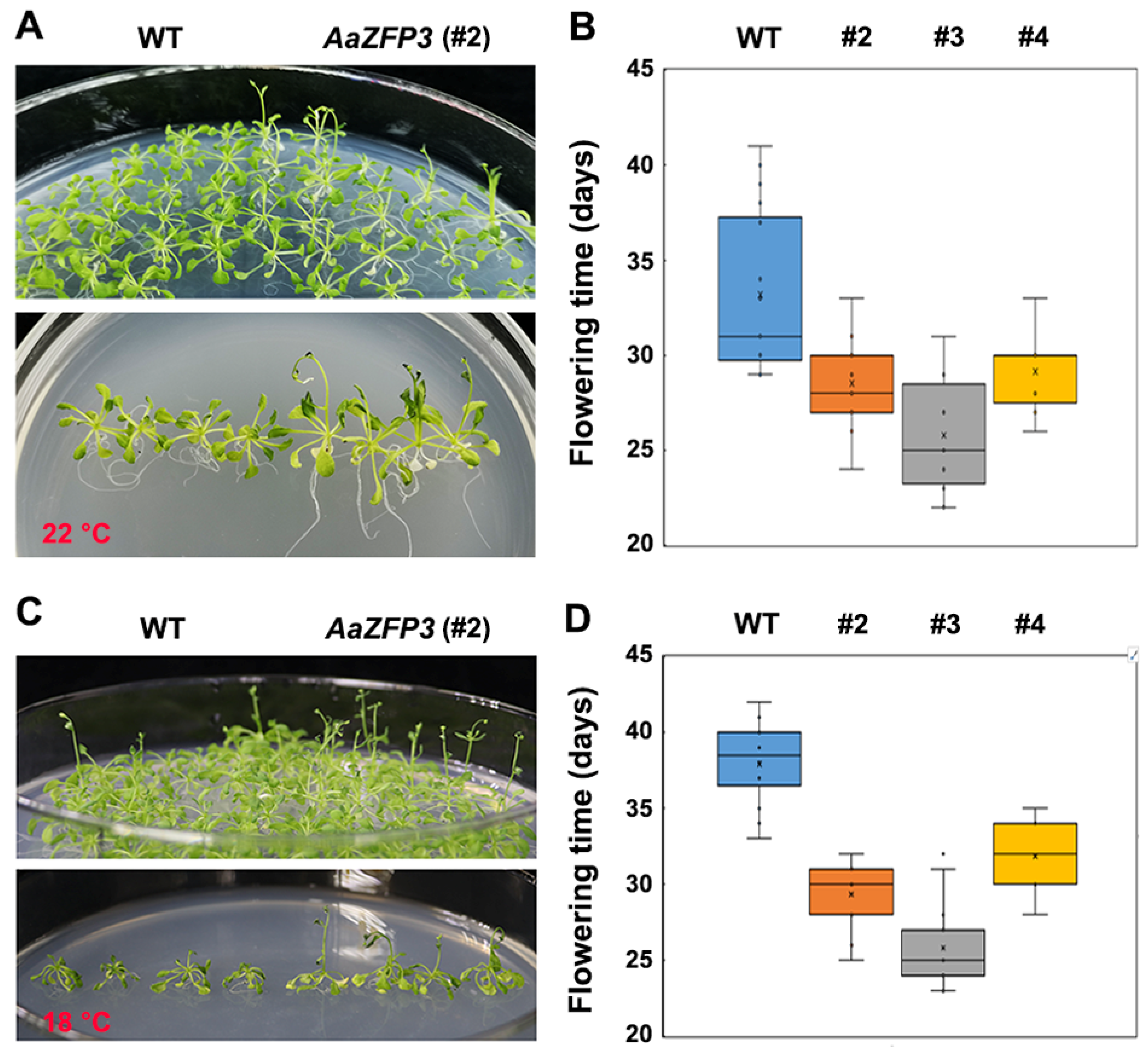
2.2. Effect of AaZFP3 Overexpression on Flowering Time in Arabidopsis
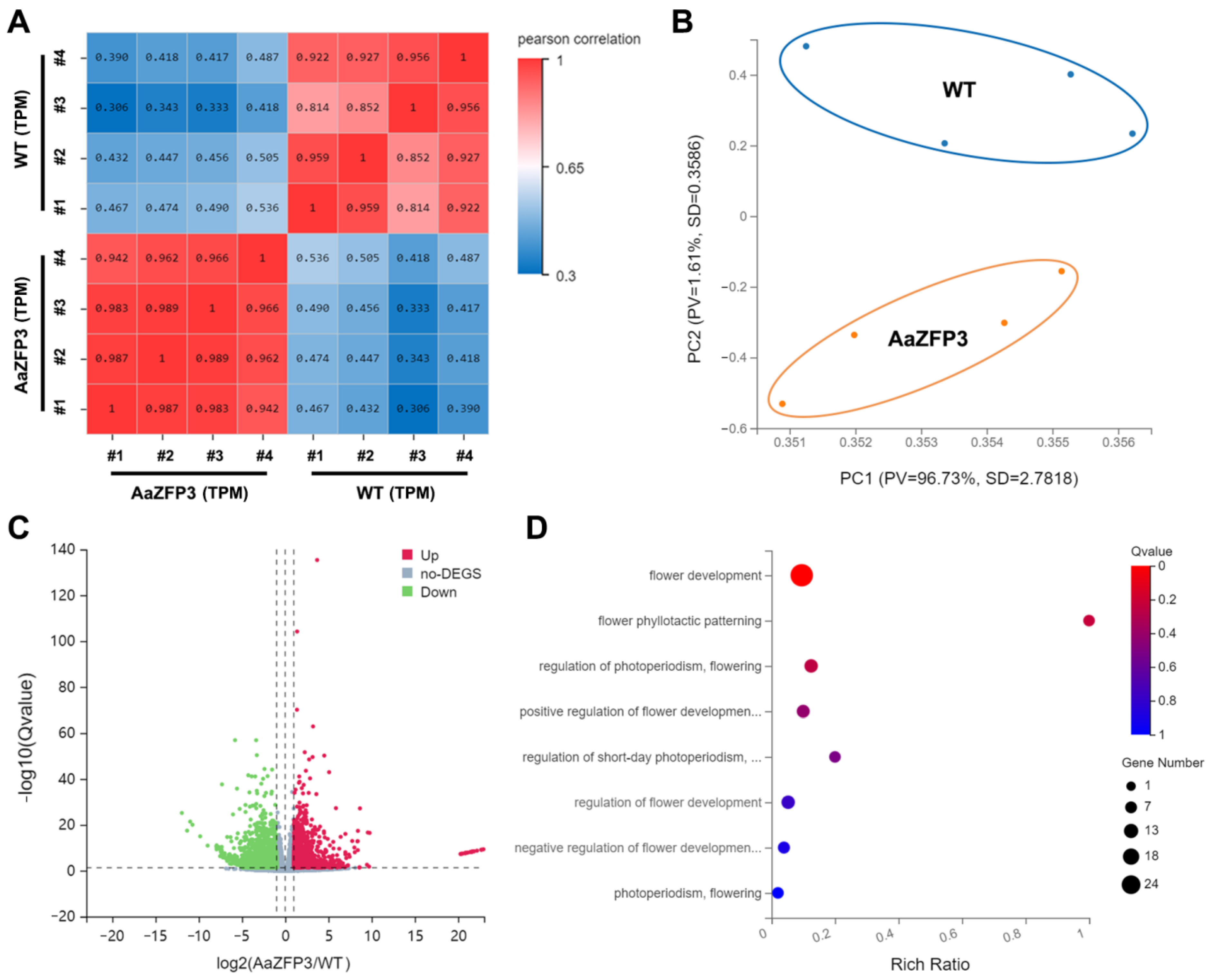
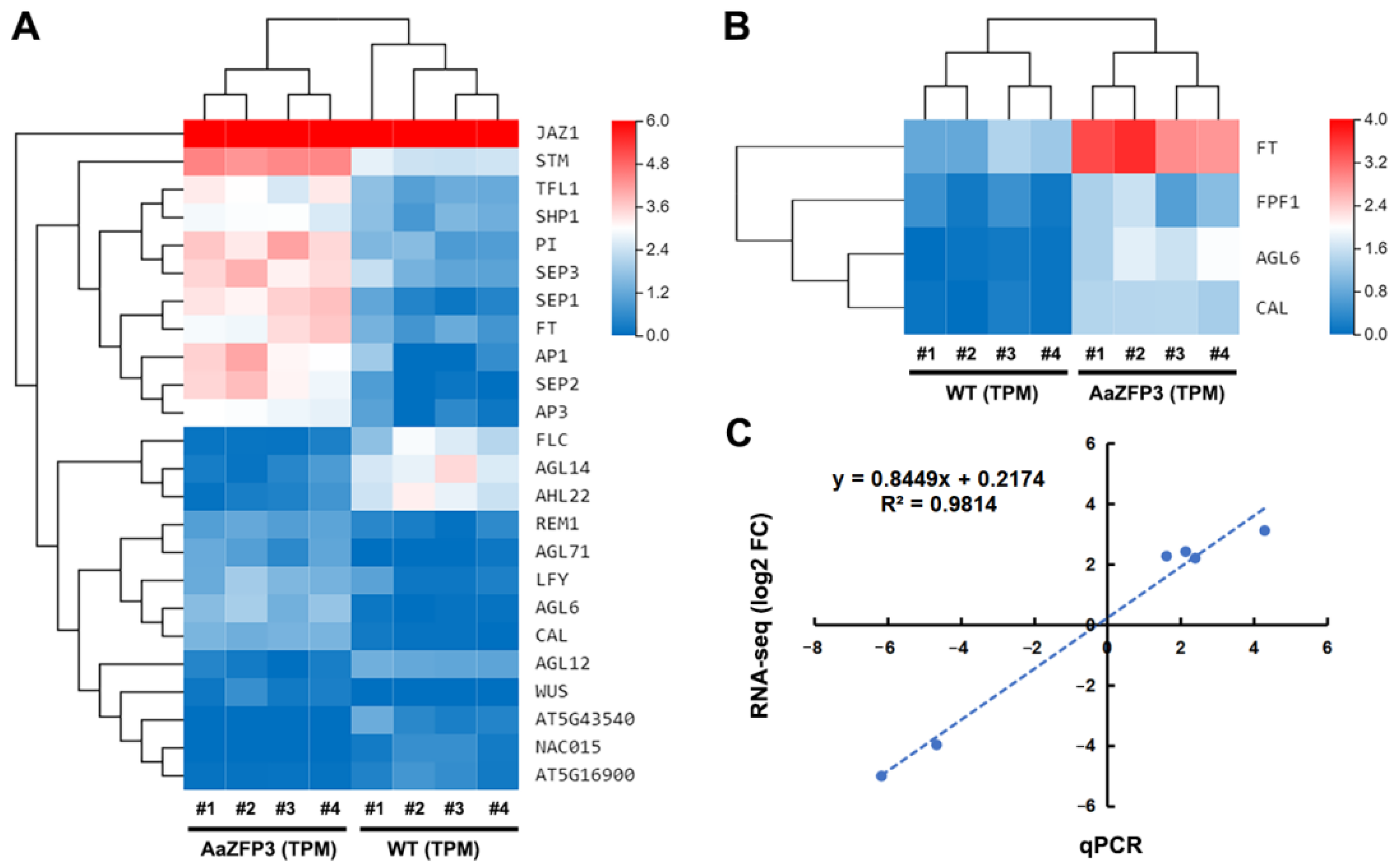
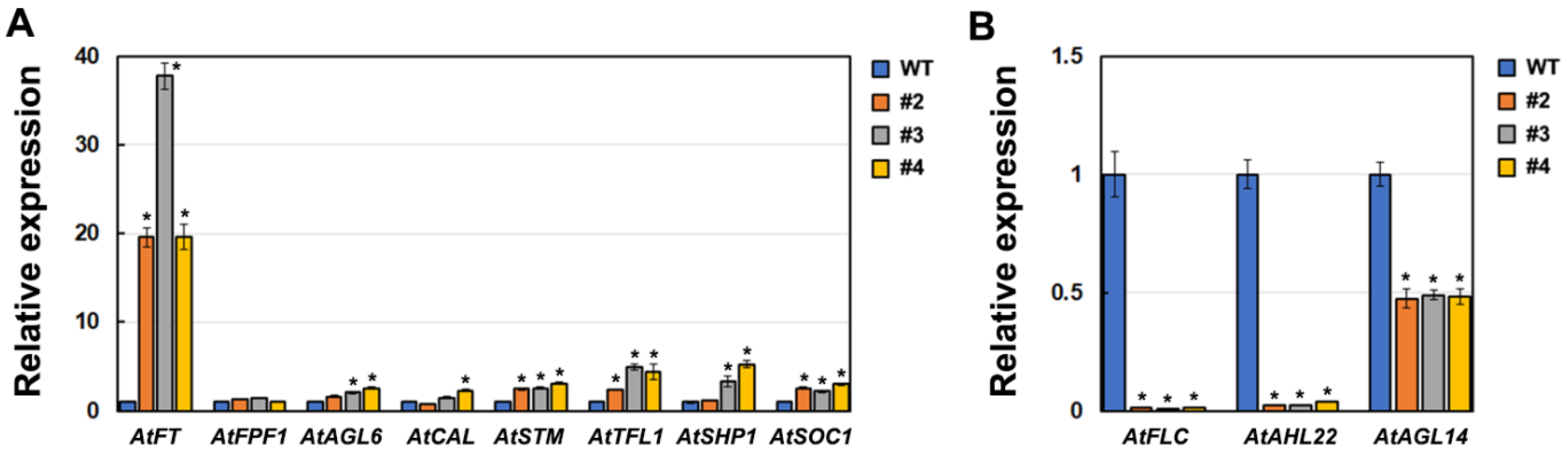
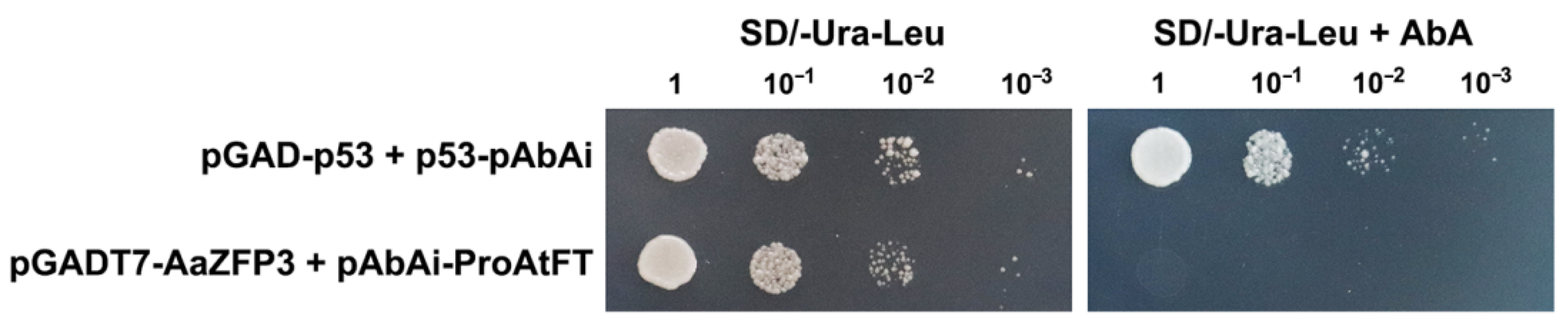
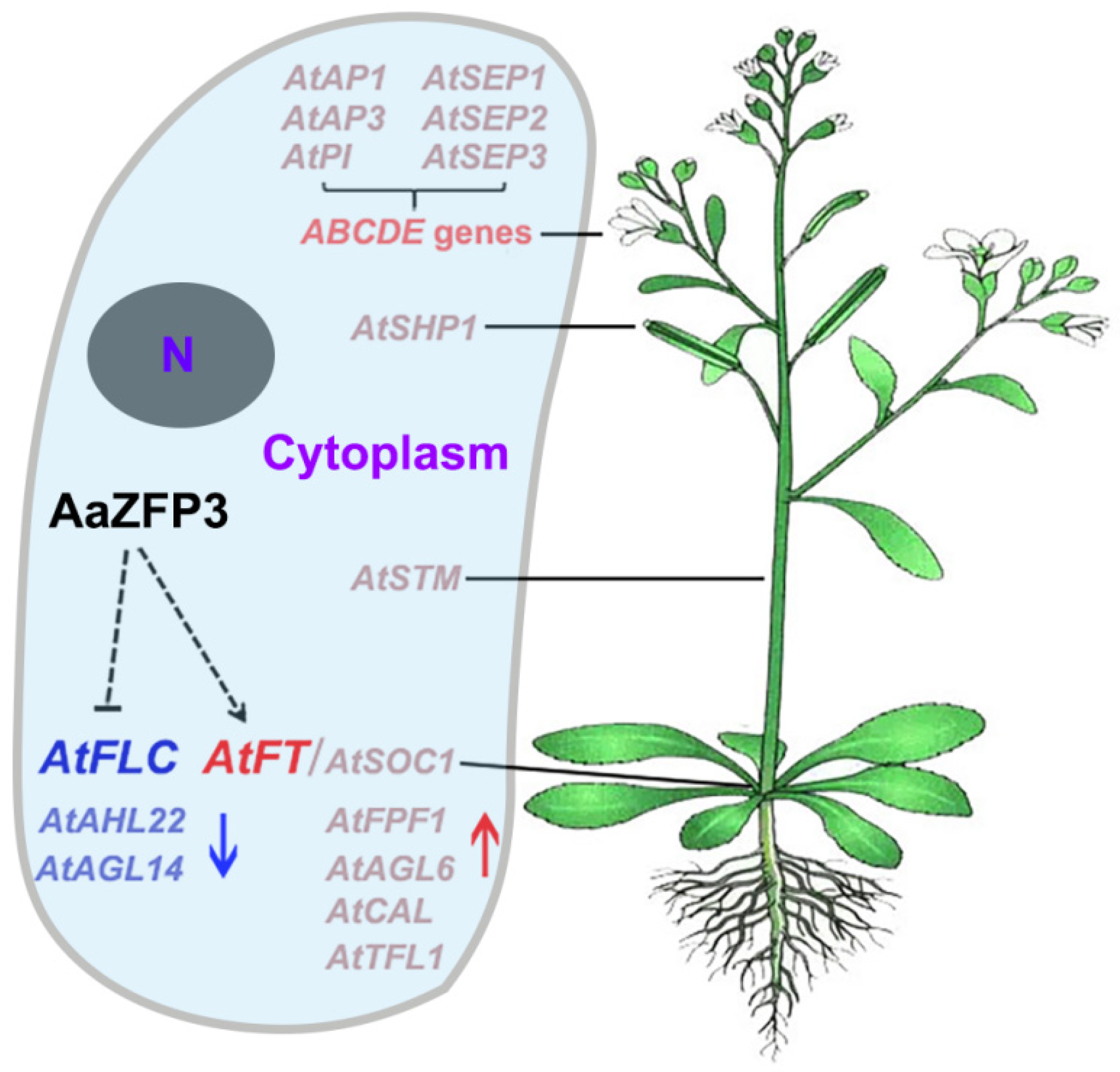
2.3. Effect of ZFP3 Overexpression on the Expression of Flowering-Related Genes in Arabidopsis
3. Discussion
4. Materials and Methods
4.1. Plant Materials
4.2. Quantitative Real-Time PCR Analysis
4.3. Gene Cloning, Vector Construction, and Plant Transformation
4.4. Subcellular Localization
4.5. RNA-Sequencing and Differential Gene Expression Analysis
4.6. Y1H Assays
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Laity, J.H.; Lee, B.M.; Wright, P.E. Zinc finger proteins: New insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001, 11, 39–46. [Google Scholar] [CrossRef]
- Takatsuji, H. Zinc-finger transcription factors in plants. Cell. Mol. Life Sci. 1998, 54, 582–596. [Google Scholar] [CrossRef] [PubMed]
- Blackshear, P.J. Tristetraprolin and other CCCH tandem zinc-finger proteins in the regulation of mRNA turnover. Biochem. Soc. T. 2002, 30, 945–952. [Google Scholar] [CrossRef]
- Wang, D.; Guo, Y.; Wu, C.; Yang, G.; Li, Y.; Zheng, C. Genome-wide analysis of CCCH zinc finger family in Arabidopsis and rice. BMC Genom. 2008, 9, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.L.; Zhong, Y.; Cheng, Z.M. Evolution and expression analysis of the CCCH zinc finger gene family in Vitis vinifera. Plant Genome 2014, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Peng, X.; Zhao, Y.; Cao, J.; Zhang, W.; Jiang, H.; Li, X.; Ma, Q.; Zhu, S.; Cheng, B. CCCH-type zinc finger family in maize: Genome-wide identification, classification and expression profiling under abscisic acid and drought treatments. PLoS ONE 2012, 7, e40120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ai, Q.; Pan, W.; Zeng, Y.; Li, Y.; Cui, L. CCCH Zinc finger genes in Barley: Genome-wide identification, evolution, expression and haplotype analysis. BMC Plant Biol. 2022, 22, 117. [Google Scholar]
- Bogamuwa, S.P.; Jang, J.C. Tandem CCCH zinc finger proteins in plant growth, development and stress response. Plant Cell Physiol. 2014, 55, 1367–1375. [Google Scholar] [CrossRef]
- Li, J.; Jia, D.; Chen, X. HUA1, a regulator of stamen and carpel identities in Arabidopsis, codes for a nuclear RNA binding protein. Plant Cell 2001, 13, 2269–2281. [Google Scholar] [CrossRef] [Green Version]
- Pomeranz, M.C.; Hah, C.; Lin, P.C.; Kang, S.G.; Finer, J.J.; Blackshear, P.J.; Jang, J.C. The Arabidopsis tandem zinc finger protein AtTZF1 traffics between the nucleus and cytoplasmic foci and binds both DNA and RNA. Plant Physiol. 2010, 152, 151–165. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.J.; Jung, H.J.; Kang, H.; Kim, S.Y. Arabidopsis zinc finger proteins AtC3H49/AtTZF3 and AtC3H20/AtTZF2 are involved in ABA and JA responses. Plant Cell Physiol. 2012, 53, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Chai, G.; Kong, Y.; Zhu, M.; Yu, L.; Qi, G.; Tang, X.; Wang, Z.; Cao, Y.; Yu, C.; Zhou, G. Arabidopsis C3H14 and C3H15 have overlapping roles in the regulation of secondary wall thickening and anther development. J. Exp. Bot. 2015, 66, 2595–2609. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, Z.Y.; Jia, J.H.; Yan, X.Y.; Shi, H.Y.; Han, Y.Z. Arabidopsis KHZ1 and KHZ2, two novel non-tandem CCCH zinc-finger and K-homolog domain proteins, have redundant roles in the regulation of flowering and senescence. Plant Mol. Biol. 2017, 95, 549–565. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Woo, D.H.; Park, H.Y.; Lee, S.Y.; Tran, H.T.; Lee, E.H.; Vu Nguyen, L.; Moon, Y.H. AtC3H17, a non-tandem CCCH zinc finger protein, functions as a nuclear transcriptional activator and has pleiotropic effects on vegetative development, flowering and seed development in Arabidopsis. Plant Cell Physiol. 2016, 57, 603–615. [Google Scholar] [CrossRef] [Green Version]
- Seok, H.Y.; Nguyen, L.V.; Park, H.Y.; Tarte, V.N.; Ha, J.; Lee, S.Y.; Moon, Y.H. Arabidopsis non-TZF gene AtC3H17 functions as a positive regulator in salt stress response. Biochem. Biophys. Res. Commun. 2018, 498, 954–959. [Google Scholar] [CrossRef]
- Kong, Z.; Li, M.; Yang, W.; Xu, W.; Xue, Y. A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol. 2006, 141, 1376–1388. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.H.; Yu, Y.P.; Wang, D.; Wu, C.A.; Yang, G.D.; Huang, J.G.; Zheng, C.C. GhZFP1, a novel CCCH-type zinc finger protein from cotton, enhances salt stress tolerance and fungal disease resistance in transgenic tobacco by interacting with GZIRD21A and GZIPR5. New Phytol. 2009, 183, 62–75. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, X.; Zhi, Y.; Li, X.; Zhang, Q.; Niu, J.; Wang, J.; Zhai, H.; Zhao, N.; Li, J.; et al. A non-tandem CCCH-type zinc-finger protein, IbC3H18, functions as a nuclear transcriptional activator and enhances abiotic stress tolerance in sweet potato. New Phytol. 2019, 223, 1918–1936. [Google Scholar] [CrossRef]
- Zhou, A.; Sun, H.; Dai, S.; Feng, S.; Zhang, J.; Gong, S.; Wang, J. Identification of transcription factors involved in the regulation of flowering in Adonis amurensis through combined RNA-seq transcriptomics and iTRAQ proteomics. Genes 2019, 10, 305. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.Q.; Zheng, H.X.; Wang, Y.; Han, G.L.; Sui, N. Overexpression of CCCH zinc finger protein gene delays flowering time and enhances salt tolerance in Arabidopsis by increasing fatty acid unsaturation. Acta Physiol. Plant 2018, 40, 196. [Google Scholar] [CrossRef]
- Lin, P.C.; Pomeranz, M.C.; Jikumaru, Y.; Kang, S.G.; Hah, C.; Fujioka, S.; Kamiya, Y.; Jang, J.C. The Arabidopsis tandem zinc finger protein AtTZF1 affects ABA- and GA-mediated growth, stress and gene expression responses. Plant J. 2011, 65, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Fornara, F.; de Montaigu, A.; Coupland, G. SnapShot: Control of flowering in Arabidopsis. Cell 2010, 141, 550.e1–550.e2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Searle, I.; He, Y.; Turck, F.; Vincent, C.; Fornara, F.; Krober, S.; Amasino, R.A.; Coupland, G. The transcription factor FLC confers a flowering response to vernalization by repressing meristem competence and systemic signaling in Arabidopsis. Genes Dev. 2006, 20, 898–912. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bouche, F.; Woods, D.P.; Amasino, R.M. Winter memory throughout the plant kingdom: Different paths to flowering. Plant Physiol. 2017, 173, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; MacGregor, D.R.; Dave, A.; Florance, H.; Moore, K.; Paszkiewicz, K.; Smirnoff, N.; Graham, I.A.; Penfield, S. Maternal temperature history activates Flowering Locus T in fruits to control progeny dormancy according to time of year. Proc. Natl. Acad. Sci. USA 2014, 111, 18787–18792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, M.; Penfield, S. Feedback regulation of COOLAIR expression controls seed dormancy and flowering time. Science 2018, 360, 1014–1017. [Google Scholar] [CrossRef] [Green Version]
- Chao, Y.; Zhang, T.; Yang, Q.; Kang, J.; Sun, Y.; Gruber, M.Y.; Qin, Z. Expression of the alfalfa CCCH-type zinc finger protein gene MsZFN delays flowering time in transgenic Arabidopsis thaliana. Plant Sci. 2014, 215–216, 92–99. [Google Scholar] [CrossRef]
- Yan, Z.Y.; Shi, H.Y.; Liu, Y.N.; Jing, M.; Han, Y.Z. KHZ1 and KHZ2, novel members of the autonomous pathway, repress the splicing efficiency of FLC pre-mRNA in Arabidopsis. J. Exp. Bot. 2020, 71, 1375–1386. [Google Scholar] [CrossRef] [Green Version]
- Hepworth, S.R.; Valverde, F.; Ravenscroft, D.; Mouradov, A.; Coupland, G. Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J. 2002, 21, 4327–4337. [Google Scholar] [CrossRef] [Green Version]
- Yun, J.; Kim, Y.S.; Jung, J.H.; Seo, P.J.; Park, C.M. The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J. Biol. Chem. 2012, 287, 15307–15316. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Li, C.; Song, S.; Teo, Z.W.N.; Shen, L.; Wang, Y.; Jackson, D.; Yu, H. FTIP-dependent STM trafficking regulates shoot meristem development in Arabidopsis. Cell Rep. 2018, 23, 1879–1890. [Google Scholar] [CrossRef] [PubMed]
- Ferrandiz, C.; Liljegren, S.J.; Yanofsky, M.F. Negative regulation of the SHATTERPROOF genes by FRUITFULL during Arabidopsis fruit development. Science 2000, 289, 436–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liljegren, S.J.; Ditta, G.S.; Eshed, Y.; Savidge, B.; Bowman, J.L.; Yanofsky, M.F. SHATTERPROOF MADS-box genes control seed dispersal in Arabidopsis. Nature 2000, 404, 766–770. [Google Scholar] [CrossRef] [PubMed]
- Theissen, G.; Saedler, H. Plant biology. Floral quartets. Nature 2001, 409, 469–471. [Google Scholar] [CrossRef]
- Gui, J.; Zheng, S.; Shen, J.; Li, L. Grain setting defect1 (GSD1) function in rice depends on S-acylation and interacts with actin 1 (OsACT1) at its C-terminal. Front. Plant Sci. 2015, 6, 804. [Google Scholar] [CrossRef] [Green Version]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, M.; Zhang, H.; Dai, S.; Feng, S.; Gong, S.; Wang, J.; Zhou, A. AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in Arabidopsis by Regulating the Expression of Flowering-Related Genes. Int. J. Mol. Sci. 2022, 23, 8166. https://doi.org/10.3390/ijms23158166
Wang M, Zhang H, Dai S, Feng S, Gong S, Wang J, Zhou A. AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in Arabidopsis by Regulating the Expression of Flowering-Related Genes. International Journal of Molecular Sciences. 2022; 23(15):8166. https://doi.org/10.3390/ijms23158166
Chicago/Turabian StyleWang, Meiqi, Haizhen Zhang, Shengyue Dai, Shuang Feng, Shufang Gong, Jingang Wang, and Aimin Zhou. 2022. "AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in Arabidopsis by Regulating the Expression of Flowering-Related Genes" International Journal of Molecular Sciences 23, no. 15: 8166. https://doi.org/10.3390/ijms23158166
APA StyleWang, M., Zhang, H., Dai, S., Feng, S., Gong, S., Wang, J., & Zhou, A. (2022). AaZFP3, a Novel CCCH-Type Zinc Finger Protein from Adonis amurensis, Promotes Early Flowering in Arabidopsis by Regulating the Expression of Flowering-Related Genes. International Journal of Molecular Sciences, 23(15), 8166. https://doi.org/10.3390/ijms23158166





