Untangling the Conformational Plasticity of V66M Human proBDNF Polymorphism as a Modifier of Psychiatric Disorder Susceptibility
Abstract
1. Introduction
2. Results and Discussion
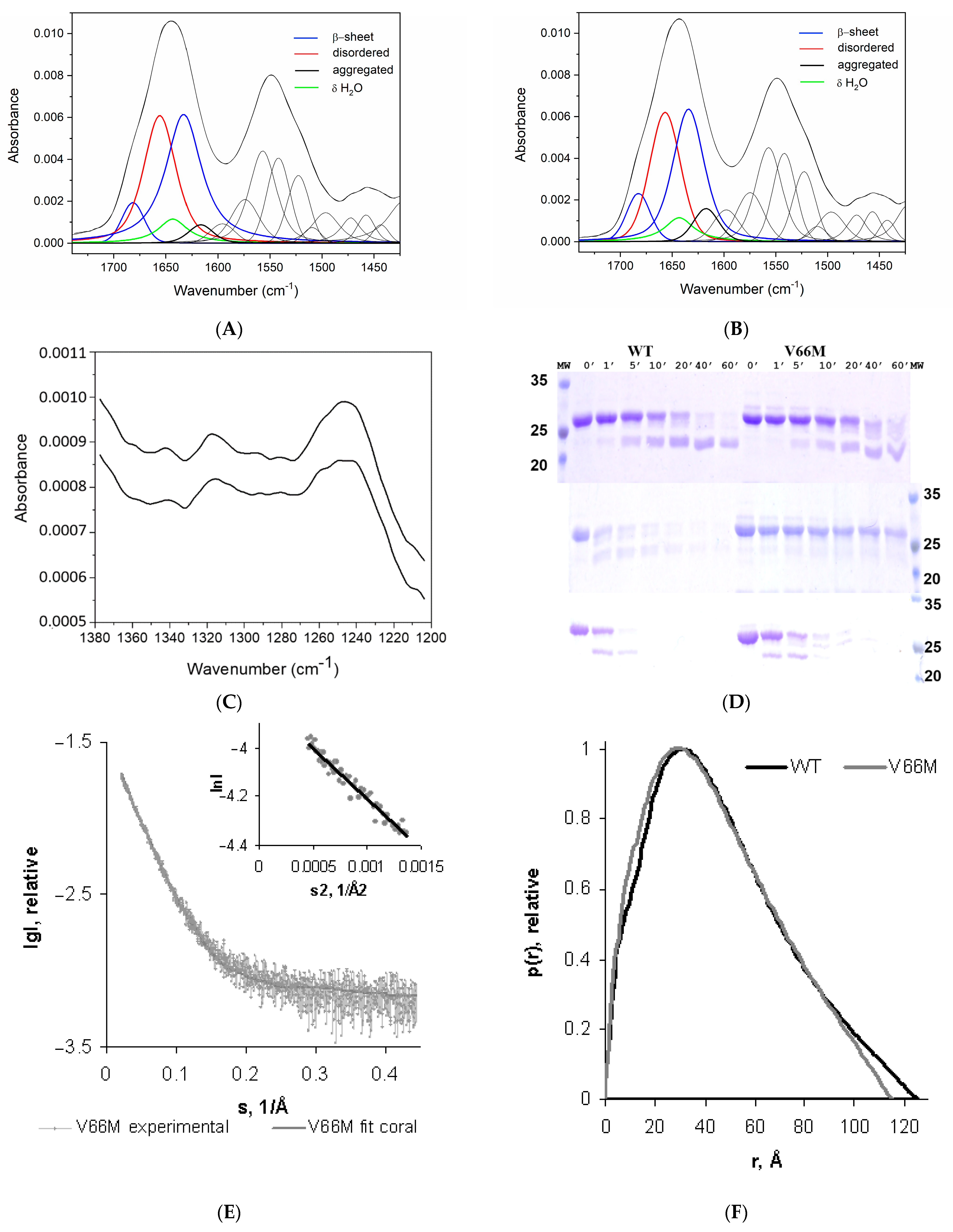
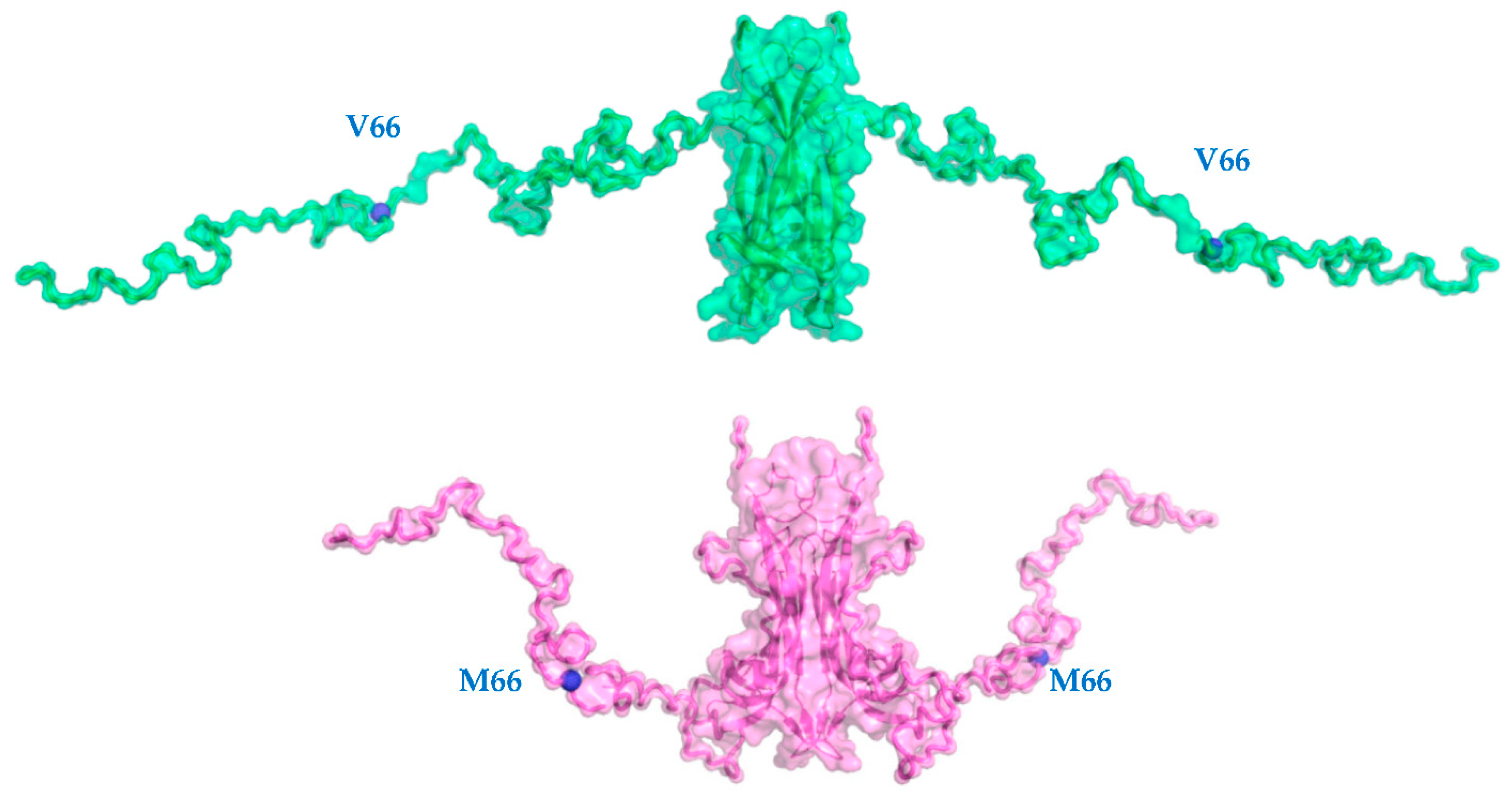
2.1. V66M Polymorphism Influences the Conformation of hproBDNF in Solution
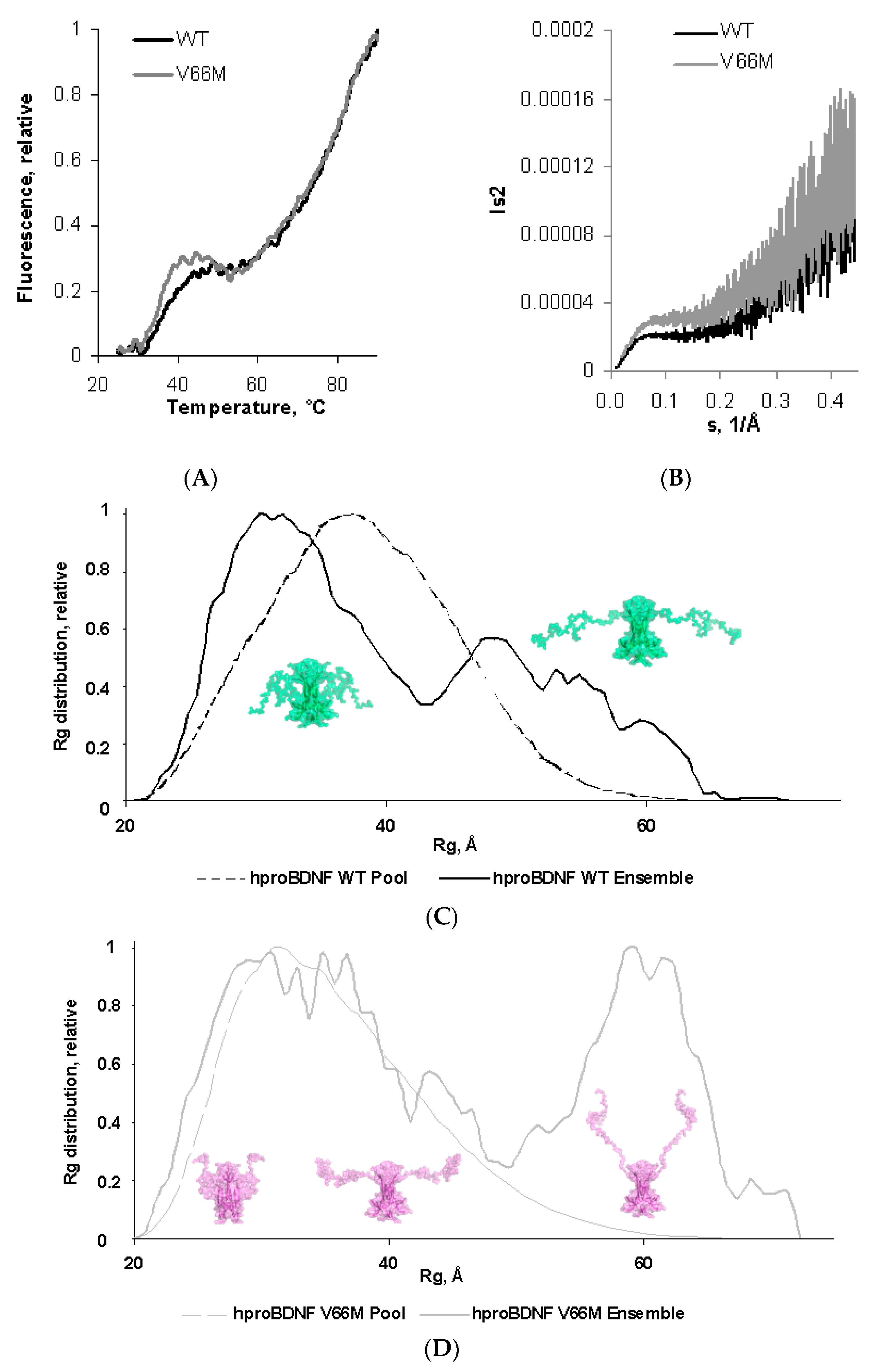
2.2. V66M Polymorphism Affects the Thermostability and the Conformational Plasticity of hproBDNF in Solution
3. Conclusions
4. Materials and Methods
4.1. Fourier Transform Infrared Spectroscopy (FT-IR)
4.2. Limited Proteolysis
4.3. Differential Scanning Fluorimetry (DSF)
4.4. Small-Angle X-ray Scattering Data Collection and Processing
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Auerbach, R.P.; Mortier, P.; Bruffaerts, R.; Alonso, J.; Benjet, C.; Cuijpers, P.; Demyttenaere, K.; Ebert, D.D.; Greif Green, J.; Hasking, P.; et al. The WHO World Mental Health Surveys International College Student Project: Prevalence and Distribution of Mental Disorders. J. Abnorm. Psychol. 2018, 127, 623–638. [Google Scholar] [CrossRef] [PubMed]
- Northoff, G.; Stanghellini, G. How to link brain and experience? Spatiotemporal psychopathology of the lived body. Front. Hum. Neurosci. 2016, 10, 76. [Google Scholar] [CrossRef] [PubMed]
- Deacon, B.J. The biomedical model of mental disorder: A critical analysis of its validity, utility, and effects on psychotherapy research. Clin. Psychol. Rev. 2013, 33, 846–861. [Google Scholar] [CrossRef] [PubMed]
- Chao, M.V. Neurotrophins and their receptors: A convergence point for many signalling pathways. Nat. Rev. Neurosci. 2003, 4, 299–309. [Google Scholar] [CrossRef]
- Lessmann, V.; Gottmann, K.; Malcangio, M. Neurotrophin secretion: Current facts and future prospects. Prog. Neurobiol. 2003, 69, 341–374. [Google Scholar] [CrossRef]
- Huang, E.J.; Reichardt, L.F. Neurotrophins: Roles in neuronal development and function. Annu. Rev. Neurosci. 2001, 24, 677–736. [Google Scholar] [CrossRef]
- Schuman, E.M. Neurotrophin regulation of synaptic transmission. Curr. Opin. Neurobiol. 1999, 9, 105–109. [Google Scholar] [CrossRef]
- Castrén, E.; Rantamäki, T. The role of BDNF and its receptors in depression and antidepressant drug action: Reactivation of developmental plasticity. Dev. Neurobiol. 2010, 70, 289–297. [Google Scholar] [CrossRef]
- Krishnan, V.; Nestler, E.J. Linking molecules to mood: New insight into the biology of depression. Am. J. Psychiatry 2010, 167, 1305–1320. [Google Scholar] [CrossRef]
- Lessmann, V.; Brigadski, T. Mechanisms, locations, and kinetics of synaptic BDNF secretion: An update. Neurosci. Res. 2009, 65, 11–22. [Google Scholar] [CrossRef]
- Wang, M.; Xie, Y.; Qin, D. Proteolytic cleavage of proBDNF to mBDNF in neuropsychiatric and neurodegenerative diseases. Brain Res. Bull. 2021, 166, 172–184. [Google Scholar] [CrossRef] [PubMed]
- Gibon, J.; Barker, P.A. Neurotrophins and Proneurotrophins: Focus on Synaptic Activity and Plasticity in the Brain. Neuroscientist 2017, 23, 587–604. [Google Scholar] [CrossRef] [PubMed]
- Petryshen, T.L.; Sabeti, P.C.; Aldinger, K.A.; Fry, B.; Fan, J.B.; Schaffner, S.F.; Waggoner, S.G.; Tahl, A.R.; Sklar, P. Population genetic study of the brain-derived neurotrophic factor (BDNF) gene. Mol. Psychiatry 2010, 15, 810–815. [Google Scholar] [CrossRef] [PubMed]
- Egan, M.F.; Kojima, M.; Callicott, J.H.; Goldberg, T.E.; Kolachana, B.S.; Bertolino, A.; Zaitsev, E.; Gold, B.; Goldman, D.; Dean, M.; et al. The BDNFV66M polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell 2003, 112, 257–269. [Google Scholar] [CrossRef]
- Pezawas, L.; Verchinski, B.A.; Mattay, V.S.; Callicott, J.H.; Kolachana, B.S.; Straub, R.E.; Egan, M.F.; Meyer-Lindenberg, A.; Weinberger, D.R. The brain-derived neurotrophic factor V66M polymorphism and variation in human cortical morphology. J. Neurosci. 2004, 24, 10099–10102. [Google Scholar] [CrossRef]
- Voineskos, A.N.; Lerch, J.P.; Felsky, D.; Shaikh, S.; Rajji, T.K.; Miranda, D.; Lobaugh, N.J.; Mulsant, B.H.; Pollock, B.G.; Kennedy, J.L. The brain-derived neurotrophic factor V66M polymorphism and prediction of neural risk for Alzheimer disease. Arch. Gen. Psychiatry 2011, 68, 198–206. [Google Scholar] [CrossRef]
- Hariri, A.R.; Goldberg, T.E.; Mattay, V.S.; Kolachana, B.S.; Callicott, J.H.; Egan, M.F.; Weinberger, D.R. Brain-derived neurotrophic factor V66M polymorphism affects human memory-related hippocampal activity and predicts memory performance. J. Neurosci. 2003, 23, 6690–6694. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Patel, P.D.; Sant, G.; Meng, C.X.; Teng, K.K.; Hempstead, B.L.; Lee, F.S. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intracellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J. Neurosci. 2004, 24, 4401–4411. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Jing, D.; Bath, K.G.; Ieraci, A.; Khan, T.; Siao, C.J.; Herrera, D.G.; Toth, M.; Yang, C.; McEwen, B.S.; et al. Genetic variant BDNF (V66M) polymorphism alters anxiety-related behavior. Science 2006, 314, 140–143. [Google Scholar] [CrossRef]
- Chen, Z.Y.; Ieraci, A.; Teng, H.; Dall, H.; Meng, C.X.; Herrera, D.G.; Nykjaer, A.; Hempstead, B.L.; Lee, F.S. Sortilin controls intracellular sorting of brain-derived neurotrophic factor to the regulated secretory pathway. J. Neurosci. 2005, 25, 6156–6166. [Google Scholar] [CrossRef]
- Chiaruttini, C.; Vicario, A.; Li, Z.; Baj, G.; Braiuca, P.; Wu, Y.; Lee, F.S.; Gardossi, L.; Baraban, J.M.; Tongiorgi, E. Dendritic trafficking of BDNF mRNA is mediated by translin and blocked by the G196A (V66M) mutation. Proc. Nat. Acad. Sci. USA 2009, 106, 16481–16486. [Google Scholar] [CrossRef] [PubMed]
- Teng, H.K.; Teng, K.K.; Lee, R.; Wright, S.; Tevar, S.; Almeida, R.D.; Kermani, P.; Torkin, R.; Chen, Z.Y.; Lee, F.S.; et al. ProBDNF induces neuronal apoptosis via activation of a receptor complex of p75NTR and sortilin. J. Neurosci. 2005, 25, 5455–5463. [Google Scholar] [CrossRef] [PubMed]
- Woo, N.H.; Teng, H.K.; Siao, C.J.; Chiaruttini, C.; Pang, P.T.; Milner, T.A.; Hempstead, B.L.; Lu, B. Activation of p75NTR by proBDNF facilitates hippocampal long-term depression. Nat. Neurosci. 2005, 8, 1069–1077. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.L.; Fan, Y.; Li, S.; Li, X.J.; Zhou, X.F. Huntingtin-associated protein-1 interacts with pro-brain-derived neurotrophic factor and mediates its transport and release. J. Biol. Chem. 2010, 285, 5614–5623. [Google Scholar] [CrossRef]
- Anastasia, A.; Deinhardt, K.; Chao, M.V.; Will, N.E.; Irmady, K.; Lee, F.S.; Hempstead, B.L.; Bracken, C. V66M polymorphism of BDNF alters prodomain structure to induce neuronal growth cone retraction. Nat. Commun. 2013, 4, 2490. [Google Scholar] [CrossRef]
- Autry, A.E.; Monteggia, L.M. Brain-derived neurotrophic factor and neuropsychiatric disorders. Pharmacol. Rev. 2012, 64, 238–258. [Google Scholar] [CrossRef]
- Williams, A.J.; Umemori, H. The best-laid plans go oft awry: Synaptogenic growth factor signaling in neuropsychiatric disease. Front. Synaptic Neurosci. 2014, 6, 4. [Google Scholar] [CrossRef]
- Neves-Pereira, M.; Mundo, E.; Muglia, P.; King, N.; Macciardi, F.; Kennedy, J.L. The brain-derived neurotrophic factor gene confers susceptibility to bipolar disorder: Evidence from a family-based association study. Am. J. Hum. Genet. 2002, 71, 651–655. [Google Scholar] [CrossRef]
- Ventriglia, M.; Bocchio Chiavetto, L.; Benussi, L.; Binetti, G.; Zanetti, O.; Riva, M.A.; Gennarelli, M. Association between the BDNF 196 A/G polymorphism and sporadic Alzheimer’s disease. Mol. Psychiatry 2002, 7, 136–137. [Google Scholar] [CrossRef]
- Masaki, T.; Matsushita, S.; Arai, H.; Takeda, A.; Itoyama, Y.; Mochizuki, H.; Kamakura, K.; Ohara, S.; Higuchi, S. Association between a polymorphism of brain derived neurotrophic factor gene and sporadic Parkinson’s disease. Ann. Neurol. 2003, 54, 276–277. [Google Scholar] [CrossRef]
- Ribases, M.; Gratacos, M.; Armengol, L.; de Cid, R.; Badia, A.; Jimenez, L.; Solano, R.; Vallejo, J.; Fernandez, F.; Estivill, X. Met66 in the brain-derived neurotrophic factor (BDNF) precursor is associated with anorexia nervosa restrictive type. Mol. Psychiatry 2003, 8, 745–751. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.; Jamra, R.A.; Becker, T.; Ohlraun, S.; Klopp, N.; Binder, E.B.; Schulze, T.G.; Deschner, M.; Schmal, C.; Hofels, S.; et al. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol. Psychiatry 2005, 58, 307–314. [Google Scholar] [CrossRef] [PubMed]
- Chepenik, L.G.; Fredericks, C.; Papademetris, X.; Spencer, L.; Lacadie, C.; Wang, F.; Pittman, B.; Duncan, J.S.; Staib, L.H.; Duman, R.S.; et al. Effects of the brain-derived neurotrophic growth factor V66M variation on hippocampus morphology in bipolar disorder. Neuropsychopharmacology 2009, 34, 944–951. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Frodl, T.; Schule, C.; Schmitt, G.; Born, C.; Baghai, T.; Zill, P.; Bottlender, R.; Rupprecht, R.; Bondy, B.; Reiser, M.; et al. Association of the brain derived neurotrophic factor V66M polymorphism with reduced hippocampal volumes in major depression. Arch. Gen. Psychiatry 2007, 64, 410–416. [Google Scholar] [CrossRef] [PubMed]
- Ho, B.C.; Andreasen, N.C.; Dawson, J.D.; Wassink, T.H. Association between brain derived neurotrophic factor V66M gene polymorphism and progressive brain volume changes in schizophrenia. Am. J. Psychiatry 2007, 164, 1890–1899. [Google Scholar] [CrossRef]
- Dorn, M.; E Silva, M.B.; Buriol, L.S.; Lamb, L.C. Three-dimensional protein structure prediction: Methods and computational strategies. Comput. Biol. Chem. 2014, 53, 251–276. [Google Scholar] [CrossRef]
- Gao, M.; Zhou, H.; Skolnick, J. Insights into disease-associated mutations in the human proteome through protein structural analysis. Structure 2016, 23, 1362–1369. [Google Scholar] [CrossRef]
- Kailainathan, S.; Piers, T.M.; Yi, J.H.; Choi, S.; Fahey, M.S.; Borger, E.; Gunn-Moore, F.J.; O’Neill, L.; Lever, M.; Whitcomb, D.J.; et al. Activation of a synapse weakening pathway by human Val66 but not Met66 pro-brain-derived neurotrophic factor (proBDNF). Pharmacol. Res. 2016, 104, 97–107. [Google Scholar] [CrossRef]
- Sneha, P.; Thirumal Kumar, D.; Saini, S.; Kajal, K.; Magesh, R.; Siva, R.; Priya, G.; Doss, C. Analyzing the Effect of V66M Mutation in BDNF in Causing Mood Disorders: A Computational Approach. Adv. Protein Chem. Struct. Biol. 2017, 108, 85–103. [Google Scholar]
- De Oliveira, C.C.S.; Pereira, G.R.C.; De Alcantara, J.Y.S.; Antunes, D.; Caffarena, E.R.; De Mesquita, J.F. In silico analysis of the V66M variant of human BDNF in psychiatric disorders: An approach to precision medicine. PLoS ONE 2019, 14, e0215508. [Google Scholar] [CrossRef]
- Zamani, M.; Eslami, M.; Nezafat, N.; Hosseini, S.V.; Ghasemi, Y. Evaluating the effect of BDNF Val66Met polymorphism on complex formation with HAP1 and Sortilin1 via structural modeling. Comput. Biol. Chem. 2019, 78, 282–289. [Google Scholar] [CrossRef] [PubMed]
- Paoletti, F.; Covaceuszach, S.; Konarev, P.V.; Gonfloni, S.; Malerba, F.; Schwarz, E.; Svergun, D.I.; Cattaneo, A.; Lamba, D. Intrinsic structural disorder of mouse proNGF. Proteins 2009, 75, 990–1009. [Google Scholar] [CrossRef] [PubMed]
- Yan, R.; Yalinca, H.; Paoletti, F.; Gobbo, F.; Marchetti, L.; Kuzmanic, A.; Lamba, D.; Gervasio, F.L.; Konarev, P.V.; Cattaneo, A.; et al. The Structure of the Pro-domain of Mouse proNGF in Contact with the NGF Domain. Structure 2019, 27, 78–89. [Google Scholar] [CrossRef] [PubMed]
- Covaceuszach, S.; Peche, L.Y.; Konarev, P.V.; Lamba, D. A combined evolutionary and structural approach to disclose the primary structural determinants essential for proneurotrophins biological functions. Comput. Struct. Biotechnol. J. 2021, 13, 2891–2904. [Google Scholar] [CrossRef] [PubMed]
- Koshimizu, H.; Kiyosue, K.; Hara, T.; Hazama, S.; Suzuki, S.; Uegaki, K.; Nagappan, G.; Zaitsev, E.; Hirokawa, T.; Tatsu, Y.; et al. Multiple functions of precursor BDNF to CNS neurons: Negative regulation of neurite growth, spine formation and cell survival. Mol. Brain 2009, 2, 27–48. [Google Scholar] [CrossRef] [PubMed]
- Goormaghtigh, E.; Raussens, V.; Ruysschaert, J.M. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim. Biophys. Acta 1999, 1422, 105–185. [Google Scholar] [CrossRef]
- Grdadolnik, J.; Mohacek-Grosev, V.; Baldwin, R.L.; Avbelj, F. Populations of the three major backbone conformations in 19 amino acid dipeptides. Proc. Natl. Acad. Sci. USA 2011, 108, 1794–1798. [Google Scholar] [CrossRef]
- Petoukhov, M.V.; Franke, D.; Shkumatov, A.V.; Tria, G.; Kikhney, A.G.; Gajda, M.; Gorba, C.; Mertens, H.D.T.; Konarev, P.V.; Svergun, D.I. New developments in the ATSAS program package for small-angle scattering data analysis. J. Appl. Cryst. 2012, 45, 342–350. [Google Scholar] [CrossRef]
- Bernadó, P.; Svergun, D.I. Structural analysis of intrinsically disordered proteins by small-angle X-ray scattering. Mol. Biosyst. 2012, 8, 151–167. [Google Scholar] [CrossRef]
- Mertens, H.D.; Svergun, D.I. Structural characterization of proteins and complexes using small-angle X-ray solution scattering. J. Struct. Biol. 2010, 172, 128–141. [Google Scholar] [CrossRef]
- Debye, P. Molecular-weight determination by light scattering. J. Phys. Chem. 1947, 51, 18–32. [Google Scholar] [CrossRef]
- Hammel, M.V. Validation of macromolecular flexibility in solution by small-angle X-ray scattering (SAXS). Eur. Biophys. J. 2012, 41, 789–799. [Google Scholar] [CrossRef] [PubMed]
- Tria, G.; Mertens, H.D.T.; Kachala, M.; Svergun, D.I. Advanced ensemble modelling of flexible macromolecules using X-ray solution scattering. IUCr J. 2015, 2, 207–217. [Google Scholar] [CrossRef] [PubMed]
- Stott, K.; Watson, M.; Howe, F.S.; Grossmann, J.G.; Thomas, J.O. Tail-mediated collapse of HMGB1 is dynamic and occurs via differential binding of the acidic tail to the A and B domains. J. Mol. Biol. 2010, 403, 706–722. [Google Scholar] [CrossRef] [PubMed]
- Uegaki, K.; Kumanogoh, H.; Mizui, T.; Hirokawa, T.; Ishikawa, Y.; Kojima, M. BDNF Binds Its Pro-Peptide with High Affinity and the Common Val66Met Polymorphism Attenuates the Interaction. Int. J. Mol. Sci. 2017, 18, 1042. [Google Scholar] [CrossRef]
- Harripaul, R.; Vasli, N.; Mikhailov, A.; Rafiq, M.A.; Mittal, K.; Windpassinger, C.; Sheikh, T.I.; Noor, A.; Mahmood, H.; Downey, S.; et al. Mapping autosomal recessive intellectual disability: Combined microarray and exome sequencing identifies 26 novel candidate genes in 192 consanguineous families. Mol. Psychiatry 2018, 23, 973–984. [Google Scholar] [CrossRef]
- Blanchet, C.E.; Spilotros, A.; Schwemmer, F.; Graewert, M.A.; Kikhney, A.; Jeffries, C.M.; Franke, D.; Mark, D.; Zengerle, R.; Cipriani, F.; et al. Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY). J. Appl. Crystallogr. 2015, 48, 431–443. [Google Scholar] [CrossRef]
- Konarev, P.V.; Volkov, V.V.; Sokolova, A.V.; Koch, M.H.J.; Svergun, D.I. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J. Appl. Crystallogr. 2003, 36, 1277–1282. [Google Scholar] [CrossRef]
- Manalastas-Cantos, K.; Konarev, P.V.; Hajizadeh, N.R.; Kikhney, A.G.; Petoukhov, M.V.; Molodenskiy, D.S.; Panjkovich, A.; Mertens, H.D.T.; Gruzinov, A.; Borges, C.; et al. ATSAS 3.0: Expanded functionality and new tools for small-angle scattering data analysis. J. Appl. Cryst. 2021, 54, 343–355. [Google Scholar] [CrossRef]
- Guinier, A. La diffraction des rayons X aux tres petits angles: Applications a l’etude de phenomenes ultramicroscopiques. Ann. Phys. 1939, 12, 161–237. [Google Scholar] [CrossRef]
- Svergun, D.I. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992, 25, 495–503. [Google Scholar] [CrossRef]
- Porod, G. Small Angle X-ray Scattering; Academic Press: London, UK, 1982; pp. 17–51. [Google Scholar]
- Kozin, M.B.; Svergun, D.I. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001, 34, 33–41. [Google Scholar] [CrossRef]
- Volkov, V.V.; Svergun, D.I. Uniqueness of ab initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2003, 36, 860–864. [Google Scholar] [CrossRef]
- Svergun, D.I.; Barberato, C.; Koch, M.H.J. CRYSOL—A Program to Evaluate X-ray Solution Scattering of Biological Macromolecules from Atomic Coordinates. J. Appl. Crystallogr. 1995, 28, 768–773. [Google Scholar] [CrossRef]
| Data Collection Parameters | hproBDNF V66M | hproBDNF |
|---|---|---|
| Instrument | P12 (PETRA III) | P12 (PETRA III) |
| Beam geometry (mm2) | 0.2 × 0.12 | 0.2 × 0.12 |
| Wavelength (Å) | 1.24 | 1.24 |
| s range (Å−1) | 0.003–0.445 | 0.003–0.445 |
| Concentration range (mg/mL) | 0.14–1.45 | 0.10–1.48 |
| Temperature (K) | 283 | 283 |
| Structural parameters | ||
| I(0) (A.U.) [from p(r)] | 0.02100 ± 0.0002 | 0.0177 ± 0.0002 |
| Rg (Å) [from p(r)] | 35.5 ± 0.05 | 37.2 ± 0.06 |
| I(0) (A.U.) [from Guinier] | 0.02263 ± 0.0002 | 0.0182 ± 0.0002 |
| Rg (Å) [from Guinier] | 35.5 ± 0.05 | 37.1 ± 0.06 |
| Dmax (Å) | 115 ± 5 | 125 ± 6 |
| Vp Porod volume estimate (Å3) | 85,675 ± 6000 | 86,750 ± 6000 |
| Molecular mass determination (Da) | ||
| MM [from I(0)] | 55,440 ± 6000 | 48,048 ± 6000 |
| MM [from Porod volume] | 50,400 ± 6000 | 51,030 ± 6000 |
| Calculated monomeric MM from sequence | 25,930 | 25,930 |
| Software employed | ||
| Primary data reduction | PRIMUS | PRIMUS |
| Data processing | GNOM | GNOM |
| Rigid body modeling | CORAL/EOM | CORAL/EOM |
| Validation and averaging | DAMAVER | DAMAVER |
| Three-dimensional graphic representations | PYMOL | PYMOL |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Covaceuszach, S.; Peche, L.Y.; Konarev, P.V.; Grdadolnik, J.; Cattaneo, A.; Lamba, D. Untangling the Conformational Plasticity of V66M Human proBDNF Polymorphism as a Modifier of Psychiatric Disorder Susceptibility. Int. J. Mol. Sci. 2022, 23, 6596. https://doi.org/10.3390/ijms23126596
Covaceuszach S, Peche LY, Konarev PV, Grdadolnik J, Cattaneo A, Lamba D. Untangling the Conformational Plasticity of V66M Human proBDNF Polymorphism as a Modifier of Psychiatric Disorder Susceptibility. International Journal of Molecular Sciences. 2022; 23(12):6596. https://doi.org/10.3390/ijms23126596
Chicago/Turabian StyleCovaceuszach, Sonia, Leticia Yamila Peche, Petr Valeryevich Konarev, Joze Grdadolnik, Antonino Cattaneo, and Doriano Lamba. 2022. "Untangling the Conformational Plasticity of V66M Human proBDNF Polymorphism as a Modifier of Psychiatric Disorder Susceptibility" International Journal of Molecular Sciences 23, no. 12: 6596. https://doi.org/10.3390/ijms23126596
APA StyleCovaceuszach, S., Peche, L. Y., Konarev, P. V., Grdadolnik, J., Cattaneo, A., & Lamba, D. (2022). Untangling the Conformational Plasticity of V66M Human proBDNF Polymorphism as a Modifier of Psychiatric Disorder Susceptibility. International Journal of Molecular Sciences, 23(12), 6596. https://doi.org/10.3390/ijms23126596







