Abstract
Zip family proteins are involved in the control of zinc (Zn) ion homeostasis. The present study cloned the promoters and investigated the transcription responses and protein subcellular localizations of three LIV-1 subfamily members (zip10, zip13, and zip14) from common freshwater teleost yellow catfish, Pelteobagrus fulvidraco, using in vitro cultured HEK293T model cells. The 2278 bp, 1917 bp, and 1989 bp sequences of zip10, zip13, and zip14 promoters, respectively, were subcloned into pGL3-Basic plasmid for promoter activity analysis. The pcDNA3.1 plasmid coding EGFP tagged pfZip10, pfZip13, and pfZip14 were generated for subsequent confocal microscope analysis. Several potential transcription factors’ binding sites were predicted within the promoters. In vitro promoter analysis in the HEK293T cells showed that high Zn administration significantly reduced the transcriptional activities of the zip10, zip13, and zip14 promoters. The −2017 bp/−2004 bp MRE in the zip10 promoter, the −360 bp/−345 bp MRE in the zip13 promoter, and the −1457 bp/−1442 bp MRE in the zip14 promoter were functional loci that were involved in the regulation of the three zips. The −606 bp/−594 bp KLF4 binding site in the zip13 promoter was a functional locus responsible for zinc-responsive regulation of zip13. The −1383 bp/−1375 bp STAT3 binding site in the zip14 promoter was a functional locus responsible for zinc-responsive regulation of zip14. Moreover, confocal microscope analysis indicated that zinc incubation significantly reduced the fluorescence intensity of pfZip10-EGFP and pfZip14-EGFP but had no significant influence on pfZip13-EGFP fluorescence intensity. Further investigation found that pfZip10 localizes on cell membranes, pfZip14 colocalized with both cell membranes and lysosome, and pfZip13 colocalized with intracellular ER and Golgi. Our research illustrated the transcription regulation of zip10, zip13, and zip14 from P. fulvidraco under zinc administration, which provided a reference value for the mechanisms involved in Zip-family-mediated control of zinc homeostasis in vertebrates.
1. Introduction
Micronutrient zinc (Zn) is an essentially functional and structural element for all living organisms, including fish [1]. Both a dietary deficiency and an excess of this micronutrient are deleterious to the growth and development of fish [2]. Zinc deficiency results in growth repression and zinc excess results in toxicity for many fish [3]. Waterborne zinc could also cause chronic toxicity for rainbow trout and other fish [4]. Thus, intracellular free Zn2+ must be maintained at a required reasonable level by various Zn transporters, buffering metallothioneins, and sequestering vesicles [5]. For example, in the zebrafish, Zip6 is required for zinc homeostasis maintenance in T-cell development [6] and pigmented cells store zinc and maintain zinc homeostasis through Znt8 [7]. In rainbow trout, MT and other zinc-binding proteins contribute to normal zinc homeostasis in liver cells [8]. Zn transporters, which are SLC39A (Zip) family members, control Zn influx that is assimilated from the extracellular space or released from the intracellular compartment to the cytoplasm. Among the Zn transporters are three LIV-1 subfamily members (Zip10, Zip13, and Zip14) that are responsible for the increment of Zn2+ concentration within the cytoplasm of many vertebrates. Each of the three Zip proteins usually forms a heterodimer with another protein and thus plays a functional role. For example, Zip10 forms a heterodimer with Zip6 to facilitate zinc flux to trigger mitosis [9]. Zip13 possesses some domains that are not found in other LIV-1 family members and Zip13 itself may form a homodimer [10]. Aydemir et al. [11] reported that Zip14 might act as a heterodimer with Zip8 to maintain an optimum zinc concentration in mitochondria. Due to these specialties of Zip10, Zip13, and Zip14, we considered it interesting and valuable to explore their transcription regulation. In some vertebrates, several studies explored the responses of these proteins to Zn. For instance, Croxford et al. [12] reported that the zip10 mRNA level was not influenced by Zn2+ deficiency, but the protein level of Zip10 were downregulated in response to zinc deficiency in mouse testes. Zheng et al. [13] reported that in zebrafish gill, zinc excess reduced the mRNA expression level of zip10 and that zebrafish Zip10 acts as a zinc importer for X. laevis oocytes. Lichten et al. [14] reported that high Zn incubation downregulated the zip10 mRNA expression level in mouse hepatocytes, the AML12 cell line, and the neuro 2a cell line. Further investigation revealed that the mouse mZip10 localized to the AML12 cell membrane and functioned as a Zn2+ importer, and that Zn deficiency enriched its localization in the cell membrane [14]. Lee and Bin [15] reported that hZip13 was located on the Golgi of hMSCs and directly mediated Zn homeostasis. Fukada et al. [16] reported that Zip13 disturbance could result in the disorder of intracellular Zn homeostasis. Jeong et al. [17] reported that Zn deficiency elevated the zip13 mRNA and protein level in HeLa cells. Xu et al. [18] reported that Zip13 released zinc from the vesicular store to the cytoplasm that is required for the normal function of ER and that 1 mM of Zn could increase the protein level of dZip13. Liuzzi et al. [19] reported that Zip14 was located in the plasma membrane of the hepatocytes and increased the Zn concentration of the hepatocytes in mice with hypozincemia. Kim et al. [20] reported that Zip14 was located in the cell membrane and mediated Zn homeostasis. Recently, in our laboratory, Chen et al. [21] found that high Zn administration reduced the zip10, zip13, and zip14 mRNA levels in both the hepatocytes and the intestinal epithelial cells of yellow catfish. Thus, all of these studies indicated that Zip10, Zip13, and Zip14 were regulated by Zn. However, the underlying regulatory mechanisms have received very little attention and remain confusing.
In the eukaryotes, transcription factors initiate and regulate gene expression via their binding locus in the promoter. For example, the TFs of NRF1 and the Smad3 binding site were found in the promoter region of the troc1 gene and play repressing roles in troc1 expression [22]. Liang et al. [23] reported that TFs of ACSL1 and ASCL2 transcriptionally regulated the fam13a gene through the binding region in its promoter. MTF-1, which possesses six Cys2-His2 zinc finger structures specializing in the binding of Zn, is a crucial transcription factor for inducing the transcription of some genes that are associated with Zn homeostasis [24]. MTF-1 first binds with Zn in the cytoplasm and then translocates into the nucleus, where it binds to a specific sequence, MRE, and regulates its target genes [25,26]. MRE elements are present in the promotor regions of many Zn transporters, such as the znt1, znt2, znt6, znt8, zip3, and zip8 transporters of P. fulvidraco [27,28], the zip10 transporter of zebrafish [14], and the znt1, znt2, and the znt5 transporter in mammals [29,30,31]. Krüppel-like factor 4 (KLF4) is another crucial zinc finger transcription factor [32]. Liuzzi et al. [33] found that Zn deprivation increased the binding of KLF4 to the zip4 promoter of mice and, thus, regulated the adaptive expression of zip4. Other studies concerning KLF4 and zip13 led us to speculate that there is an enigmatic connection between KLF4 and Zn homeostasis that is mediated by zip13 [16,34]. The signal transducer and activator of transcription 3 (STAT3) belongs to the JAK/STAT signaling pathway family. STAT3 possesses DNA-binding activity and is capable of interacting with enhancer elements in the promoters of downstream genes [35]. Studies suggested that Zn increased the gene expression level of STAT3 [36] but suppressed the STAT3 signaling cascade [37]. Cousins et al. [38] reported that phosphorylated STAT are capable of regulating the transcription of zip14. Thus, it is valuable to investigate whether these transcription factors mediate the transcriptional regulation of zip members zip10, zip13, and zip14.
Compared with terrestrial animals that sequester Zn from their diets, aquatic fish possess two pathways for obtaining Zn from diets or from a water environment [39]. Thus, it is worth exploring the regulatory mechanisms in fish through which Zn homeostasis is regulated. Yellow catfish, Pelteobagrus fulvidraco, are found broadly across Asian countries and provide an excellent model for nutrition research, due to their delicious flavor, high nutritional value, and economic value [40,41]. Recently, we characterized the full-length cDNA sequences of zip10, zip13, and zip14 of yellow catfish [21]. The current study investigates their transcriptional regulation, their responsive mechanisms, and their protein subcellular localization in response to Zn. We used HEK293T as our host cells, because they are reliable model cells for investigating the regulatory mechanisms of the promoters [41], the gene 3′-UTR [42], ER stress [43], and the lipid metabolism [44] of P. fulvidraco. Our study illustrated the zinc-responsive transcription regulation mechanisms of zip10, zip13, and zip14 and provides some views regarding Zip-family-mediated zinc homeostatic control in vertebrates.
2. Results
2.1. Cloning and Sequence Analysis of zip10, zip13, and zip14 Promoters of Yellow Catfish
The 2278 bp, 1917 bp, and 1989 bp sequences of yellow catfish zip10, zip13 and zip14 promoters, respectively, were cloned successfully, and several putative TFBSs were predicted, including the binding sites of MRE, PPARα:RXR, FOXO4, FOXO6, SREBP, CREB1, AP1, ZEB1, RREB1, SP1, KLF4, STAT3, STAT5a:STAT5b, STAT6, CEBPα, ATF4, CCAAT-box, and TATA-box. The relative positions of the predicted putative TFBSs are listed in Table 1. The nucleotide sequence of the zip10, zip13, and zip14 promoters are presented in Figure 1.

Table 1.
Summary of the relative location of the predicted putative TFBS in the zip10, zip13, and zip14 promoters. “None” indicates that there were no predicted corresponding TFBS by setting a threshold of 0.85 in MatInspestor and a threshold of 9 in Jaspar.


Figure 1.
The nucleotide sequences of the zip10, zip13, and zip14 promoters of P. fulvidraco. (A) zip10; (B) zip13; (C) zip14. Numbers represent the relative distance from the corresponding position to the transcription start site (TSS). The putative TFBSs are boxed in the rectangle. The sequences above the red characters represent the MRE, the KLF4 binding site, and the STAT3 binding site of these promoters.
2.2. 5′-Sequence Deletion Mutation Assays of zip10, zip13, and zip14 Promoters of Yellow Catfish
We investigated the activities of the zip10, zip13, and zip14 promoters using in vitro cultured HEK293T cells and characterized their functional regions (Figure 2). The deletion of the sequence from −2182 bp to −1767 bp significantly decreased the luciferase activity of the zip10 promoter, while the deletion of the sequence from −1767 bp to −1228 bp and from −992 bp to −262 bp increased that activity (Figure 2A). The deletion of the sequence from −861 bp to −489 bp significantly enhanced the luciferase activity of the zip13 promoter (Figure 2B). The sequence deletion from −1854 bp to −1409 bp and from −654 bp to −266 bp increased the luciferase activity of the zip14 promoter (Figure 2C).
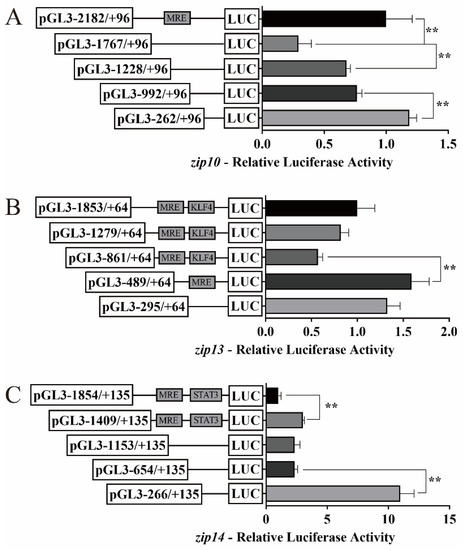
Figure 2.
5′-unidirectional deletion assays of zip10, zip13, and zip14 promoters of P. fulvidraco. (A) zip10; (B) zip13; (C) zip14. Values mean the ratio of Firefly to Renilla luciferase activities, normalized to the control plasmid. The putative MRE, KLF4 binding site, and STAT3 binding site boxed in the rectangle indicate that they were not deleted by mutation. Results are presented as mean ± SEM (n = 3). Student t-test was used to evaluate the difference. Asterisks (**) mean significant differences between the two groups (p < 0.05).
Next, we explored the responses of the zip10, zip13, and zip14 promoters to Zn2+ administration (Figure 3). For the zip10 promoter, compared with the control, Zn incubation significantly decreased the luciferase activity from −2182 bp to +96 bp and from −1228 bp and +96 bp. However, compared with the wildtype plasmid, the sequence deletion from −2182 bp to −1767 bp significantly alleviated the Zn-responsive regulatory effect and the sequence deletion from −1767 bp to −1228 bp significantly abolished that effect. These results indicated that Zn has a negative regulatory effect on the zip10 promoter (Figure 3A). For the zip13 promoter, compared with the control, Zn incubation markedly decreased the luciferase activity from −1853 bp to +64 bp and from −489 bp to +64 bp. Compared with the wild-type plasmid, Zn incubation markedly alleviated the Zn-induced regulatory effect from −1853 bp to −861 bp. These results suggested that Zn negatively regulates the two areas on the zip13 promoter (Figure 3B). For the zip14 promoter, compared with the control, Zn significantly decreased the relative luciferase activity of −1854 bp to +135 bp and −1409 bp to +135 bp. Compared with wild type plasmid, Zn incubation significantly alleviated the negative regulatory effect from −1854 bp to −1409 bp and from −1409 bp to −1153 bp. These results suggest that the two areas on the zip14 promoter negatively responded to Zn (Figure 3C).
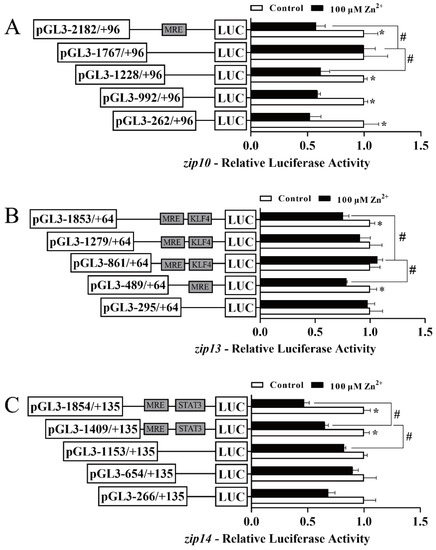
Figure 3.
5′-unidirectional deletion assays of the zip10, zip13, and zip14 promoters of P. fulvidraco after Zn2+ administration. (A) zip10; (B) zip13; (C) zip14. Values show the ratio of Firefly to Renilla luciferase activities, normalized to the control. The putative MRE, the KLF4 binding site, and the STAT3 binding site boxed in the rectangle indicate that they were not deleted by mutation. Results are presented as mean ± SEM (n = 3). Student t-test was used to evaluate the difference. Asterisk (*) indicates significant differences between the control and the Zn-incubated group (p < 0.05); hashtag (#) indicates significant differences between two 5′-unidirectional deletion plasmids under the same treatment (p < 0.05).
2.3. Site-Mutation Assays of the MTF-1, KLF4, and STAT3 Binding Sites in the zip10, zip13 and zip14 Promoters of Yellow Catfish
We carried out site-mutation analysis to determine the functional TFBS in the zip10, zip13, and zip14 promoters of yellow catfish using in vitro cultured HEK293T cells (Figure 4). The sequences of the selected predicted TFBSs are shown in Figure S1. The bigger a deoxynucleotide is, the more conservative the deoxynucleotide is among vertebrates. For the zip10 promoter, compared with the wild-type plasmid, the mutation of the putative MRE site (−2004 bp/−2017 bp) markedly increased the luciferase activity of the zip10 promoter after 100 µM Zn2+ incubation (Figure 4A), suggesting the negative regulatory effect by this MRE site in response to high Zn2+ administration. For the zip13 promoter, compared with the wild-type plasmid, the mutation of the putative MRE site (−345 bp/−360 bp) or the KLF4 binding site (−640 bp/−651 bp) significantly increased the luciferase activity of the zip13 promoter after 100µM Zn2+ incubation (Figure 4B), indicating that the MRE and KLF4 binding sites negatively regulated the transcriptional activity of the zip13 promoter in response to high Zn2+ administration. For the zip14 promoter, compared with the wild-type plasmid, the mutation of the putative MRE (−1442 bp/−1457 bp) site or the mutation of the STAT3 (1) binding site (−1383 bp/−1375 bp) markedly increased the luciferase activity of the zip14 promoter after 100 µM Zn2+ incubation. However, the mutation of the STAT3 (2) binding site (−1358 bp/−1348 bp) showed no remarkable effects (Figure 4C). Surprisingly, we found that the decreased percentage of the full-length promoter activity in Figure 4C was lower than that in Figure 3C. Based on the findings of Miller et al. [45], we thought that both full-length promoter activities in Figure 3C and Figure 4C showed a significant decrease under zinc administration, with a confidence level of 95%. Therefore, these results showed that the MRE and the STAT3 (1) binding site can negatively regulate zip14 promoter activity. However, the STAT3 (2) binding site could not participate in the Zn-responsive zip14 transcription.
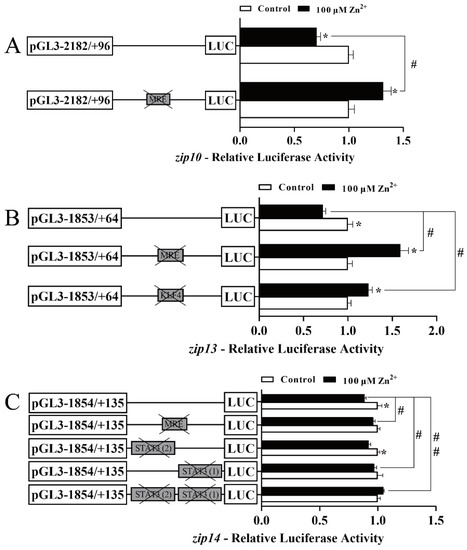
Figure 4.
Assays of the predicted MTF-1, KLF4, and STAT3 binding sites in the zip10, zip13, and zip14 promoters of P. fulvidraco after site-directed mutagenesis. (A) Site mutagenesis of MTF-1 on the pGl3-zip10-2078 vector; (B) site mutagenesis of MTF-1 and KLF4 on the pGl3-zip13-1917 vector; (C) site mutagenesis of MTF-1 and STAT3 on the pGl3-zip14-1989 vector. Values mean the ratio of activities of Firefly to Renilla luciferase, normalized to the control. Results are presented as mean ± SEM (n = 3). Student t-test was used to evaluate the difference. Asterisk (*) indicates significant differences between the control and the Zn group (p < 0.05); hashtag (#) indicates significant differences between two 5′-unidirectional deletion plasmids under the same treatment (p < 0.05).
2.4. Electrophoretic Mobility Shift Assay
Next, we carried out EMSA to verify whether these MREs and the binding sites of KLF4 and STAT3 were involved in the regulation on the three zips promoters. For the MRE from −2017 bp to −2004 bp on the zip10 promoter, when the MRE sequences were labeled by the biotin as the probe, we found a bright binding band (lane 2, Figure 5A). Unexpectedly, the 100-fold unlabeled MRE slightly competed for the binding with the NP (lane 3, Figure 5A), and the 100-fold unlabeled unspecific probe alleviated the competition (lane 4, Figure 5A), indicating that −2017 bp/−2004 bp MRE of the zip10 promoter could bind with the nuclear protein. Zn enhances the brightness of the band (lane 5, Figure 5A), suggesting that this MRE of zip10 was functional in response to zinc. For the MRE from −345 to −360 bp on the zip13 promoter, when the MRE sequence was labeled by the biotin as the probe, we found an apparently single binding band (lane 2, Figure 5B). The 100-fold unlabeled MRE probe competed for its binding with the NP (lane 3, Figure 5B), and the 100-fold unlabeled unspecific MRE probe alleviated the competition (lane 4, Figure 5B), implying that −345 bp/−360 bp MRE of the zip13 promoter could bind with the nuclear protein. However, Zn slightly decreased the brightness of the band, compared with the control (lane 5, Figure 5B). For the MRE from −1457 bp to −1442 bp on the zip14 promoter, when the MRE sequence was labeled by the biotin as the probe, we found an obvious single binding band (lane 2, Figure 5C). The 100-fold unlabeled MRE probe competed for its binding with the NP (lane 3, Figure 5C), and the 100-fold unlabeled mutated probe alleviated their competition (lane 4, Figure 5C), implying that the −1457 bp/−1442 bp MRE of the zip14 promoter could bind with the nuclear protein. However, similar to the zip13 promoter, Zn also decreased the brightness of the binding band, compared with the control (lane 5, Figure 5C). A similar phenomenon was also reported in the promoter region of mt [40] and zip3 [28]. Those results suggested that these putative MRE sites may be involved in the zinc-responsive transcription regulation of the zip10, zip13 and zip14 promoters For the KLF4 binding sequence from −651 bp to −640 bp on the zip13 promoter, when the sequence was labeled by the biotin as the probe, we found an apparent binding band (lane 2, Figure 5D). The 100-fold unlabeled sequence competed for the binding with the NP (lane 3, Figure 5D), and the 100-fold unlabeled unspecific probe alleviated their competition (lane 4, Figure 5C), indicating that the −651 bp/−640 bp KLF4 binding site of the zip13 promoter could bind with the nuclear protein. Meanwhile, Zn also increased the brightness of the band compared with the control (lane 5, Figure 5D), suggesting that the putative KLF4 binding site might participate in the zinc-responsive transcription regulation of the zip13 promoter. For the STAT3 binding sequence from −1383 bp to −1374 bp on the zip14 promoter, when the sequence was labeled by the biotin as the probe, we found an apparent binding band (lane 2, Figure 5E). The 100-fold unlabeled MRE competed for the binding with the NP (lane 3, Figure 5E), and the 100-fold unlabeled unspecific probe alleviated their competition (lane 4, Figure 5E), implying that the −1383 bp/−1374 bp STAT3 binding site of the zip14 promoter could bind with the nuclear protein. Meanwhile, Zn also decreased the brightness of the band, compared with the control (lane5, Figure 5E). The result suggested that the putative STAT3 binding site may participate in zinc-responsive zip14 transcription regulation. Taken together, our study suggested that these MREs and the binding sites of KLF4 and STAT3 were involved in the regulation on the three zip promoters.
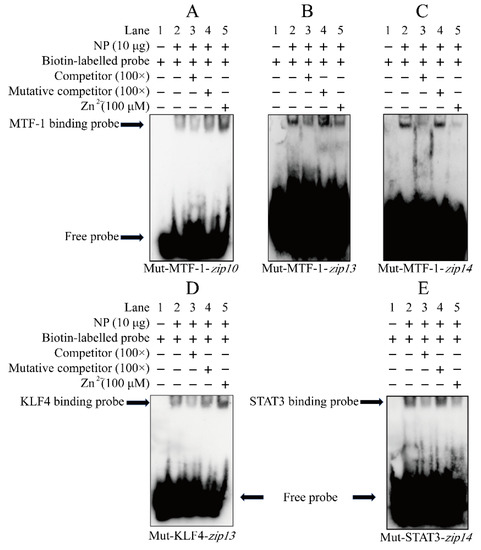
Figure 5.
EMSA of the predicted MTF-1, KLF4, and STAT3 binding sequences on the zip10, zip13, and zip14 promoters of P. fulvidraco. (A) MTF-1 binding sites between −2017 bp and −2004 bp in zip10 promoter; (B) MTF-1 binding sites between −360 bp and −345 bp in the zip13 promoter; (C) MTF-1 binding sites between −1457 bp and −1442 bp in the zip14 promoter; (D) KLF4 binding site between −606 bp and −594 bp in the zip10 promoter; (E) STAT3 binding site between −1383 bp and −1375 bp of the zip14 promoter. NP: nuclear protein. The numbers 1–5 represent the five lanes, respectively.
2.5. Response of pfZip10-EGFP, pfZip13-EGFP, and pfZip14-EGFP to Zn Administration in HEK293T Cells
Next, we explored whether the pfZip10, pfZip13, and pfZip14 protein expressions were post-translationally regulated by Zn. Due to the absence of available antibodies, we tagged them with the EGFP, which can release the green fluorescence after being excited by ultraviolet light. The analysis method was adopted according to Shihan et al. [46]. In brief, we first used a confocal microscope to catch fluorescent images. Then, the images were inverted into 8-bit one and gray background via Image J. After setting identical upper and lower thresholds for a single EGFP-tagged protein, the images were subjected to calculate the mean fluorescent intensity (Mean). A similar method was also used in other studies [47,48]. The administration of 100 μM Zn significantly reduced the relative green fluorescent intensities of the pfZip10-EGFP and the pfZip14-EGFP, but not that of the pfZip13-EGFP, indicating that the pfZip10 and pfZip14 proteins were post-translationally regulated by Zn (Figure 6).
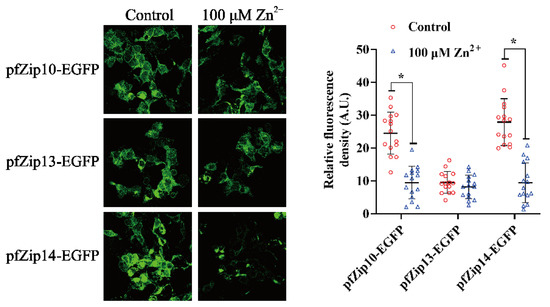
Figure 6.
The statistical analysis of the relative fluorescent density of pfZip10-EGFP, pfZip13-EGFP, and pfZip14-EGFP after Zn2+ administration. The left panels show the representative image under microscope, with or without Zn incubation. Only one of the fifteen independent experimental results is presented for each group. The right panels show the results, after statistical analysis of the relative fluorescent density. Data were calculated using Image J by setting the upper and lower thresholds. The results were presented as mean ±SEM (n = 15) and by scatter diagram. Student t-test was used to evaluate the difference. Asterisk (*) indicates significant differences between the control and the Zn group (p < 0.05).
2.6. The Subcellular Localization of the Zip10, Zip13, and Zip14 Proteins of Yellow Catfish in HEK293T Cells
Next, we explored the subcellular localization of pfZip10, pfZip13, and pfZip14 of P. fulvidraco by using HEK293T cells. The C-terminal EGFP-tagged pfZip10, pfZip13, and pfZip14 were transfected into human HEK293T cells, and the green fluorescence represented the position of pfZip10, pfZip13, and pfZip14. The membrane tracker (red), the lysosome tracker (red), the ER tracker (red), and the Golgi tracker were applied to mark the positions of the cell membrane, the lysosome, the endoplasmic reticulum, and the Golgi apparatus, respectively. The green fluorescence from the pfZip10-EGFP overlapped with the red fluorescence released by the membrane tracker (red), indicating that pfZip10 was localized to the plasma membrane (Figure 7A). The green fluorescence from the pfZip13-EGFP partially overlapped with the red fluorescence released by both ER tracker (red) and Golgi tracker (red) (Figure 7B,C), indicating that the pfZip13 colocalize with the Golgi apparatus and the endoplasmic reticulum. The green fluorescence released by pfZip14-EGFP also overlapped with the red fluorescence released by the membrane tracker (red) (Figure 7D) and with the red fluorescence released by the lysosome tracker (red) (Figure 7E), indicating that the pfZip14 localize to the plasma membrane and colocalize with the subcellular lysosome.
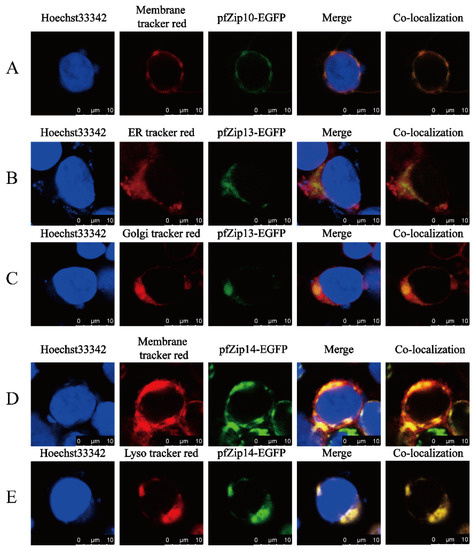
Figure 7.
Laser confocal microscope imaging of pfZip10-EGFP, pfZip13-EGFP, and pfZip14-EGFP. Blue, red, and green represents the cell nucleus, the corresponding cell compartment or cytomembrane, and pfZips-EGFP, respectively. The yellow represents the red merged with the green. (A) The green overlap with the red indicates the localization of pfZip10 to the cell membrane; (B,C) The green overlap with the red indicates that pfZip13 colocalizes with the ER and the Golgi; (D,E) The green overlap with the red indicates that pfZip14 localizes to the cell membrane and colocalizes with the lysosome. Scale labels are 10 μm.
3. Discussion
The promoter region is a short 100–1000 bp DNA fragment upstream from the transcription start site [49]. Chen et al. [40] and Yan et al. [50] reported approximately 2000 bp gene promoters of fish species. Similarly, in our study, 2278 bp, 1917 bp, and 1989 bp zip10, zip13 and zip 14 promoters of P. fulvidraco were cloned. The earlier studies suggested that the core promoters of genes played central roles in initiating the transcriptions [51,52]. In our study, we predicted one GC-box (SP1), one CCAAT-box (NF-Y), and one TATA-box in the core region of the zip10 promoter. Similarly, Zang et al. [53] reported that SP1 can bind to the human zip8 promoter and modulate its expression. Langmade et al. [54] reported one SP1 in the mouse mt1 promoter, and four SP1 in the mouse znt1 promoter, which are required for their basal expression. Bird et al. [55] reported that the yeast homologous Zn transporter zrt2 possesses a TATA-box that is required for restraining its expression. The CCAAT-box is a significant component regulated by NF-Y [56]. Our study also indicated that the zip13 promoter possesses one CCAAT-box and one TATA-box, in agreement with the results reported for the zip8 and mt promoters of P. fulvidraco [28,40]. In addition, our study found only one SP1 in the zip14 promoter. Furthermore, our study predicted some putative TFBSs in their promoters, such as CEBPα, SREBP, PPARα, MTF-1, ATF, KLF, STAT3, RREB1, and ZEB1, which may possess a regulatory function for the three promoters (zip10, zip13, and zip14). These TFBSs have been reported in previously published studies as being within the promoter regions of other genes [27,31,57,58]. In the present study, we chose MTF-1, KLF4, and STAT3 binding sites to explore whether they are functional sites for the regulation of the zip10, zip13, and zip14 promoters.
MTF-1 is an intracellular Zn sensor that can bind to the MRE of the promoters of its target genes and activate target genes associated with Zn homeostasis [29]. Recently, Chen et al. [40] found that the MRE existed in the region of the mt promoter and was essential for the upregulation of mt expression. Song et al. [41] found that MTF-1 can bind to the MREs of znt1, znt2, znt6. and znt8 and upregulate their expression. As in our study, the site mutation analysis implied that the mutation of MRE alleviated the decrease in luciferase activity that was caused by high Zn administration. Here, we found that the −2017 bp/−2004 bp MRE in the zip10 promoter, the −360 bp/−345 bp MRE in the zip13 promoter, and the −1457 bp /−1442 MRE in the zip14 promoter were indispensable for the downregulation of zip10, zip13, and zip14 expression induced by high Zn administration. Similarly, Chen et al. [21] reported the decreased relative gene expression of the zip10, zip13, and zip14 promoters in both hepatocytes and intestinal epithelial cells. Interestingly, in the zip10 promoter, the MRE deletion showed different activity with the mutated MRE. Deletion mutation deleted a sequence of approximately 400 bp, while site mutation disrupted only the putative binding sequence. Therefore, we speculated that the rest sequence in the deleted 400 bp~ was functionally antagonistic to the MRE in response to zinc. Furthermore, our EMSA results showed that those MREs were capable of binding nuclear protein. High Zn enhanced the binding effect in the zip10 promoter. However, high-Zn administration showed a relatively low binding affinity in the zip13 and zip14 promoters, and a similar phenomenon was observed in the zip3 and mt promoters [28,40]. Lichten et al. [14] showed that knockout of the MTF-1 gene in mice increased the gene expression of the zip10 promoter, implying that MTF-1 was a functional repressor of the zip10 promoter under high-Zn treatment. Similarly, Chen et al. [28] found a negative regulatory effect of the MRE on the zip3 and zip8 promoters. Taken together, these results indicated the zinc-responsive regulation roles of these MREs in the transcriptional regulation of the zip10, zip13, and zip14 promoters.
In our study, we predicted −606 bp/−594 bp KLF4 binding sites within the zip13 promoter. We found that the mutation of the KLF4 binding site alleviated the Zn-induced decrease in the activity of the zip13 promoter, suggesting that this KLF4 binding site is necessary for the decrease in zip13 expression activity in a high-Zn environment. Our EMSA showed that high Zn significantly enhanced the binding effect between nuclear proteins and the KLF4 binding sites. Thus, we confirmed that the KLF4 binding sites could regulate the zip13 expression in response to a high zinc environment, implying a crucial mechanism whereby KLF4 maintains Zn homeostasis. It was also reported that the KLF4 binding site is responsible for the upregulation of mtf-1 expression [40]. It seems reasonable to speculate that the KLF4 binding site mediated the MTF-1-induced downregulation of the zip13 promoter. Liuzzi et al. [33] reported that knockdown of klf4 restrained the upregulation of the zip4 promoter in a zinc-depleted environment. Similarly, our results indicated that KLF4 play crucial roles in maintaining intracellular Zn homeostasis in a high-Zn environment through regulating zip13 expression.
We also predicated the −1383 bp /−1375 bp STAT3 (1) and the −1358 bp /−1348 bp STAT3 (2) binding sites within the zip14 promoter. The site mutation analysis indicated that STAT3 (1), not STAT3 (2), was responsible for the downregulation of the activity of the zip14 promoter, and our EMSA confirmed that the effect of the −1383 bp /−1375 bp STAT3 (1) binding site on the zip14 promoter was the functional one. Thus, our results indicated that STAT3 mediated the maintenance of Zn homeostasis by regulating the zip14 expression via the −1373/−1383 bp STAT3 (1) binding site on its promoter. Interestingly, Du et al. [59] discovered that STAT3 was activated in response to Zn deprivation and mediated Zn deficiency-induced upregulation of the zip2 expression. Kitabayashi et al. [37] reported that Zn could attenuate STAT3 activation. Thus, we speculated that Zn alleviated the binding effect of STAT3 to the zip14 promoter. In addition, other studies suggested that the zip6 promoter served as the STAT3 target [60,61]. Cousins et al. [38] mentioned that phosphorylated STAT accounted for the expression of the zip14 promoter Thus, these results, together with ours, suggest that STAT3 plays a crucial role in maintaining Zn homeostasis and that the STAT3 binding site participated in the control of the zip14 promoter of P. fulvidraco.
Within the body, there could be enhancer (or silencer) DNA fragments far away from the transcription start site, and the formation of a chromosome loop makes it possible for the enhancer (or silencer) to participate in enhancing (or repressing) transcription. In our study, we confirmed that the predicted zinc-related TF binding sites were functional and involved in zinc-responsive regulation. Most TF binding sites are a 4 bp- to 30-bp wide integral DNA sequence [62]. Furthermore, the core consensus sequence of the MRE is TGCRCNC, in which R represents A or G and N represents any nucleotide [63]. STAT3-binding sites are characterized by clusters of conserved motifs with a core sequence of TTCT/CNA/GGAA, in which N represents any nucleotide [64]. KLF4 binding sites possess a DNA bind domain of CACCC-box [65]. Therefore, site-mutation was carried out by substituting the predicted conservative sequences with non-conservative ones, according to Jaspar. This may result in low specificity of the experiment. Further study is required to identify the function of a single nucleotide. We highlighted that the promoters are functional in the regulation of the zinc homeostasis of the host HEK293T cells. Referring to our previous study [21], we also speculate that the promoter might participate in the zinc homeostasis control of its native organism, P. fulvidraco.
The SLC39A Zn transporters carry Zn into cytoplasm from intracellular organelles or extracellular space, and their specific subcellular localization determines the specific function of the Zn transport. For example, Kong et al. [66] found that maternally derived Zip10 localize to oocytes membrane and help to drive the oocyte-to-egg transition. Jeong et al. [17] reported that the vesicular-localized Zip13 could adjust the dynamic of cytosolic labile Zn levels, which help accumulate Zn into vesicles, causing ER dysfunction and stress. Fukunaka et al. [67] found that the Zip13 located on the Golgi could suppress the biogenesis of beige adipocytes and energy expenditure by regulating the C/EBP-β expression. Kim et al. [20] pointed out that Zn transported by the plasma-membrane-located Zip14 could subdue apoptosis and steatosis by restraining the activity of PTP1B and ER stress. Thus, it is worth exploring the subcellular localization of Zip10, Zip13, and Zip14. In our study, the pfZip10 located on the cell membrane, in agreement with other reports [14,66,68]. Our study indicated that the pfZip13 colocalizes with both the Golgi apparatus and the endoplasmic reticulum. Similarly, Xiao et al. [69] found that the Drosophila dZip13 partially colocalize with the ER/Golgi. In our study, the pfZip14 not only localize to the plasma membrane, but also colocalize with intracellular lysosome. Similarly, Zhao et al. [70] reported that the mouse mZip14 localizes to the basolateral membrane of enterocytes, early endosomes, and lysosomes.
Several studies have established that high Zn reduced the mRNA and protein expression of the zip10, zip13, and zip14 promoters [13,14,17,18,21]. Similarly, our fluorescence statistics experiments showed that Zn administration reduced both pfZip10 and pfZip14, but not pfZip13. Lysosome is an organelle involved in the degradation of cell debris and unnecessary protein via endocytosis. Wang et al. [71] reported that Zn repletion could lead exogenously-expressed mouse mZip1-HA and mZip3-HA protein to the endocytosis pathway. Kim et al. [72] reported that mZip4-HA in HEK293 was endocytosed in response to zinc treatment. Their studies suggested that mZip1-HA, mZip3-HA, and mZip4-HA are post-translationally regulated by zinc, which led us to speculate that high zinc could regulate pfZip10, pfZip13, and pfZip14 from the protein level.
4. Materials and Methods
4.1. Animals, Cells, and Reagents
The tail fins of P. fulvidraco, used for genomic DNA extraction, were purchased from a local fishery farm (Wuhan, China). The HEK293T cell lines were from the Cell Resource Center in the Fishery College of Huazhong Agricultural University (HZAU, Wuhan, China). The Dulbecco’s modified eagles medium (DMEM), the 0.25% trypsin-EDTA, and the fetal bovine serum (FBS) were from Gibco (Thermo Fisher Scientific, Waltham, MA, USA). Lipo293 for transfecting plasmid into HEK293T were from Beyotime Biotechnology, Wuhan, China. Passive lysis buffer and the dual luciferase activity assay reagents were from Promega, Madison, WI, USA. Nuclear protein extraction kits were from Thermo Fisher Scientific, Waltham, MA, USA). We made sure that the protocols for animal experiments and the use of cell lines followed the institutional ethical guidelines of HZAU for the care and use of laboratory animals, and that they were approved by the university’s ethics committee (I.D. code: Fish-2020-09-24).
4.2. Promoters Cloning and Plasmids Construction
Based on the full-length complementary DNA (cDNA) of the zip10, zip13, and zip14 promoters obtained in our laboratory [21], we identified their transcription start sites (TSS) through the RNA ligase-mediated rapid amplification of 5′cDNA ends (RLM-5′ RACE) method that was described in Xu et al. [73]. For constructs for promoters, the specific primers used for nested PCR-generating promoters are listed in Table S1. Then, the zip10, zip13 and zip14 promoters were subcloned into the pGL3-Basic between the Sac Ⅰ and Hind Ⅲ restriction cloning sites with Clone press II One Step Cloning Kit (Vyzame, Piscataway, NJ, USA), according to the distance from the TSS. Those full-length promoters (wild type) were pGL3-2182/+96 (zip10), pGL3-1853/+64 (zip13), and pGL3-1854/+135 (zip14). The unidirectional deletion mutation products of corresponding promoters were randomly generated using the full-length promoters as templates. The primer sequences are listed in Table S2. According to the distance from the TSS, those deletion mutation promoters were pGL3-1767/+96, pGL3-1228/+96, pGL3-992/+96, and pGL3-262/+96 for zip10 promoter; pGL3-1279/+64, pGL3-861/+64, pGL3-489/+64, and pGL3-295/+64 for zip13 promoter; and pGL3-1409/+135, pGL3-1153/+135, pGL3-654/+135, and pGL3-266/+135 for zip14 promoter. For constructs coding EGFP-tagged pfZips (pfZip10, pfZip13, and pfZip14), the complete cDNA sequences of the zip10, zip13 and zip14 promoters were generated by the same nested PCR principle. The primers are listed in Table S3. Then, the pfZips’ cDNA end with sequences coding EGFP were subcloned into pcDNA3.1 with the CMV promoter between the Himd III and Xhol restriction cloning sites with a Clone press II One Step Cloning Kit (Vyzame, Piscataway, NJ, USA), and the constructs were pcDNA3.1-pfZip10-EGFP, pcDNA3.1-pfZip13-EGFP, and pcDNA3.1-pfZip14-EGFP.
4.3. Promoter Sequence Analysis
For the function characterization of these promoters, we predicted the putative transcription factor bind sites (TFBSs) with the online database MatInspector (http://www.genomatix.de/, accessed on 31 August 2021), with a minimum threshold of 0.85, and Jaspar (http://jaspar.genereg.net/, accessed on 31 August 2021), with minimum threshold of 9. The MegAlign Pro Sequence Alignment Software was used to evaluate whether the sequences were successfully cloned, according to NCBI. (Accession numbers: pfZip10, MK448212.1; pfZip13, MK448215.1; and pfZip14, MK448216.1).
4.4. Dual Luciferase Activities Assays
The transfection of the constructed promoter plasmids into the HEK293T cells and the assay of the dual luciferase activities were carried out according to the methods of Xu et al. [73]. Briefly, the HEK293T cells were seeded into 24-well plates and maintained in the DMEM medium with 10% FBS at 5% CO2 and 37 °C in a humidified atmosphere until the relative density reached 70% to 80% confluence. Then, the maintaining DMEM was replaced with fresh DMEM (10% FBS) or DMEM (10% FBS) + 100 μM Zn2+. The Zn concentration was selected according to our recent study [40]. Four hundred and fifty ng constructed promoter plasmids and 50 ng pRL-TK as control were co-transfected into the HEK293T cells using Lipo 293. Each transfection was conducted in triplicate. Then, after 24 h incubation, the cells were harvested and lysed with the passive lyse buffer for analysis of the dual luciferase activities according to the protocols of the dual-luciferase reporter assay system. We measured the ratio of the Firefly to Renilla luciferase activity.
4.5. Site Mutation Analysis of TFBS in the zip10, zip13, and zip14 Promoters
To identify the corresponding binding sites on the zip10, zip13 and zip14 promoters, we used the QuickChange II Site-Directed Mutagenesis Kit (Vazyme, Piscataway, NJ, USA) to direct the site mutation. The corresponding full-length promoters were used as templates. The specific primers for the site mutation are listed in Table S4, and the plasmids were Mut-MTF-1-zip10, Mut-MTF-1-zip13, Mut-KLF4-pGL3-zip14, Mut-MTF-1-pGL3-zip13, and Mut-STAT3-pGL3-zip14. Then, the plasmids were transfected and the cells were harvested for dual luciferase activity analysis.
4.6. Electrophoretic Mobility-Shift Assay
An EMSA was conducted to verify the MTF-1, KLF4, and STAT3 binding on the zip10, zip13, and zip14 promoters. Briefly, the HEK293T cells were seeded into 6-cm dishes and maintained in the DMEM with or without 100 μM Zn2+. The incubation lasted for 24 h. Then, the cells were lysed for the extraction of nucleoprotein (NP) and the protein concentration was determined with a bicinchoninic acid assay (BCA) method. Ten μg of nucleoprotein was added and mixed with each oligonucleotide duplex of the binding sequence of the MTF-1, KLF4, and STAT3, using the EMSA kit according to manufacturer’s protocols (Invitrogen, Carlsbad, CA, USA). The oligonucleotide sequences of the EMSA are presented in Table S5.
4.7. Fluorescence Intensity Analysis
The transfection protocols of the constructed pcDNA3.1-pfZip10-EGFP, pcDNA3.1-pfZip13-EGFP, and pcDNA3.1-pfZip14-EGFP were the same as that of the promoter plasmids indicated in the Section 2.4; the 500 ng constructed plasmids were transfected. Then, after 24 h, the cells were washed carefully three times with PBS containing 1 mm MgCl2 and 0.1 mm CaCl2. They were fixed with 4% paraformaldehyde for 5 min at room temperature. Next, the cells were again washed carefully three times with PBS. A Leica TCS DMI8 fluorescent microscope was used to catch the green fluorescence images. Image J software was used to quantify the relative fluorescence intensity according to the methods Shihan et al. [46] and Ruggiero et al. [47].
4.8. Subcellular Localization Investigation of Zip10, Zip13, and Zip14
We further investigated the pfZips’ subcellular localization. The HEK293T cells were seeded on coverslips, and the transfection of the constructed pcDNA3.1-pfZip10-EGFP, pcDNA3.1-pfZip13-EGFP, and pcDNA3.1-pfZip14-EGFP and the cell-washing were as indicated in the Section 2.4. Then, the membrane tracker (red) (Dil, 1:400), the lysosome tracker (red) (DND-99, 1:1000), the ER tracker (red) (Glipalamides, 1:1000), or the Golgi tracker (red) (C5-ceramide, 1:100) were used to track the cell membrane, the lysosome, the endoplasmic reticulum (ER), or the Golgi apparatus, respectively. To stain the nuclei, the cells were washed with PBS and then incubated in 10 g/mL Hoechst for 10 min. After several washes via PBS, coverslips were mounted on the microscope slides for the confocal imaging, using a Leica TCS SP8 laser-scanning confocal microscope.
4.9. Statistical Analysis
We carried out the statistical analysis with SPSS 22.0 software (Armonk, NY, USA). All of the data were presented as means ± standard error of mean (SEM). First, we analyzed the data for normality via the Kolmogorov–Smirnov test, and for the homogeneity of the variances among the treatments via Bartlett’s test. Then, we used one-factor ANOVA and Duncan’s multiple range test to analyze the significance of the differences among ≥3 treatments, and Student’s t-test between two treatments; p < 0.05 was considered to be statistically significant.
5. Conclusions
In summary, for the first time, we characterized the structure and function of the zip10, zip13, and zip14 promoters from P. fulvidraco in the heterologous expression host HEK293T. We discovered that a high concentration of Zn decreased the activities of the three zip promoters. The MRE in the zip10 promoter, the MRE and KLF4 binding sites in the zip13 promoter, and the MRE and STAT3 binding sites in the zip14 promoter mediated the zinc responsive transcription regulation of their activities. We found that the pfZip10 located on the cell membrane, the pfZip13 colocalized with ER and Golgi, and the pfZip14 colocalized with lysosome. A high Zn administration reduced the protein level of pfZip10 and pfZip14. We illustrated the transcription regulation of zip10, zip13, and zip14 promoters under zinc administration and provided a novel mechanism for Zip-family-mediated zinc homeostatic control in vertebrates.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/ijms23148034/s1.
Author Contributions
Z.L.: conceptualization; writing—review and editing; project administration; supervision; and funding acquisition. S.-Z.L.: methodology; formal analysis; investigation; visualization; writing—review and editing. Y.-C.X.: validation; writing—review and editing. X.-Y.T.: conceptualization; writing—review and editing. T.Z.: formal analysis; writing—review and editing. D.-G.Z.: methodology; visualization; writing—review and editing. H.Y.: methodology; writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This work received funding from the National Natural Science Foundation of China (32030111, 31872585) and the National Key R&D Program of China (2018YFD0900400). The authors sincerely thank all the staff in our laboratory who contributed to the experiments.
Institutional Review Board Statement
The protocols for animal experiments and the use of cells lines followed the institutional ethical guidelines of Huazhong Agricultural University for the care and use of laboratory animals and were approved by the university’s ethics committee (I.D. code: Fish-2020-09-24).
Informed Consent Statement
Informed consent was obtained from all subjects involved in the study.
Data Availability Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Conflicts of Interest
The authors declare no conflict of interest.
Abbreviations
LZT: LIV-1 subfamily of zinc transporter; MRE: metal responsive element; PPARα: peroxisome proliferator-activated receptor alpha; FOXO: forkhead box protein; SREBP: sterol regulatory element-binding protein; CREB1: cAMP responsive element-binding protein 1; ZEB1: zinc finger E-box-binding homeobox 1; SP1: GC-box; STAT3: signal transducer and activator of transcription 3; KLF4: Krüppel-like factor 4; CEBPα: CCAAT enhancer binding protein alpha; RREB1: ras responsive element-binding protein-1; MTF-1: metal-responsive transcription factor-1; TFBS: transcription factor binding sites; TSS: transcription start sites; DMEM: Dulbecco’s modified eagles medium; FBS: fetal bovine serum; RLM-5′ RACE: RNA ligase-mediated rapid amplification of 5′cDNA ends; EMSA: electrophoretic mobility shift assay; NE: nuclear extract; EGFP, enhanced green fluorescent protein; SEM: standard error of mean; ER: endoplasmic reticulum; Zn: zinc; SLC39A: solute carrier 39A, Zip: protein; zip: gene.
References
- Roh, H.C.; Collier, S.; Guthrie, J.; Robertson, J.D.; Kornfeld, K. Lysosome-related organelles in intestinal cells are a zinc storage site in C. elegans. Cell Metab. 2012, 15, 88–99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Watanabe, T.; Kiron, V.; Satoh, S. Trace minerals in fish nutrition. Aquaculture 1997, 151, 185–207. [Google Scholar] [CrossRef]
- Lall, S.P.; Kaushik, S.J. Nutrition and Metabolism of Minerals in Fish. Animals 2021, 11, 2711. [Google Scholar] [CrossRef] [PubMed]
- De Schamphelaere, K.A.C.; Janssen, C.R. Bioavailability and Chronic Toxicity of Zinc to Juvenile Rainbow Trout (Oncorhynchus mykiss): Comparison with Other Fish Species and Development of a Biotic Ligand Model. Environ. Sci. Technol. 2004, 38, 6201–6209. [Google Scholar] [CrossRef] [PubMed]
- Kambe, T.; Tsuji, T.; Hashimoto, A.; Itsumura, N. The Physiological, Biochemical, and Molecular Roles of Zinc Transporters in Zinc Homeostasis and Metabolism. Physiol. Rev. 2015, 95, 3. [Google Scholar] [CrossRef]
- Zhao, L.; Tan, J.Q.; Li, D.; Jiang, L.J.; Li, T.J.; Yang, Y.; Wang, G.X.; Shang, Z.; Wang, J.; Zhou, J.F. SLC39A6/ZIP6 is essential for zinc homeostasis and T-cell development in zebrafish. Biochem. Biophys. Res. Commun. 2019, 511, 896–902. [Google Scholar] [CrossRef]
- Mackay, E.W.; Molero, S.I.; Tirathdas, L.H.; Peterson-Maduro, J.; Zang, J.J.; Neuhauss, S.C.F.; Schulte-Merker, S.; Wilson, S.W. Zinc dysregulation in slc30a8 (znt8) mutant zebrafish leads to blindness and disrupts bone mineralisation. bioRxiv 2020. [Google Scholar] [CrossRef]
- Muylle, F.A.R.; Adriaensen, D.; Coen, W.D.; Jean-Pierre Timmermans, J.P.; Blust, R. Tracing of labile zinc in live fish hepatocytes using FluoZin-3. BioMetals 2006, 19, 437–450. [Google Scholar] [CrossRef]
- Nimmanon, T.; Ziliotto, S.; Ogle, O.; Burt, A.; Gee, J.M.W.; Andrews, G.K. The ZIP6/ZIP10 heteromer is essential for the zinc-mediated trigger of mitosis. Cell Mol. Life Sci. 2021, 78, 1781–1798. [Google Scholar] [CrossRef]
- Bin, B.H.; Fukada, T.; Hosaka, T.; Nishida, K.; Yokoyama, S.; Hirano, T. Biochemical Characterization of Human ZIP13 Protein: A homo-dimerized zinc transporter involved in the spondylocheiro dysplastic ehlers-danlos syndrome. J. Biol. Chem. 2011, 286, 40255–40265. [Google Scholar] [CrossRef] [Green Version]
- Aydemir, T.B.; Troche, C.; Kim, M.H.; Cousins, R.J. Hepatic ZIP14-mediated Zinc Transport Contributes to Endosomal Insulin Receptor Trafficking and Glucose. J. Biol. Chem. 2016, 291, 23939–23951. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Croxford, T.P.; McCormick, N.H.; Kelleher, S.L. Moderate Zinc Deficiency Reduces Testicular Zip6 and Zip10 Abundance and Impairs Spermatogenesis in Mice. J. Nutr. 2011, 41, 359–365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, D.; Feeney, G.P.; Kille, P.; Hogstrand, C. Reglation of ZIP and ZnT zinc transporters in zebrafish gill: Zinc repression of ZIP10 transcription by an intronic MRE cluster. Physiol. Genom. 2008, 34, 205–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lichten, L.A.; Ryu, M.S.; Guo, L.; Embury, J.; Cousins, R.J. MTF-1-mediated repression of the zinc transporter Zip10 is alleviated by zinc restriction. PLoS ONE 2011, 6, e21526. [Google Scholar] [CrossRef]
- Lee, M.G.; Bin, B.H. Different actions of intracellular zinc transporters ZIP7 and ZIP13 are essential for dermal development. Int. J. Mol. Sci. 2019, 20, 3941. [Google Scholar] [CrossRef] [Green Version]
- Fukada, T.; Asada, Y.; Mishima, K.; Shimoda, S.; Saito, I. Slc39a13/Zip13, a crucial zinc transporter involved in tooth development and inherited disorders. J. Oral Biosci. 2011, 531, 1–12. [Google Scholar] [CrossRef]
- Jeong, J.; Walker, J.M.; Wang, F.; Park, J.G.; Palmer, A.E.; Giunta, C.; Rohrbach, M.; Steinmann, B.; Eidea, D.J. Promotion of vesicular zinc efflux by ZIP13 and its implications for spondylocheiro dysplastic Ehlers–Danlos syndrome. Proc. Natl. Acad. Sci. USA 2012, 109, E3530–E3538. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.X.; Wan, Z.H.; Zhou, B. Drosophila ZIP13 is post-translationally regulated by iron-mediated stabilization. BBA–Mol. Cell Res. 2019, 1866, 1487–1497. [Google Scholar] [CrossRef]
- Liuzzi, J.P.; Lichten, L.A.; Rivera, S.; Blanchard, R.K.; Aydemir, T.B.; Knutson, M.D.; Ganz, T.; Cousins, R.J. Interleukin-6 regulates the zinc transporter Zip14 in liver and contributes to the hypozincemia of the acute-phase response. Proc. Natl. Acad. Sci. USA 2005, 102, 6843–6848. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.H.; Aydemir, T.B.; Kim, J.; Cousins, R.J. Hepatic ZIP14-mediated zinc transport is required for adaptation to endoplasmic reticulum stress. Proc. Natl. Acad. Sci. USA 2017, 114, E5805–E5814. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.W.; Wu, K.; Lv, W.H.; Song, C.C.; Luo, Z. Molecular characterization of ten zinc Zn transporter genes and their regulation to Zn metabolism in freshwater teleost yellow catfish Pelteobagrus fulvidraco. J. Trace Elem. Med. Bio. 2020, 59, 126433. [Google Scholar] [CrossRef] [PubMed]
- Khan, R.; Abbas Raza, S.H.; Schreurs, N.; Wang, X.Y.; Wang, H.B.; Ullah, I.; AbdurRahman, A.; Suhail, S.M.; Khan, S.; Linsen, Z. Bioinformatics analysis and transcriptional regulation of TORC1 gene through transcription factors NRF1 and Smad3 in bovine preadipocytes. Genomics 2020, 112, 1575–1587. [Google Scholar] [CrossRef] [PubMed]
- Liang, C.; Li, A.; Raza, S.H.A.; Khan, R.; Wang, X.; Wang, S.; Wang, G.; Zhang, Y.; Zan, L. The Molecular Characteristics of the FAM13A Gene and the Role of Transcription Factors ACSL1 and ASCL2 in Its Core Promoter Region. Genes 2019, 10, 981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laity, J.H. and Andrews, G.K. Understanding the mechanisms of zinc-sensing by metal response element binding transcription factor-1 (MTF-1). Arch. Biochem. Biophys. 2007, 463, 201–210. [Google Scholar] [CrossRef]
- Kimura, T.; Itoh, N.; Andrews, G.K. Mechanisms of heavy metal sensing by metal response element-binding transcription factor-1. J. Health Sci. 2009, 55, 484–494. [Google Scholar] [CrossRef] [Green Version]
- Giedroc, D.P.; Chen, X.H.; Apuy, J.L. Metal response element (MRE)-binding transcription factor-1 (MTF-1), structure, function, and regulation. Antioxid. Redox. Sign. 2001, 3, 577–596. [Google Scholar] [CrossRef]
- Song, C.C.; Chen, G.H.; Zhong, C.C.; Chen, F.; Chen, S.W.; Luo, Z. Transcriptional responses of four slc30a/znt family members and their roles in Zn homeostatic modulation in yellow catfish Pelteobagrus fulvidraco. BBA-Gene Regul. Mech. 2021, 1864, 194723. [Google Scholar] [CrossRef]
- Chen, S.W.; Wu, K.; Lv, W.H.; Chen, F.; Song, C.C.; Luo, Z. Functional analysis of two zinc Zn, ransporters ZIP3 and ZIP8, promoters and their distinct response to MTF1 and RREB1 in the regulation of Zn metabolism. Int. J. Mol. Sci. 2020, 21, 6135. [Google Scholar] [CrossRef]
- Hardyman, J.E.; Tyson, J.; Jackson, K.A.; Aldridge, C.; Cockell, S.J.; Wakeling, L.A.; Valentine, R.A.; Ford, D. Zinc sensing by metal-responsive transcription factor 1 (MTF1) controls metallothionein and ZnT1 expression to buffer the sensitivity of the transcriptome response to zinc. Metallomics 2016, 8, 337–343. [Google Scholar] [CrossRef] [Green Version]
- Jackson, K.A.; Helston, R.M.; McKay, J.A.; O’Neill, E.D.; Mathers, J.C.; Ford, D. Splice variants of the human zinc transporter ZnT5 SLC30A5 are differentially localized and regulated by zinc through transcription and mRNA stability. J. Biol. Chem. 2007, 282, 10423–10431. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.J.; Liu, Y.C.; Lin, M.C.; Chen, Y.T.; Lin, L.Y. Coordinative modulation of human zinc transporter 2 gene expression through active and suppressive regulators. J. Nutr. Biochem. 2015, 26, 351–359. [Google Scholar] [CrossRef]
- Ghaleb, A.M. and Yang, V.W. Krüppel-like factor 4 (KLF4): What we currently know. Gene 2017, 611, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Liuzzi, J.P.; Guo, L.; Chang, S.M.; Cousins, R.J. Krüppel-like factor 4 regulates adaptive expression of the zinc transporter Zip4 in mouse small intestine. Am. J. Physiol.-Gastr. Liver Physiol. 2009, 296, G517–G523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Segre, J.A.; Bauer, C.; Fuchs, E. KLF4 is a transcription factor required for establishing the barrier function of the skin. Nat. Genet. 1999, 22, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Levy, D.E.; Lee, C.K. What does Stat3 do? J. Clin. Investig. 2002, 109, 1143–1148. [Google Scholar] [CrossRef]
- Liu, J.; Zhou, Z.X.; Zhang, W.; Bell, M.W.; Waalkes, M.P. Changes in hepatic gene expression in response to hepatoprotective levels of zinc. Liver Int. 2009, 29, 1222–1229. [Google Scholar] [CrossRef] [Green Version]
- Kitabayashi, C.; Fukada, T.; Kanamoto, M.; Ohashi, W.; Hojyo, S.; Atsumi, T.; Ueda, N.; Azuma, I.; Hirota, H.; Murakami, M.; et al. Zinc suppresses Th17 development via inhibition of STAT3 activation. Int. Immunol. 2010, 22, 375–386. [Google Scholar] [CrossRef] [Green Version]
- Cousins, R.J.; Liuzzi, J.P.; Lichten, L.A. Mammalian zinc transport, trafficking, and signals. J. Biol. Chem. 2006, 281, 24085–24089. [Google Scholar] [CrossRef] [Green Version]
- Bury, N.R.; Walker, P.A.; Glover, G.N. Nutritive metal uptake in teleost fish. J. Exp. Biol. 2003, 206, 11–23. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.H.; Lv, W.H.; Xu, Y.H.; Wei, X.L.; Xu, Y.C.; Luo, Z. Functional analysis of MTF-1 and MT promoters and their transcriptional response to zinc Zn and copper Cu in yellow catfish Pelteobagrus fulvidraco. Chemosphere 2020, 246, 125792. [Google Scholar] [CrossRef]
- Song, C.C.; Wu, L.X.; Chen, G.H.; Lv, W.H.; Chen, S.W.; Luo, Z. Six members of SLC30A/ZnTs family related with the control of zinc homeostasis, characterization, mRNA expression and their responses to dietary ZnO nanoparticles in yellow catfish. Aquaculture 2020, 528, 735570. [Google Scholar] [CrossRef]
- Chen, G.H.; Zhao, T.; Wei, X.L.; Zhang, D.G.; Zhuo, M.Q.; Luo, Z. miR-101b Regulates Lipid Deposition and Metabolism of Primary Hepatocytes in Teleost Yellow Catfish Pelteobagrus fulvidraco. Genes 2020, 11, 861. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.G.; Zhao, T.; Xu, X.J.; Lv, W.H.; Luo, Z. Dietary marginal and excess selenium increased triglycerides deposition, induced endoplasmic reticulum stress and differentially influenced selenoproteins expression in the anterior and middle intestines of yellow catfish Pelteobagrus fulvidraco. Antioxidants 2021, 10, 535. [Google Scholar] [CrossRef]
- Wu, L.X.; Xu, Y.C.; Hogstrand, C.; Zhao, T.; Wu, K.; Xu, Y.H.; Liu, W.; Luo, Z. Lipophagy mediated glucose-induced changes of lipid deposition and metabolism via ROS dependent AKT-Beclin1 activation. J. Nutr. Biochem. 2022, 100, 108882. [Google Scholar] [CrossRef] [PubMed]
- Miller, D.A. ‘Significant’ and ‘Highly Significant’. Nature 1966, 210, 1190. [Google Scholar] [CrossRef] [PubMed]
- Shihan, M.H.; Novo, S.G.; Le Marchand, S.J.; Wang, Y.; Duncan, M.K. A simple method for quantitating confocal flu-orescent images. Biochem. Biophys. Rep. 2021, 25, 100916. [Google Scholar] [CrossRef] [PubMed]
- Ruggiero, I.; Terracciano, M.; Martucci, N.M.; Stefano, L.D.; Migliaccio, N.; Tatè, R.; Rendina, I.; Arcari, P.; Lamberti, A.; Rea, I. Diatomite silica nanoparticles for drug delivery. Nanoscale Res. Lett. 2014, 9, 329. [Google Scholar] [CrossRef] [Green Version]
- Shihan, M.H.; Kanwar, M.; Wang, Y.; Jackson, E.E.; Faranda, A.P.; Duncan, M.K. Fibronectin has multifunctional roles in posterior capsular opacification (PCO). Matrix Biol. 2020, 90, 79–108. [Google Scholar] [CrossRef]
- Khanh Le, N.Q.; Yee Yapp, E.K.; Nagasundaram, N.; Yeh, H.Y. Classifying Promoters by Interpreting the Hidden In-formation of DNA Sequences via Deep Learning and Combination of Continuous FastText N-Grams. Front. Bioeng. Biotechnol. 2019, 7, 305. [Google Scholar] [CrossRef] [Green Version]
- Yan, C.H.M.; Chan, K.M. Cloning of zebrafish metallothionein gene and characterization of its gene promoter region in HepG2 cell line. Biochim. Biophys. Acta (BBA)—Gene Struct. Expr. 2004, 1679, 47–58. [Google Scholar] [CrossRef]
- Sloutskin, A.; Shir-Shapira, H.; Freiman, R.N.; Gershon, T.J. The core promoter is a regulatory hub for developmental gene expression. Front. Cell. Dev. Biol. 2021, 9, 666508. [Google Scholar] [CrossRef] [PubMed]
- Goodrich, J.A.; Tjian, R. Unexpected roles for core promoter recognition factors in cell-type-specific transcription and gene regulation. Nat. Rev. Genet. 2010, 11, 549–558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zang, Z.S.; Xu, Y.M.; Lau Andy, T.Y. Molecular and pathophysiological aspects of metal ion uptake by the zinc transporter ZIP8 (SLC39A8). Toxicol. Res. 2016, 5, 987–1002. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langmade, S.J.; Ravindra, R.; Daniels, P.J.; Andrews, G.K. The transcription factor MTF-1 mediates metal regulation of the mouse ZnT1 gene. J. Biol. Chem. 2000, 275, 34803–34809. [Google Scholar] [CrossRef] [Green Version]
- Bird, J.A.; Blankman, E.; Stillman, D.J.; Eide, D.J.; Winge, D.R. The Zap1 transcriptional activator also acts as a repressor by binding downstream of the TATA box in ZRT2. EMBO J. 2004, 23, 1123–1132. [Google Scholar] [CrossRef] [Green Version]
- Dolfini, D.; Zambelli, F.; Pavesi, G.; Mantovani, R.A. perspective of promoter architecture from the CCAAT-box. Cell Cycle 2009, 8, 4127–4137. [Google Scholar] [CrossRef] [Green Version]
- Nishito, Y.; Kambe, T. Zinc transporter 1 ZNT1 expression on the cell surface is elaborately controlled by cellular zinc levels. J. Biol. Chem. 2019, 294, 15686–15697. [Google Scholar] [CrossRef]
- Guo, L.; Lichten, L.A.; Ryu, M.S.; Liuzzi, J.P.; Wang, F.; Cousins, R.J. STAT5-glucocorticoid receptor interaction and MTF-1 regulate the expression of Znt2 (SLC30A2) in pancreatic acinar cells. Proc. Natl. Acad. Sci. USA 2010, 107, 2818–2823. [Google Scholar] [CrossRef] [Green Version]
- Du, L.; Zhang, H.; Zhao, H.; Cheng, X.; Qin, J.; Teng, T.; Yang, Q.; Xu, Z. The critical role of the zinc transporter Zip2 SLC39A2 in ischemia/reperfusion injury in mouse hearts. J. Mol. Cell Cardiol. 2019, 132, 136–145. [Google Scholar] [CrossRef]
- Hogstrand, C.; Kille, P.; Ackland, M.L.; Hiscox, S.; Taylor, K.M. A mechanism for epithelial-mesenchymal transition and anoikis resistance in breast cancer triggered by zinc channel ZIP6 and STAT3 (signal transducer and activator of transcription 3). Biochem. J. 2013, 455, 229–237. [Google Scholar] [CrossRef]
- Yamashita, S.; Miyagi, C.; Fukada, T.; Kagara, N.; Che, Y.S.; Hirano, T. Zinc transporter LIVI controls epithelial mesenchymal transition in zebrafish gastrula organizer. Nature 2004, 429, 298–302. [Google Scholar] [CrossRef] [PubMed]
- Tan, G.; Lenhard, B. TFBSTools: An R/Bioconductor package for transcription factor binding site analysis. Bioinformatics 2016, 32, 1555–1556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, R.R.; Song, Z.X.; Lin, J.M.; Li, Z.G.; Shan, G.; Huang, C. Gawky modulates MTF-1-mediated transcription activation and metal discrimination. Nucleic Acids Res. 2021, 49, 6296–6314. [Google Scholar] [CrossRef] [PubMed]
- Wingelhofer, B.; Neubauer, H.A.; Valent, P. Implications of STAT3 and STAT5 signaling on gene regulation and chromatin remodeling in hematopoietic cancer. Leukemia 2018, 32, 1713–1726. [Google Scholar] [CrossRef] [Green Version]
- Ilsley, M.D.; Gillinder, K.R.; Magor, G.W.; Huang, S.; Bailey, T.L.; Crossley, M.; Perkins, A.C. Krüppel-like factors compete for promoters and enhancers to fine-tune transcription. Nucleic Acids Res. 2017, 45, 6572–6588. [Google Scholar] [CrossRef] [Green Version]
- Kong, B.Y.; Duncan, F.E.; Que, E.L.; Kim, A.M.; O’Halloran, T.V. Maternally-derived zinc transporters ZIP6 and ZIP10 drive the mammalian oocyte-to-egg transition. Mol. Hum. Reprod. 2014, 20, 1077–1089. [Google Scholar] [CrossRef]
- Fukunaka, A.; Fukada, T.; Bhin, J.; Suzuki, L.; Tsuzuki, T.; Takamine, Y.; Bin, B.H.; Yoshihara, T.; Sekine, N.I.; Naito, H.; et al. Zinc transporter ZIP13 suppresses beige adipocyte biogenesis and energy expenditure by regulating C/EBP-β expression. PLoS Genet. 2017, 138, e1006950. [Google Scholar] [CrossRef] [Green Version]
- Landry, G.M.; Furrow, E.; Holmes, H.L.; Hirata, T.; Kato, A.; Williams, P.; Strohmaier, K.; Gallo, C.J.R.; Chang, M.; Pandey, M.K.; et al. Cloning, function, and localization of human, canine and Drosophila ZIP10 (SLC39A10), a Zn2+ transporter. Am. J. Physiol.-Renal. 2019, 316, F263–F273. [Google Scholar] [CrossRef]
- Xiao, G.R.; Wan, Z.H.; Fan, Q.W.; Tang, X.N.; Zhou, B. The metal transporter ZIP13 supplies iron into the secretory pathway in Drosophila melanogaster. eLife 2014, 3, e03191. [Google Scholar] [CrossRef]
- Zhao, N.N.; Gao, J.W.; Enns, C.A.; Knutson, M.D. ZRT/IRT-like Protein 14 (ZIP14) Promotes the cellular assimilation of iron from transferrin. J. Biol. Chem. 2010, 285, 32141–32150. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Beattie, J.D.; Kim, B.F.; Petris, M.J.; Andrews, G.; Eide, D.J. Zinc-stimulated endocytosis controls activity of the mouse ZIP1 and ZIP3 zinc uptake transporters. J. Biol. Chem. 2004, 279, 24631–24639. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, B.E.; Wang, F.; Dufner-Beattie, J.; Andrews, G.K.; Eide, D.J.; Petris, M.J. Zn2+-stimulated endocytosis of the mZIP4 zinc transporter regulates its location at the plasma membrane. J. Biol. Chem. 2004, 279, 4523–4530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.H.; Luo, Z.; Wu, K.; Song, Y.F.; You, W.J.; Zhang, L.H. Structure and functional analysis of promoters from two liver isoforms of CPT I in grass carp Ctenopharyngodon idella. Int. J. Mol. Sci. 2017, 18, 2405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).