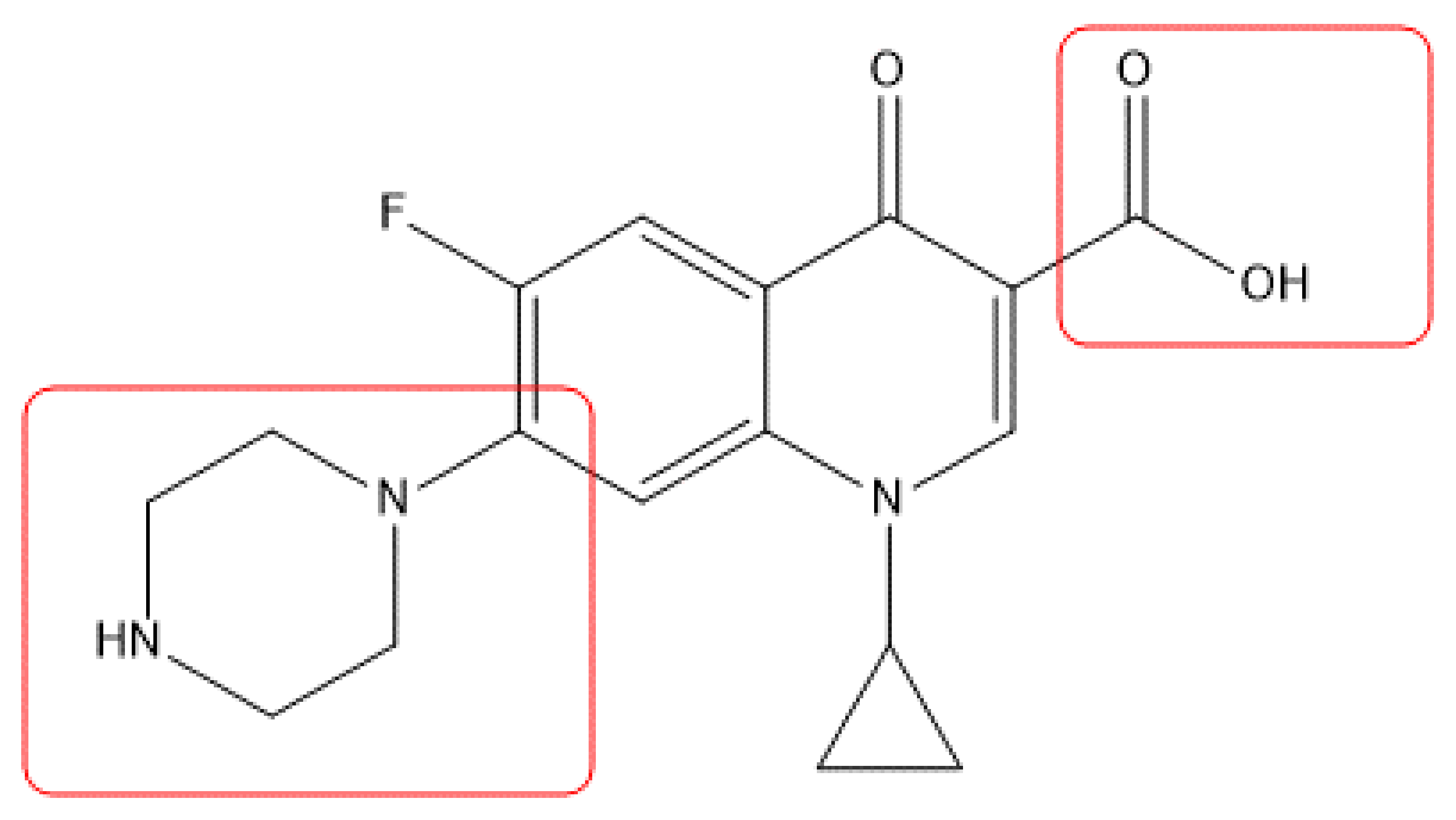
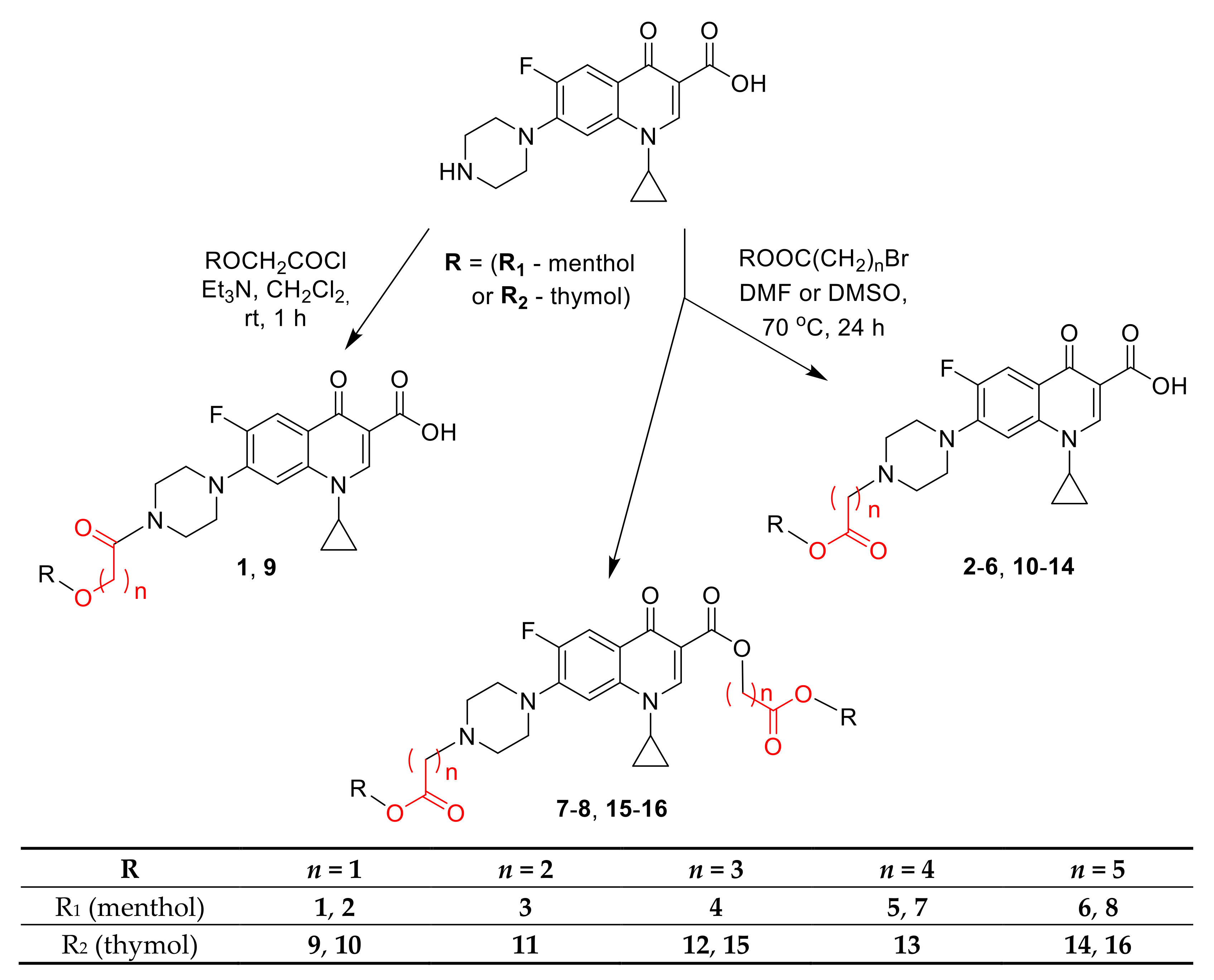
3.2.1. General Procedure for Synthesis of Ciprofloxacin Amides 1 and 9
To a magnetically stirred at room temperature suspension of Ciprofloxacin (0.318 g; 0.96 mmol, 1 eqv) in CH2Cl2 (50 mL), triethylamine (0.254 mL; 1.82 mmol, 1.9 eqv) was added and next, a solution of carboxylic acid chloride (1 eqv) in CH2Cl2 (2 mL) was dropped in over 2 min. After 1h of the reaction, a mixture of water (30 mL) and 3% HClaq solution were added to a pH equal to 3–4 and after separation of the phases, the water layer was extracted with CH2Cl2 (30 mL). The combined organic layers were washed with water (30 mL) and dried over Na2SO4. After evaporation of the solvent under reduced pressure, the product was isolated using column chromatography on silica gel and CH2Cl2:MeOH mixture (0–6% MeOH) as an eluent.
1-cyclopropyl-6-fluoro-7-{4-[2-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)acetyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (1)
White solid. Yield 92%. Mp = 185.7–189.6 °C.
1H NMR (CDCl3, 300 MHz) δ (ppm): 0.72 (d, J = 6.6 Hz, 3H), 0.89–0.93 (m, 9H), 1.15–1.34 (m, 6H), 1.53–1.61 (m, 2H), 2.06–2.17 (m, 2H), 3.01–3.18 (m, 1H), 3.27–3.30 (m, 4H), 3.51 (bs, 1H), 3.71–3.81 (m, 4H), 4.05–4.24 (m, 2H), 7.29 (d, J = 7.2 Hz, 1H), 7.85 (d, J = 12.9 Hz, 1H), 8.60 (s, 1H), 14.81 (s, 1H). 13C NMR (CDCl3, 75 MHz) δ (ppm): 8.2 (2xC), 16.1, 21.0, 22.3, 23.1, 25.5, 31.4, 34.3, 35.3, 40.0, 41.3, 45.2, 48.3, 49.3, 50.1, 68.3, 80.1, 105.1 (d, 3JC–F = 3.0 Hz), 107.9, 112.3 (d, 2JC–F = 23.3 Hz), 120.0 (d, 3JC–F = 7.5 Hz), 139.9, 145.4 (d, 2JC–F = 10.5 Hz), 147.4, 153.5 (d, 1JC–F = 249.8 Hz), 166.7, 168.6, 176.8 (d, 4JC–F = 2.3 Hz). HRMS (ESI) m/z 550.2693 calc. for C29H38FN3O5Na [M+Na]+; found 550.2681.
1-cyclopropyl-6-fluoro-7-{4-[2-(2-isopropyl-5-methylphenoxy)acetyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (9)
White solid. Yield 81%. Mp = 232.8–234.0 °C.
1H NMR (CDCl3\CD3OD, 9:1 mixture, 300 MHz) δ (ppm): 1.18–1.20 (m, 2H), 1.21 (d, J = 7.0 Hz, 6H), 1.39–143 (m, 2H), 2.33 (s, 3H), 3.37–3.93 (m, 5H), 3.54–3.61 (m, 1H), 3.90 (t, J = 5.1 Hz, 4H), 4.76 (s, 2H), 6.72 (bs, 1H), 6.82 (d, J = 7.8 Hz, 1H), 7.14 (d, J = 7.5 Hz, 1H), 7.39 (d, J = 6.9 Hz, 1H), 8.04 (d, J = 12.9 Hz, 1H), 8.79 (s, 1H). 13C NMR (CDCl3\CD3OD, 9:1 mixture, 125 MHz) δ (ppm): 8.0 (2xC), 21.1, 22.8 (2xC), 26.2, 35.4, 41.7 (2xC), 45.2 (2xC), 67.9, 105.2 (d, 3JC–F = 3.0 Hz), 107.6, 112.2, 112.4 (d, 2JC–F = 23.3 Hz), 120.2 (d, 3JC–F = 7.5 Hz), 122.4, 126.1, 133.8, 136.6, 138.9, 145.2 (d, 2JC–F = 9.8 Hz), 147.7, 153.5 (d, 1JC–F = 249.8 Hz), 154.5, 167.3, 167.4, 177.0 (d, 4JC–F = 2.3 Hz). HRMS (ESI) m/z 544.2224 calc. for C29H32FN3O5Na [M+Na]+; found 544.2237.
3.2.2. General Procedure for Synthesis of Menthol Derivatives of Ciprofloxacin
To a magnetically stirred at room temperature solution of appropriate menthol ester (1.2 mmol, 2 eqv) in DMF (10mL), Ciprofloxacin (0.60 mmol, 1 eqv) and NaHCO3 (1.2 mmol, 2 eqv) were added. The resulting suspension was stirred and heated at 70 °C (oil bath) for 20 h. The reaction mixture was evaporated under reduced pressure to dryness and to the residue, CH2Cl2 (20 mL) and water (80 mL) were added. Next, the 3% HClaq solution was added to a pH equal to 3–4 and after separation of the phases, the water layer was extracted with CH2Cl2 (20 mL). The combined organic layers were washed with water (10 mL) and dried over Na2SO4. After evaporation of the solvent under reduced pressure, the product was isolated using column chromatography on silica gel and CH2Cl2:MeOH mixture (0–4% MeOH) as an eluent.
1-cyclopropyl-6-fluoro-7-{4-[2-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-2-oxoethyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (2)
White solid. Yield 63%. Mp = 214.5–215.4 °C.
1H NMR (CDCl3, 300 MHz) δ (ppm): 0.78 (d, J = 6.9 Hz, 3H), 0.84–0.96 (m, 1H), 0.90 (d, J = 2.4 Hz, 3H), 0.92 (d, J = 2.1 Hz, 3H), 0.96–1.14 (m, 2H), 1.18–1.23 (m, 2H), 1.37–1.55 (m, 4H), 1.69–1.72 (m, 2H), 1.82–1.89 (m, 1H), 1.99–2.04 (m, 1H), 2.84 (bs, 4H), 3.30(d, J = 2.7 Hz, 2H), 3.41 (t, J = 4.5 Hz, 4H), 3.51–3.61 (m, 1H), 4.77(dt, J = 4.5 Hz, 10.8 Hz, 1H), 7.37 (d, J = 7.2 Hz, 1H), 7.95 (d, J = 12.9 Hz, 1H), 8.72 (s, 1H), 15.00 (s, 1H). 13C NMR (CDCl3, 75 MHz) δ (ppm): 8.2 (2xC), 16.3, 20.7, 22.0, 23.4, 26.4, 31.4, 34.1, 35.3, 40.9, 46.9, 49.6 (d, 4JC–F = 4.5 Hz, 2xC), 52.5 (2xC), 59.3, 74.8, 104.9 (d, 3JC–F = 3.0 Hz), 108.0, 112.2 (d, 2JC–F = 23.3 Hz), 119.7 (d, 3JC–F = 7.5 Hz),139.0, 145.8 (d, 2JC–F = 10.5 Hz), 147.3,153.6 (d, 1JC–F = 247.5 Hz), 166.9, 169.6, 177.0 (d, 4JC–F = 2.3 Hz). HRMS (ESI) m/z 550.2693 calc. for C29H38FN3O5Na [M+Na]+; found 550.2685.
1-cyclopropyl-6-fluoro-7-{4-[3-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-3-oxopropyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (3)
White solid. Yield 38%. Mp = 217.5–219.1 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 0.78 (d, J = 7.0 Hz, 3H), 0.83–0.89 (m, 1H), 0.90 (d, J = 3.0 Hz, 3H), 0.91 (d, J = 2.5 Hz, 3H), 0.95–1.02 (m, 1H), 1.02–1.11 (m, 1H), 1.19–1.22 (m, 2H), 1.35–1.41 (m, 3H), 1.46–1.54 (m, 1H), 1.66–1.72 (m, 2H), 1.90–1.96 (m, 1H), 1.98–2.03 (m, 1H), 2.53 (t, J = 7.0 Hz, 2H), 2.70–2.72 (m, 4H), 2.79 (t, J = 7.0 Hz, 2H), 3.33(t, J = 5.0 Hz, 4H), 3.52–3.57 (m, 1H), 4.73(dt, J = 4.5 Hz, 11.0 Hz, 1H), 7.35 (d, J = 7.0 Hz, 1H), 7.98 (d, J = 13.0 Hz, 1H), 8.75 (s, 1H), 15.01 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.2 (2xC), 16.4, 20.9, 22.1, 23.4, 26.2, 31.4, 32.9, 34.2, 35.3, 41.0, 47.0, 49.8 (d, 4JC–F = 4.5 Hz, 2xC), 52.5 (2xC), 53.6, 74.3, 104.7 (d, 3JC–F = 3.8 Hz), 108.1, 112.4 (d, 2JC–F = 22.5 Hz), 119.8 (d, 3JC–F = 8.8 Hz),139.1, 145.9 (d, 2JC–F = 10.0 Hz), 147.4,153.7 (d, 1JC–F = 250.0 Hz),167.0, 171.9, 177.1 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 564.2850 calc. for C30H40FN3O5Na [M+Na]+; found 564.22863.
1-cyclopropyl-6-fluoro-7-{4-[4-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-4-oxobutyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (4)
White solid. Yield 70%. Mp = 269.4–270.8 °C.
1H NMR (CDCl3\CD3OD, 9:1 mixture, 500 MHz) δ (ppm):0.77 (d, J = 7.0 Hz, 3H), 0.86–0.89 (m, 1H), 0.91 (d, J = 5.0 Hz, 3H), 0.92 (d, J = 4.5 Hz, 3H), 0.97–1.02 (m, 1H), 1.03–1.10 (m, 1H), 1.22–1.25 (m, 2H), 1.38–1.42 (m, 1H), 1.46–1.48 (m, 2H), 1.50–1.54 (m, 1H), 1.68–1.72 (m, 2H), 1.80–1.86 (m, 1H), 1.95–1.99 (m, 1H), 2.22–2.28 (m, 2H), 2.51 (t, J = 6.0 Hz, 2H),3.24 (t, J = 5.5 Hz, 2H), 3.37 (bs, 4H), 3.65–3.69 (m, 1H), 3.82 (bs, 4H), 4.71(dt, J = 4.5 Hz, 11.0 Hz, 1H), 7.52 (d, J = 7.0 Hz, 1H), 7.91 (d, J = 12.5 Hz, 1H), 8.75 (s, 1H). 13C NMR (CDCl3\CD3OD, 9:1 mixture, 125 MHz) δ (ppm): 8.1 (2xC), 16.1, 20.5, 21.8, 23.2, 26.1, 26.2, 30.8, 31.3, 33.9, 35.6, 40.7, 46.5 (2xC), 46.7, 51.6 (2xC), 56.5, 75.1, 106.3 (d, 3JC–F = 2.5 Hz), 107.6, 112.1 (d, 2JC–F = 23.8 Hz), 120.8 (d, 3JC–F = 7.5 Hz), 138.8, 143.8 (d, 2JC–F = 10.0 Hz), 147.8,153.3 (d, 1JC–F = 248.8 Hz), 167.1, 171.7, 176.8 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 578.3006 calc. for C31H42FN3O5Na [M+Na]+; found 578.3021.
1-cyclopropyl-6-fluoro-7-{4-[5-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-5-oxopentyl)piperazin-1-yl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (5)
Pale beige solid. Yield 79%. Mp = 239.2–240.7 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 0.77 (d, J = 7.0 Hz, 3H), 0.84–0.89 (m, 1H), 0.90 (d, J = 4.0 Hz, 3H), 0.92 (d, J = 3.5 Hz, 3H), 0.95–1.02 (m, 1H), 1.02–1.10 (m, 1H), 1.22–1.25 (m, 2H), 1.35–1.41 (m, 1H), 1.42–1.46 (m, 2H), 1.47–1.54 (m, 1H), 1.67–1.71 (m, 2H), 1.73–1.77 (m, 2H), 1.82–1.88 (m, 1H), 1.93–2.00 (m, 3H), 2.38 (t, J = 7.0 Hz, 2H), 3.01 (t, J = 8.5 Hz, 2H), 3.32 (bs, 4H), 3.57–3.61 (m, 1H), 3.77 (bs, 4H), 4.69 (dt, J = 4.0 Hz, 10.5 Hz, 1H), 7.37 (d, J = 7.0 Hz, 1H), 7.77 (d, J = 13.0 Hz, 1H), 8.65 (s, 1H), 14.85 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.3 (2xC), 16.3, 20.7, 22.0, 22.3, 23.4, 23.7, 26.3, 31.4, 33.6, 34.1, 35.5, 40.9, 46.9, 47.0 (2xC), 51.8 (2xC), 57.4, 74.4, 105.6, 107.9, 112.0 (d, 2JC–F = 23.8 Hz), 120.1 (d, 3JC–F = 7.5 Hz), 138.8, 144.2 (d, 2JC–F = 10.0 Hz), 147.5, 153.2 (d, 1JC–F = 250.0 Hz), 166.5, 172.5, 176.6 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 592.3163 calc. for C32H44FN3O5Na [M+Na]+; found 592.3185.
1-cyclopropyl-6-fluoro-7-{4-[6-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-6-oxohexyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (6)
Pale beige solid. Yield 85%. Mp = 210.8–212.4 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 0.76 (d, J = 7.0 Hz, 3H), 0.83–0.89 (m, 1H), 0.90 (d, J = 3.5 Hz, 3H), 0.91 (d, J = 3.0 Hz, 3H), 0.94–1.01 (m, 1H), 1.02–1.10 (m, 1H), 1.22–1.25 (m, 2H), 1.35–1.41 (m, 1H), 1.42–1.46 (m, 4H), 1.47–1.53 (m, 1H), 1.66–1.72 (m, 4H), 1.80–1.87 (m, 1H), 1.87–1.98 (m, 3H), 2.33 (t, J = 7.0 Hz, 2H), 2.95 (t, J = 7.0 Hz, 2H), 3.28 (bs, 4H), 3.57–3.61 (m, 1H), 3.75 (bs, 4H), 4.68 (dt, J = 4.5 Hz, 11.0 Hz, 1H), 7.37 (d, J = 7.0 Hz, 1H), 7.78 (d, J = 13.0 Hz, 1H), 8.66 (s, 1H), 14.88 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.3 (2xC), 16.3, 20.7, 22.0, 23.4, 24.1, 24.4, 26.3, 26.4, 31.4, 34.2, 34.3, 35.5, 40.9, 46.9, 47.2 (2xC), 51.9 (2xC), 57.7, 74.2, 105.6, 107.9, 111.9 (d, 2JC–F = 23.8 Hz), 120.1 (d, 3JC–F = 7.5 Hz), 138.8, 144.3 (d, 2JC–F = 11.3 Hz), 147.5, 153.2 (d, 1JC–F = 248.8 Hz), 166.6, 172.8, 176.6 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 606.3319 calc. for C33H46FN3O5Na [M+Na]+; found 606.3327.
5-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-5-oxopentyl-1-cyclopropyl-6-fluoro-7-{4-[5-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-5-oxopentyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylate (7)
Solidifying oil. Yield 7%.
1H NMR (CDCl3, 300 MHz) δ (ppm): 0.75 (d, J = 4.8 Hz, 3H), 0.77 (d, J = 4.8 Hz, 3H), 0.84–0.92 (m, 14H), 0.95–1.16 (m, 6H), 1.28–1.51 (m, 6H), 1.64–1.72 (m, 8H), 1.78–1.90 (m, 6H), 1.94–2.02 (m, 2H), 2.31–2.39 (m, 4H), 2.45 (t, J = 4.2 Hz, 2H), 2.65 (t, J = 4.8 Hz, 4H), 3.29 (t, J = 4.8 Hz, 4H), 3.38–3.46 (m, 1H), 4.32 (t, J = 6.0 Hz, 2H), 4.63–4.74 (m, 2H), 7.27 (d, J = 7.2 Hz, 1H), 8.04 (d, J = 13.5 Hz, 1H), 8.52 (s, 1H). 13C NMR (CDCl3, 75 MHz) δ (ppm): 8.1 (2xC), 16.3, 16.3, 20.7, 20.8, 21.7, 22.0, 23.0, 23.4, 23.4, 26.2, 26.2, 26.3, 28.2, 31.4, 34.2, 34.2, 34.5, 34.5, 40.9, 41.0, 47.0 (d, 4JC–F = 3.0 Hz), 49.9 (d, 4JC–F = 3.8 Hz), 52.9, 58.0, 64.3, 74.0, 104.7 (d, 4JC–F = 2.3 Hz), 110.3, 113.3 (d, 2JC–F = 23.3 Hz), 122.9 (d, 3JC–F = 7.5 Hz), 138.0, 144.6 (d, 2JC–F = 10.5 Hz), 148.0, 153.4 (d, 1JC–F = 247.5 Hz), 165.7, 173.0, 173.0 (d, 4JC–F = 1.5 Hz), 173.1. HRMS (ESI) m/z 830.5095 calc. for C47H70FN3O7Na [M+Na]+; found 830.5072.
6-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-6-oxohexyl-1-cyclopropyl-6-fluoro-7-{4-[6-((1R,2S,5R)-2-isopropyl-5-methylcyclohexyloxy)-6-oxohexyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylate (8)
Solidifying oil. Yield 8%.
1H NMR (CDCl3, 300 MHz) δ (ppm): 0.74 (d, J = 4.8 Hz, 3H), 0.77 (d, J = 4.8 Hz, 3H), 0.84–0.94 (m, 14H), 0.97–1.19 (m, 6H), 1.30–1.43 (m, 6H), 1.46–1.59 (m, 6H), 1.62–1.72 (m, 8H), 1.79–1.89 (m, 4H), 1.94–2.02 (m, 2H), 2.28–2.34 (m, 4H), 2.43 (t, J = 4.5 Hz, 2H), 2.67 (t, J = 4.2 Hz, 4H), 3.29 (t, J = 4.5 Hz, 4H), 3.40–3.46 (m, 1H), 4.31 (t, J = 6.6 Hz, 2H), 4.62–4.73 (m, 2H), 7.26 (d, J = 6.3 Hz, 1H), 8.03 (d, J = 13.2 Hz, 1H), 8.51 (s, 1H). 13C NMR (CDCl3, 75 MHz) δ (ppm): 8.1 (2xC), 16.3, 16.3, 20.7, 20.8, 22.0, 22.0, 23.4, 24.8, 25.0, 25.7, 26.2, 26.3, 26.5, 27.0, 28.4, 31.3, 34.2, 34.4, 34.5, 34.6, 40.9, 40.9, 47.0 (d, 4JC–F = 2.3 Hz), 49.9 (d, 4JC–F = 4.5 Hz), 52.9, 58.3, 64.6, 73.9, 73.9, 104.7 (d, 4JC–F = 3.0 Hz), 110.3, 113.2 (d, 2JC–F = 23.3 Hz), 122.9 (d, 3JC–F = 6.8 Hz), 138.0, 144.5 (d, 2JC–F = 10.5 Hz), 148.0, 153.4 (d, 1JC–F = 247.5 Hz), 165.8, 173.0 (d, 4JC–F = 1.5 Hz), 173.1, 173.2. HRMS (ESI) m/z 858.5408 calc. for C49H74FN3O7Na [M+Na]+; found 858.5427.
3.2.3. General Procedure for Synthesis of Thymol Derivatives of Ciprofloxacin
To a magnetically stirred at room temperature solution of appropriate menthol ester (1.2 mmol, 2 eqv) in DMF (10 mL) Ciprofloxacin (0.60 mmol, 1 eqv) and NaHCO3 (1.2 mmol, 2 eqv) were added. The resulting suspension was stirred and heated at 70 °C (oil bath) for 24 h. The reaction mixture was evaporated under reduced pressure to dryness and to residue CH2Cl2 (20 mL) and water (80 mL) were added. Next, the 3% HClaq solution was added to a pH equal to 3–4 and after separation of the phases, the water layer was extracted with CH2Cl2 (20 mL). The combined organic layers were washed with water (10 mL) and dried over Na2SO4. After evaporation of the solvent under reduced pressure, the product was isolated using column chromatography on silica gel and CH2Cl2:MeOH mixture (0–6% MeOH) as an eluent.
1-cyclopropyl-6-fluoro-7-{4-[2-(2-isopropyl-5-methylphenoxy)acetyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (10)
Pale beige solid. Yield 65%. Mp = 240.2–241.8 °C.
1H NMR (CDCl3\CD3OD, 9:1 mixture, 500 MHz) δ (ppm): 1.18–1.20 (m, 2H), 1.21 (d, J = 7.0 Hz, 6H), 1.39–143 (m, 2H), 2.33 (s, 3H), 2.95 (t, J = 4.0 Hz, 4H), 3.38–3.40 (m, 1H), 3.45 (t, J = 4.5 Hz, 4H), 3.56–3.60 (m, 1H), 3.63 (s, 2H), 6.83 (bs, 1H), 7.04–7.06 (m, 1H), 7.22 (d, J = 8.0 Hz, 1H), 7.40 (d, J = 7.0 Hz, 1H), 7.98 (d, J = 13.0 Hz, 1H), 8.76 (s, 1H). 13C NMR (CDCl3\CD3OD, 9:1 mixture, 125 MHz) δ (ppm): 8.1 (2xC), 20.7, 22.9 (2xC), 27.1, 35.4, 49.5 (d, 4JC–F = 2.0 Hz, 2xC), 52.4 (2xC), 58.8, 105.0 (d, 3JC–F = 3.8 Hz), 107.7, 112.2 (d, 2JC–F = 23.8 Hz), 119.8 (d, 3JC–F = 7.5 Hz), 122.4, 126.5, 127.3, 136.6, 136.8, 139.1, 145.7 (d, 2JC–F = 10.0 Hz), 147.3, 147.5, 153.6 (d, 1JC–F = 250.0 Hz), 167.3, 168.9, 177.0 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 544.2224 calc. for C29H32FN3O5Na [M+Na]+; found 544.2240.
1-cyclopropyl-6-fluoro-7-{4-[3-(2-isopropyl-5-methylphenoxy)-3-oxopropyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (11)
White solid. Yield 45%. Mp = 171.6–173.0 °C.
1H NMR (CDCl3, 300 MHz) δ (ppm): 1.17–1.23 (m, 2H), 1.20 (d, J = 6.9 Hz, 6H), 1.36–143 (m, 2H), 2.31 (s, 3H), 2.77–2.84 (m, 6H), 2.94 (t, J = 6.0 Hz, 2H), 3.03–3.13 (m, 1H), 3.38 (t, J = 4.8 Hz, 4H), 3.52–3.59 (m, 1H), 6.83 (bs, 1H), 7.01–7.05 (m, 1H), 7.21 (d, J = 7.8 Hz, 1H), 7.36 (d, J = 7.2 Hz, 1H), 7.95 (d, J = 13.2 Hz, 1H), 8.72 (s, 1H), 15.00 (s, 1H). 13C NMR (CDCl3, 75 MHz) δ (ppm): 8.2 (2xC), 20.8, 23.2 (2xC), 26.8, 32.7, 35.3, 49.8 (d, 4JC–F = 5.3 Hz, 2xC), 52.6 (2xC), 67.1, 104.8 (d, 3JC–F = 3.8 Hz), 108.0, 112.3 (d, 2JC–F = 23.3 Hz), 119.7 (d, 3JC–F = 8.3 Hz), 122.6, 126.4, 127.2, 136.6, 137.0, 139.0, 145.8 (d, 2JC–F = 9.8 Hz), 147.3, 147.8, 153.6 (d, 1JC–F = 249.8 Hz), 166.9, 171.0, 177.0 (d, 4JC–F = 2.3 Hz). HRMS (ESI) m/z 558.2380 calc. for C30H34FN3O5Na [M+Na]+; found 558.22364.
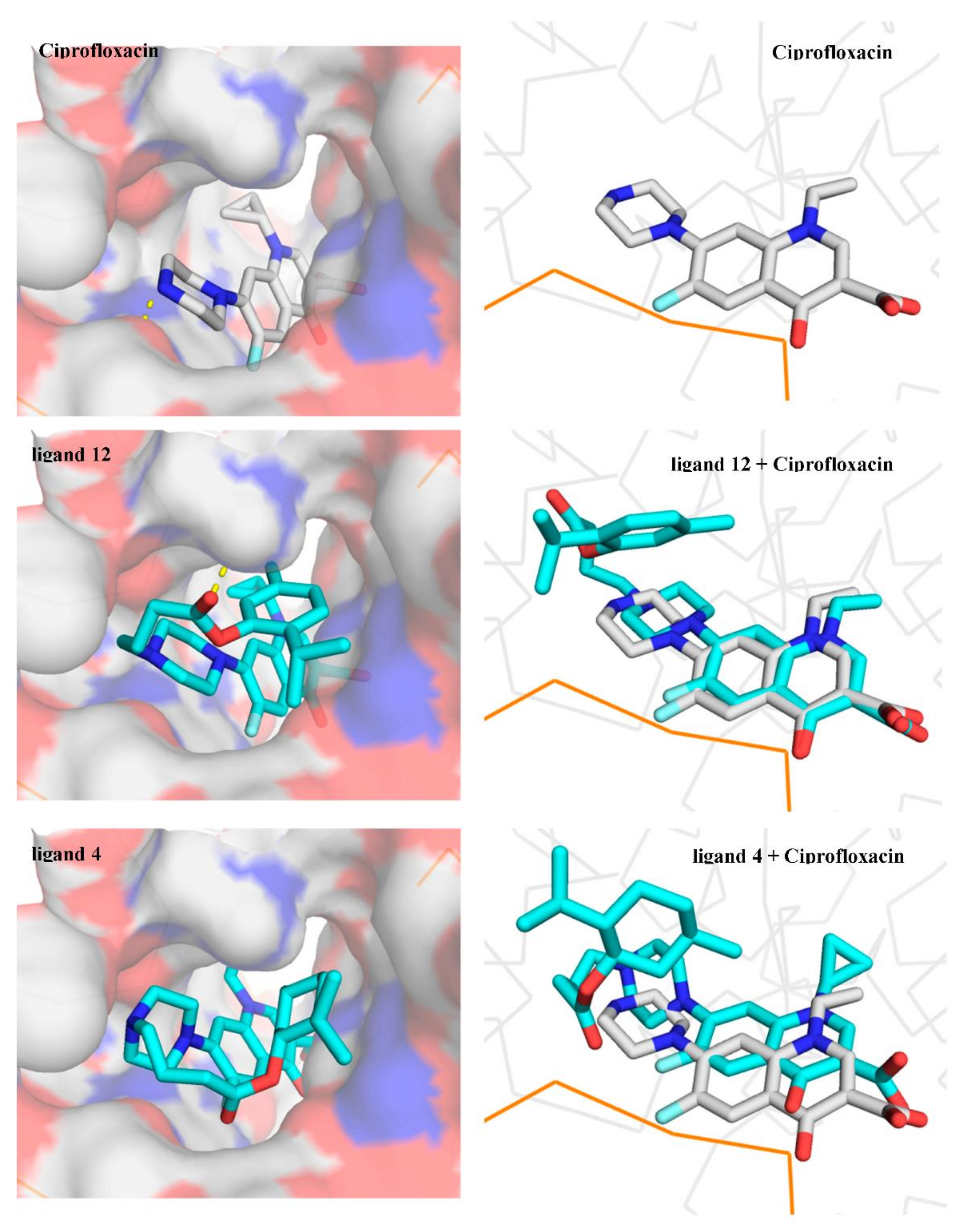
1-cyclopropyl-6-fluoro-7-{4-[4-(2-isopropyl-5-methylphenoxy)-4-oxobutyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (12)
White solid. Yield 59%. Mp = 244.1–245.7 °C.
1H NMR (CDCl3\CD3OD, 9:1 mixture, 500 MHz) δ (ppm): 1.20 (d, J = 6.5 Hz, 6H), 1.22–1.24 (m, 2H), 1.42–1.43 (m, 2H), 2.29–2.32 (m, 5H), 2.82 (t, J = 6.5 Hz, 2H), 2.91–2.96 (m, 1H), 3.12–3.21 (m, 2H), 3.29–3.44 (m, 4H), 3.59–3.62 (m, 1H), 3.73 (bs, 4H), 6.82 (s, 1H), 7.05 (d, J = 8.0 Hz, 1H), 7.21 (d, J = 8.0 Hz, 1H), 7.45 (d, J = 7.5 Hz, 1H), 7.90 (d, J = 12.5 Hz, 1H), 8.73 (s, 1H). 13C NMR (CDCl3\CD3OD, 9:1 mixture, 125 MHz) δ (ppm): 8.2 (2xC), 19.5, 20.7, 23.0 (2xC), 27.1, 31.0, 35.5, 47.2 (2xC), 51.9 (2xC), 56.6, 105.9, 107.7, 112.2 (d, 2JC–F = 23.8 Hz), 120.6 (d, 3JC–F = 7.5 Hz), 122.4, 126.5, 127.3, 136.6, 136.7, 138.2, 144.3 (d, 2JC–F = 11.3 Hz), 147.5, 147.7, 153.4 (d, 1JC–F = 250.0 Hz), 167.0, 171.4, 176.8 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 550.2717 calc. for C31H37FN3O5 [M+H]+; found 550.2736.
1-cyclopropyl-6-fluoro-7-{4-[5-(2-isopropyl-5-methylphenoxy)-5-oxopentyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (13)
White solid. Yield 81%. Mp = 212.3–213.9 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 1.19 (d, J = 6.5 Hz, 6H), 1.21–1.23 (m, 2H), 1.40–1.46 (m, 2H), 1.83–1.95 (m, 4H), 2.11–2.19 (m, 2H), 2.32 (s, 3H), 2.68–2.73 (m, 2H), 2.90–2.96 (m, 1H), 3.23 (bs, 4H), 3.59–3.65 (m, 1H), 3.88 (bs, 4H), 6.81 (s, 1H), 7.03 (d, J = 7.5 Hz, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.38 (bs, 1H), 7.71 (d, J = 12.5 Hz, 1H), 8.61 (s, 1H), 14.85 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.3 (2xC), 20.8, 21.9, 23.0 (2xC), 27.0, 27.1, 33.1, 35.6, 46.3 (d, 4JC–F = 2.5 Hz, 2xC), 51.5 (2xC), 57.1, 105.9, 107.7, 111.9 (d, 2JC–F = 25.0 Hz), 120.2 (d, 3JC–F = 7.5 Hz), 122.6, 126.4, 127.2, 136.6, 136.8, 138.7, 143.6 (d, 2JC–F = 10.0 Hz), 147.5, 147.6, 153.1 (d, 1JC–F = 248.8 Hz), 166.4, 171.7, 176.5 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 586.2693 calc. for C32H38FN3O5Na [M+Na]+; found 586.2679.
1-cyclopropyl-6-fluoro-7-{4-[6-(2-isopropyl-5-methylphenoxy)-6-oxohexyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylic acid (14)
White solid. Yield 64%. Mp = 185.4–187.1 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 1.19 (d, J = 6.5 Hz, 6H), 1.21–1.23 (m, 2H), 1.40–1.44 (m, 2H), 1.51–1.58 (m, 2H), 1.81–1.87 (m, 2H), 1.88–1.93 (m, 2H), 2.31 (s, 3H), 2.63 (t, J = 7.5 Hz, 2H), 2.88 (bs, 2H), 2.92–2.98 (m, 1H), 3.17 (bs, 4H), 3.54–3.59 (m, 1H), 3.67 (bs, 4H), 6.80 (s, 1H), 7.02 (d, J = 8.0 Hz, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.36 (d, J = 7.0 Hz, 1H), 7.82 (d, J = 12.5 Hz, 1H), 8.67 (s, 1H), 14.85 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.3 (2xC), 20.8, 23.0 (2xC), 24.4, 24.6, 26.5, 27.1, 33.9, 35.4, 47.7 (2xC), 52.1 (2xC), 57.7, 105.4 (d, 3JC–F = 2.5 Hz), 107.9, 112.1 (d, 2JC–F = 23.8 Hz), 120.0 (d, 3JC–F = 7.5 Hz), 122.6, 126.4, 127.1, 136.5, 136.9, 138.9, 144.6 (d, 2JC–F = 10.0 Hz), 147.5, 147.7, 153.3 (d, 1JC–F = 250.0 Hz), 166.6, 172.1, 176.7 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 578.3030 calc. for C33H41FN3O5 [M+H]+; found 578.3049.
2-(2-isopropyl-5-methylphenoxy)-2-oxobutyl-1-cyclopropyl-6-fluoro-7-{4-[2-(2-isopropyl-5-methylphenoxy)-2-oxobutyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylate (15)
White solid. Yield 21%. Mp = 105.5–107.2 °C.
1H NMR (CDCl3, 500 MHz) δ (ppm): 1.06–1.09 (m, 2H), 1.16 (d, J = 7.0 Hz, 6H), 1.20 (d, J = 7.0 Hz, 6H), 1.24–1.28 (m, 2H), 1.98–2.04 (m, 2H), 2.23 (s, 3H), 2.24–2.28 (m, 2H), 2.31 (s, 3H), 2.56 (t, J = 7.0 Hz, 2H), 2.68 (t, J = 7.0 Hz, 2H), 2.71 (t, J = 4.0 Hz, 4H), 2.83 (t, J = 7.5 Hz, 2H), 2.93–3.01 (m, 2H), 3.30 (t, J = 4.5 Hz, 4H), 3.33–3.38 (m, 1H), 4.43 (t, J = 6.5 Hz, 2H), 6.78 (s, 1H), 6.82 (s, 1H), 6.99 (d, J = 8.0 Hz, 1H), 7.02 (d, J = 7.5 Hz, 1H), 7.17 (d, J = 7.5 Hz, 1H), 7.20 (d, J = 8.0 Hz, 1H), 7.26 (d, J = 7.5 Hz, 1H), 8.02 (d, J = 13.5 Hz, 1H), 8.48 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.0 (2xC), 20.7, 20.8, 22.0, 23.0 (2xC), 23.0 (2xC), 24.3, 27.0, 27.1, 31.2, 32.0, 34.5, 49.9 (d, 4JC–F = 5.0 Hz, 2xC), 52.9 (2xC), 57.4, 63.7, 104.7 (d, 3JC–F = 2.5 Hz), 110.0, 113.2 (d, 2JC–F = 22.5 Hz), 122.6, 122.7, 122.9 (d, 3JC–F = 7.5 Hz), 126.3, 126.4, 127.0, 127.1, 136.4, 136.5, 136.9, 137.0, 138.0, 144.5 (d, 2JC–F = 10.0 Hz), 147.8 (2xC), 148.2, 153.4 (d, 1JC–F = 246.3 Hz), 165.6, 171.9, 172.2, 173.1 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 768.4024 calc. for C45H55FN3O7 [M+H]+; found 768.4011.
4-(2-isopropyl-5-methylphenoxy)-4-oxohexyl-1-cyclopropyl-6-fluoro-7-{4-[4-(2-isopropyl-5-methylphenoxy)-4-oxohexyl]piperazin-1-yl}-4-oxo-1,4-dihydroquinoline-3-carboxylate (16)
Solidifying oil. Yield 14%.
1H NMR (CDCl3, 500 MHz) δ (ppm): 1.09–1.12 (m, 2H), 1.18 (d, J = 7.0 Hz, 6H), 1.19 (d, J = 7.0 Hz, 6H), 1.26–1.30 (m, 2H), 1.47–1.53 (m, 2H), 1.58–1.66 (m, 4H), 1.80–1.90 (m, 6H), 2.29 (s, 3H), 2.31 (s, 3H), 2.47 (t, J = 7.5 Hz, 2H), 2.59–2.63 (m, 4H), 2.68 (t, J = 5.0 Hz, 4H), 2.92–2.99 (m, 2H), 3.30 (t, J = 5.0 Hz, 4H), 3.36–3.41 (m, 1H), 4.35 (t, J = 6.5 Hz, 2H), 6.79 (s, 1H), 6.80 (s, 1H), 6.99–7.03 (m, 2H), 7.17–7.20 (m, 2H), 7.26 (d, J = 7.0 Hz, 1H), 7.26 (d, J = 7.5 Hz, 1H), 8.02 (d, J = 13.0 Hz, 1H), 8.50 (s, 1H). 13C NMR (CDCl3, 125 MHz) δ (ppm): 8.1 (2xC), 20.8, 20.8, 23.0 (2xC), 23.0 (2xC), 24.7, 24.9, 25.6, 26.5, 27.0, 27.0, 27.1, 28.5, 34.2, 34.2, 34.4, 49.9 (d, 4JC–F = 5.0 Hz, 2xC), 53.0 (2xC), 58.3, 64.5, 104.7 (d, 3JC–F = 2.5 Hz), 110.3, 113.2 (d, 2JC–F = 23.8 Hz), 122.7, 122.7, 122.9 (d, 3JC–F = 7.5 Hz), 126.3, 126.4, 127.0, 127.0, 136.4, 136.5, 136.9, 137.0, 138.0, 144.5 (d, 2JC–F = 11.3 Hz), 147.8, 147.9, 148.1, 153.4 (d, 1JC–F = 247.5 Hz), 165.9, 172.3, 172.3, 173.1 (d, 4JC–F = 2.5 Hz). HRMS (ESI) m/z 846.4469 calc. for C49H62FN3O7Na [M+Na]+; found 846.4437.