Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings
Abstract
:1. Introduction
2. Results
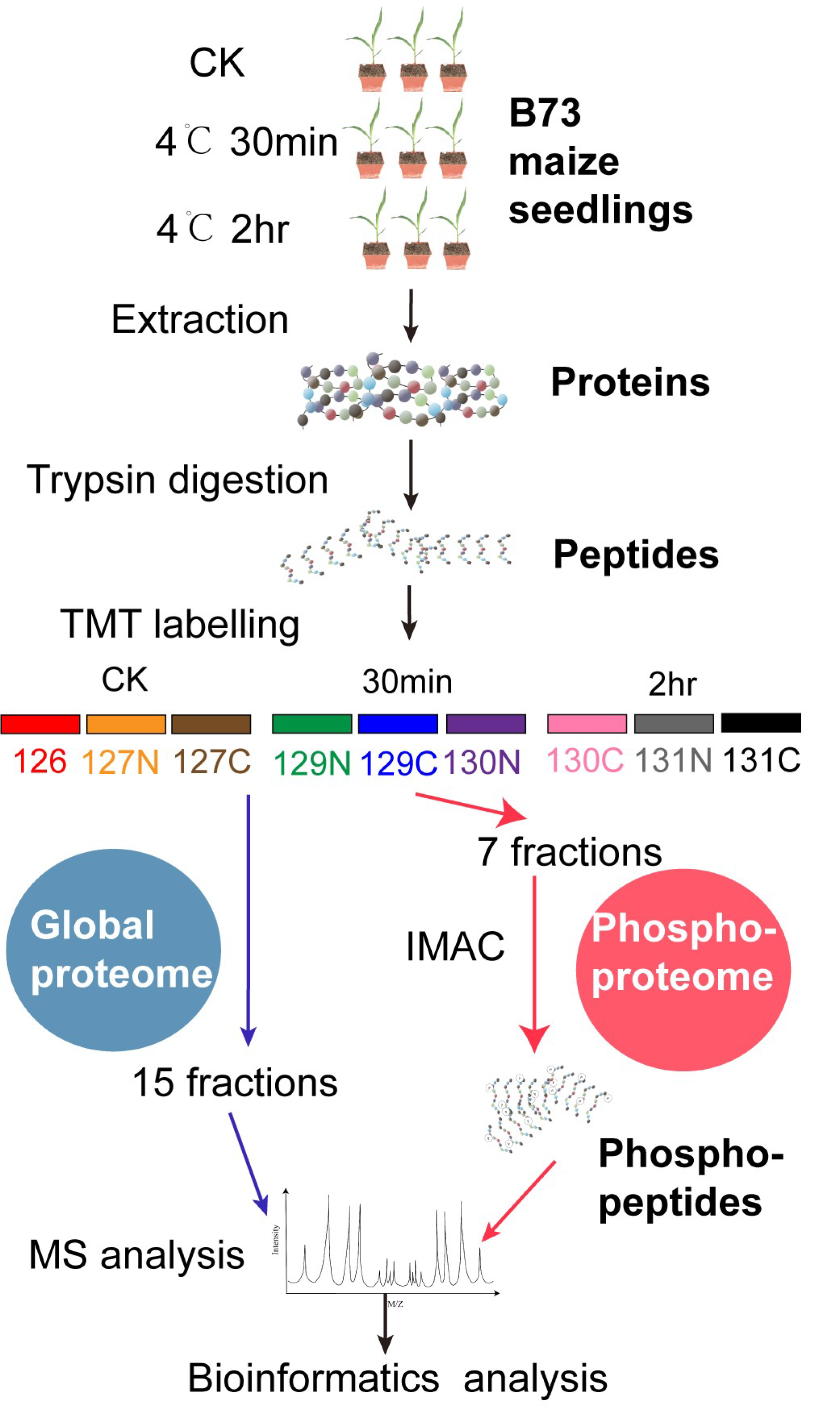
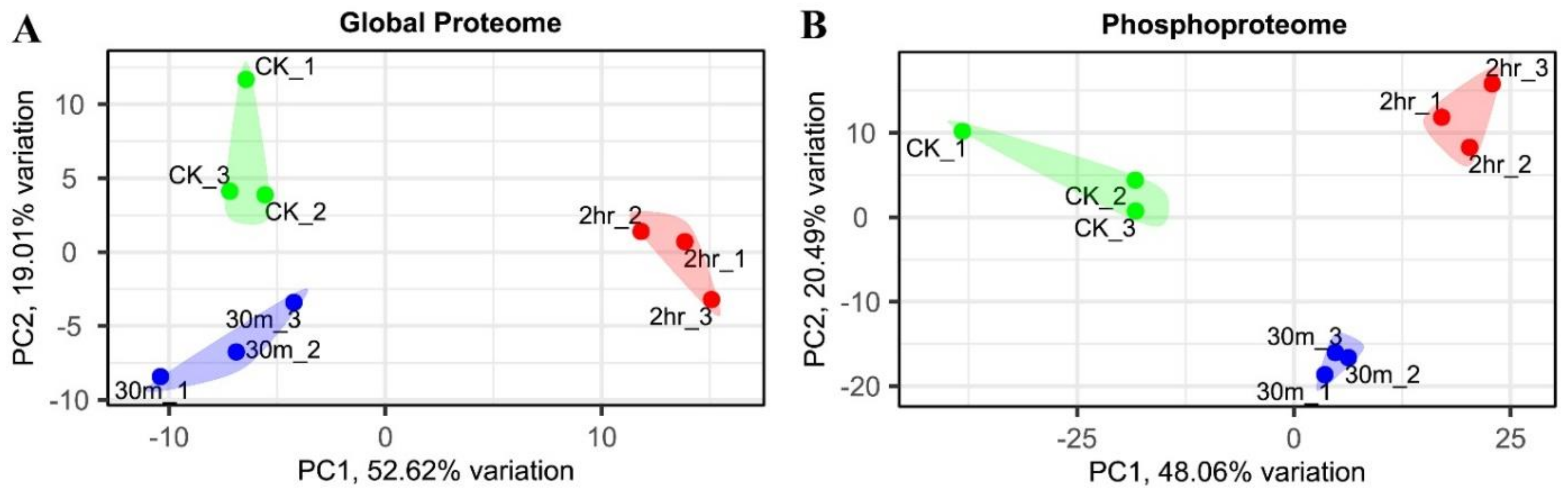
2.1. Proteome and Phosphoproteome Analyses of Maize Seedlings upon Short Time Cold Stress
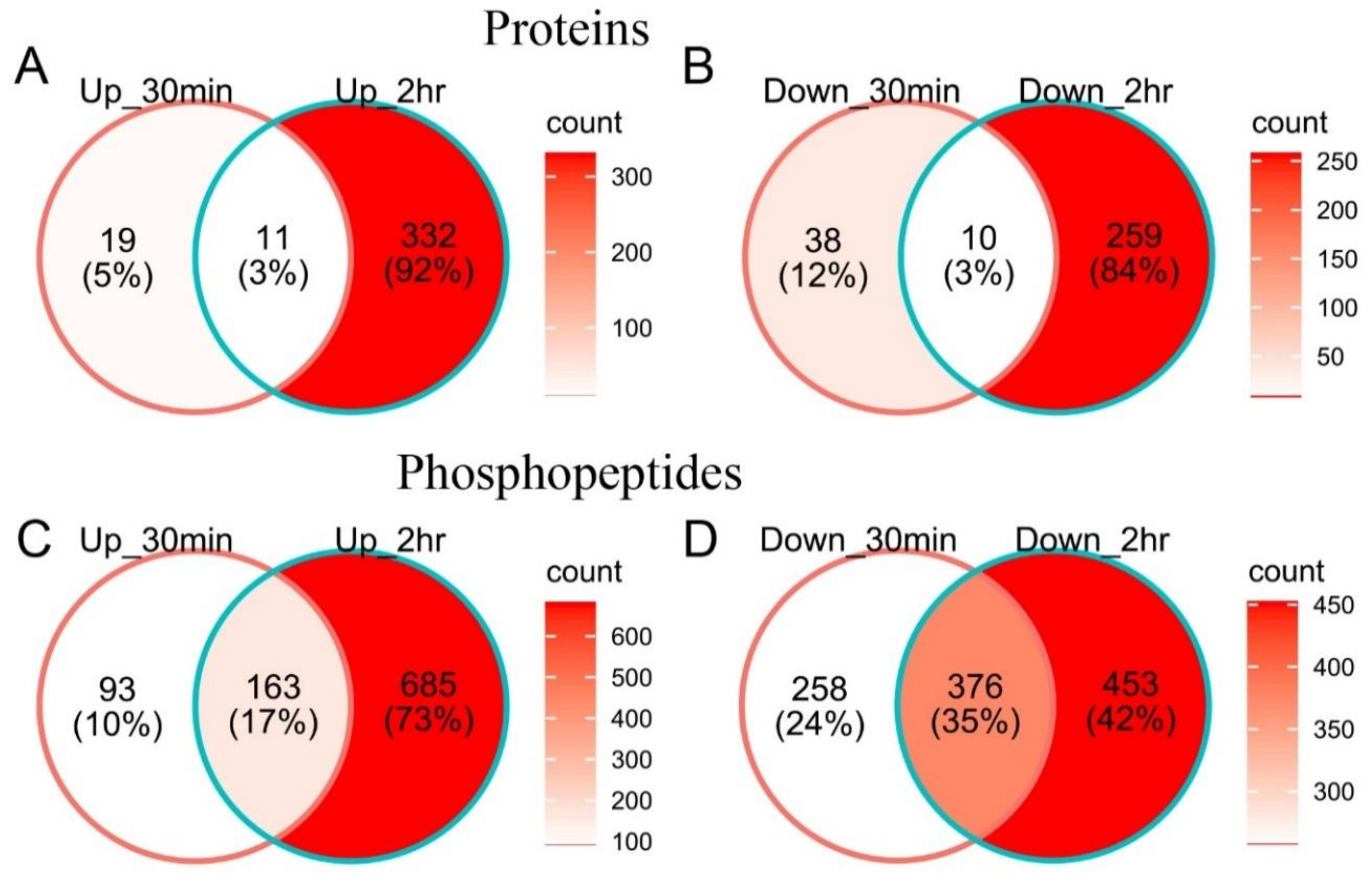
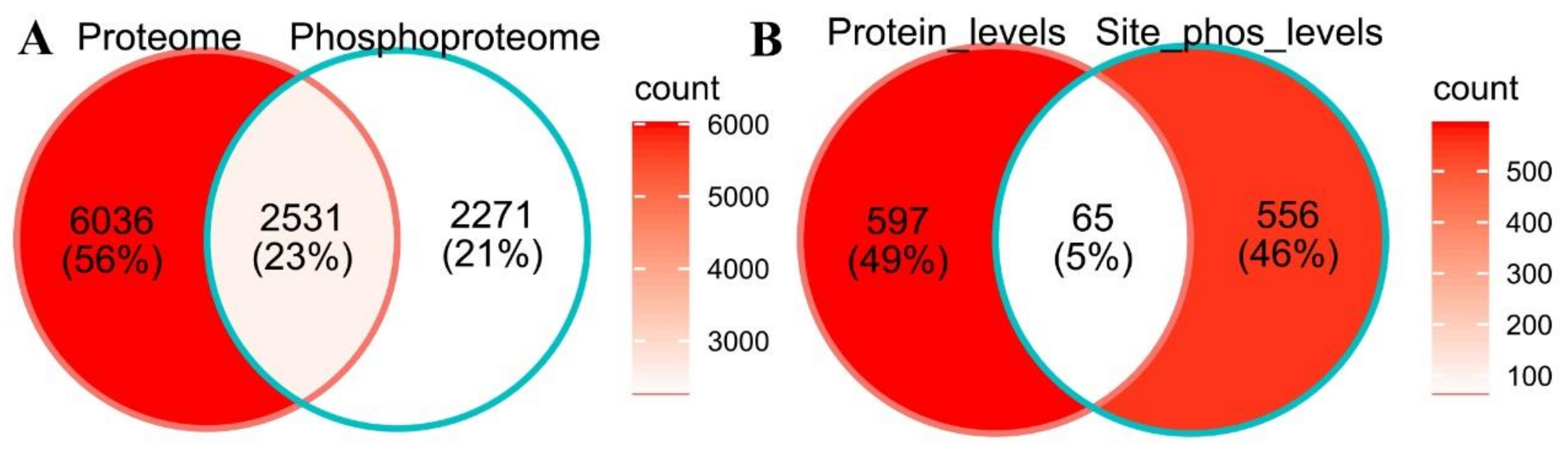
2.2. Cold-Responsive Proteins and Phosphopeptides
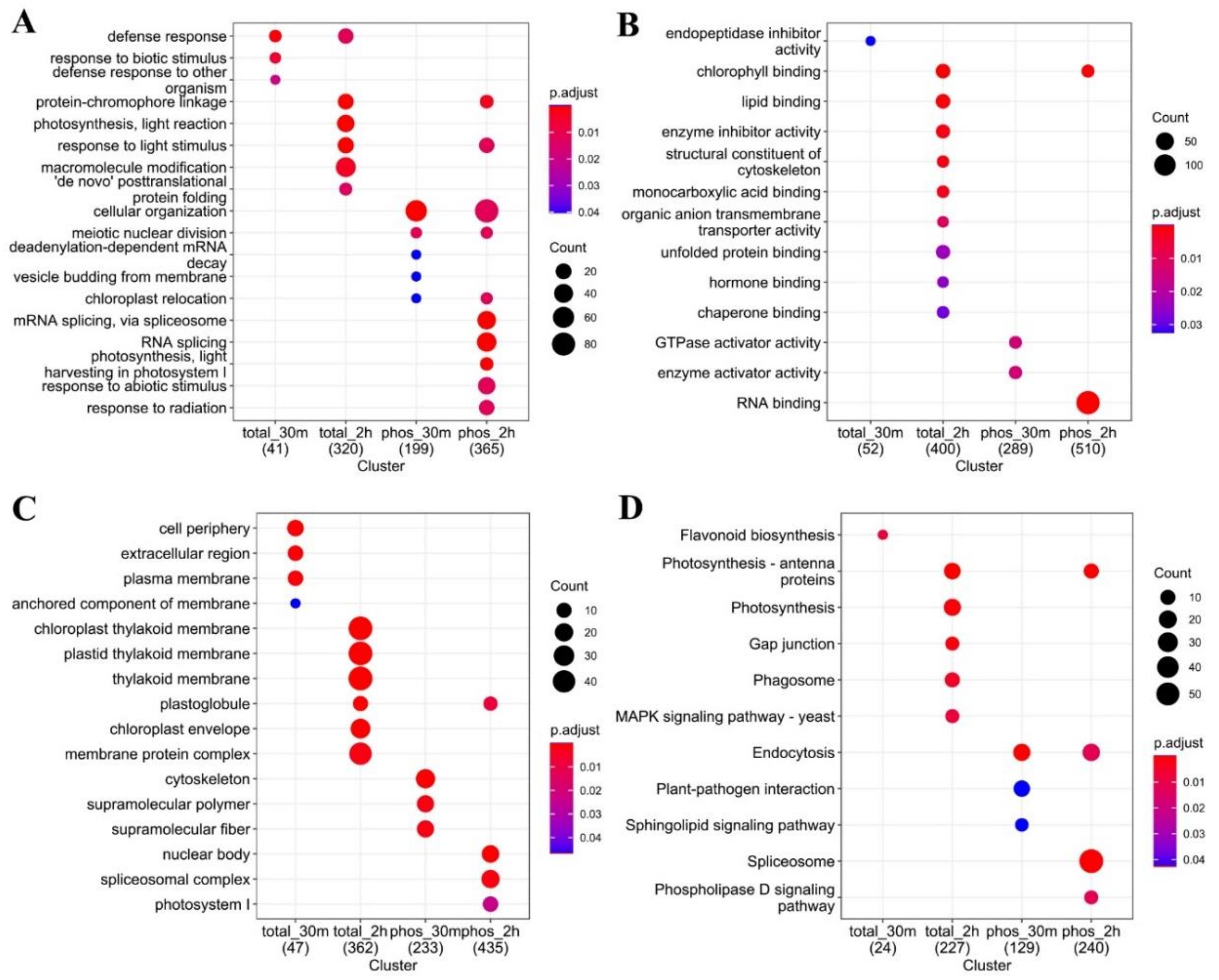
2.3. Functional Analysis of the Cold-Responsive Proteome and Phosphoproteme Reveals Different Groups of Functional Terms
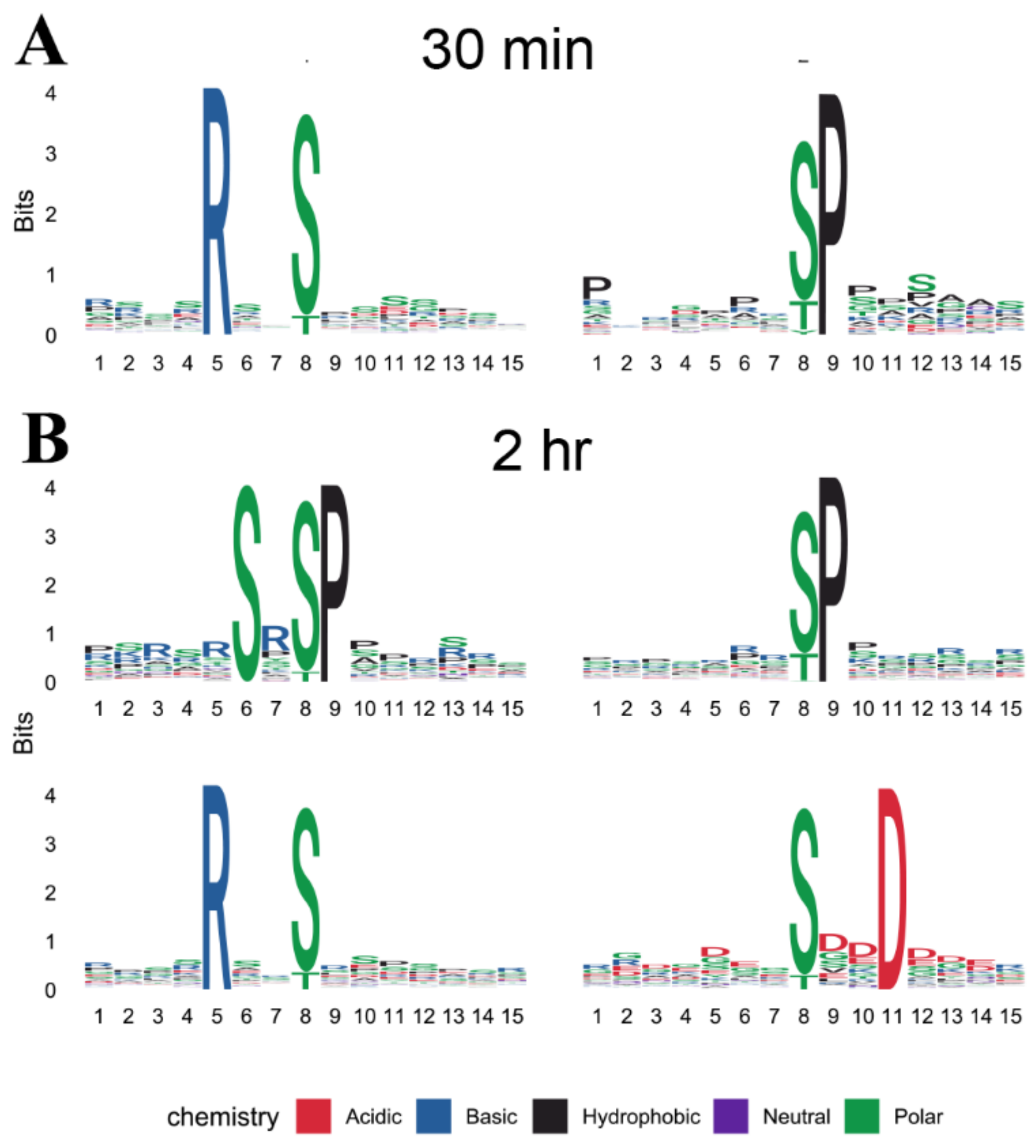
2.4. Site-Specific Phosphorylation Modification during Cold Treatment
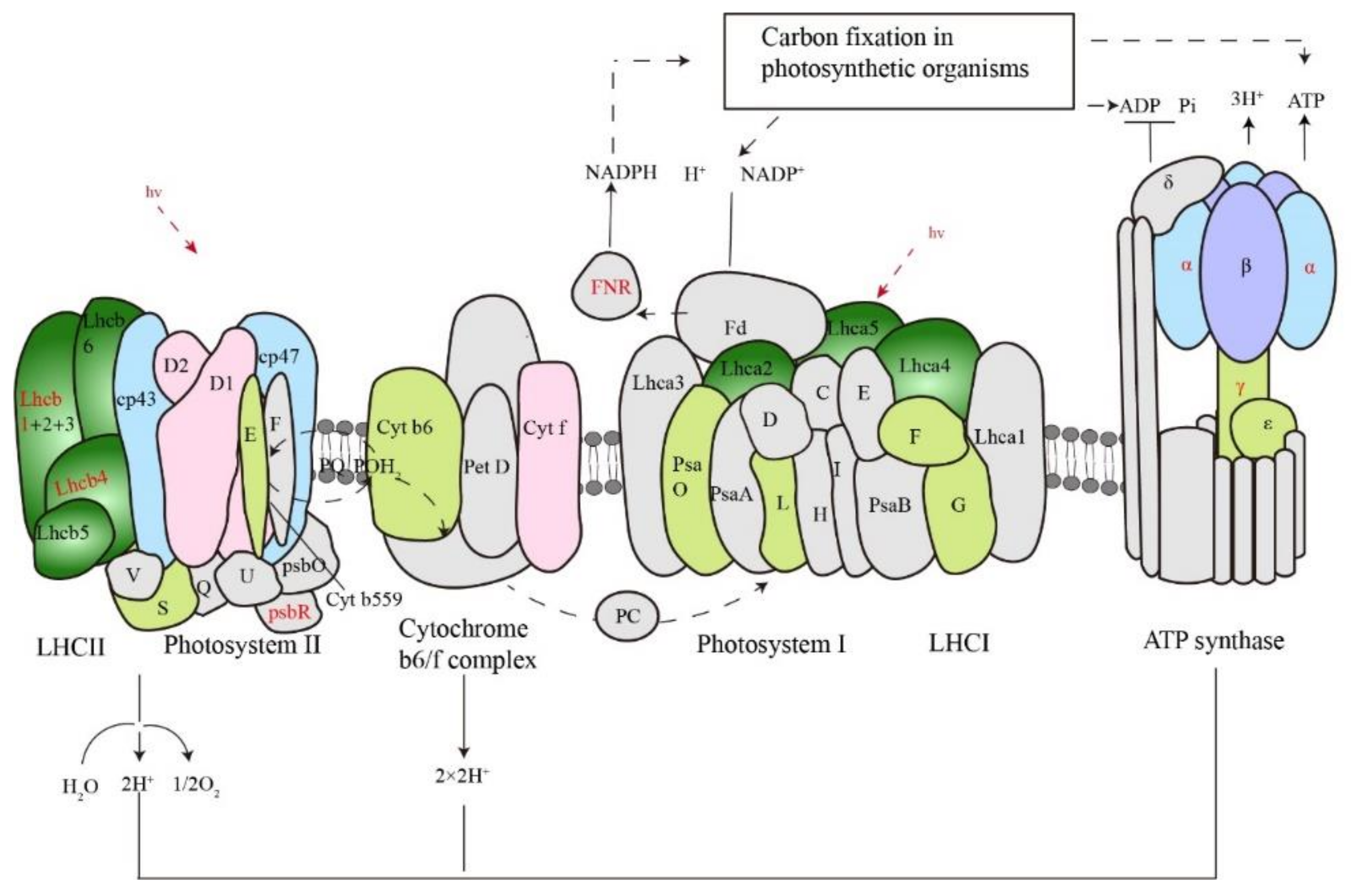
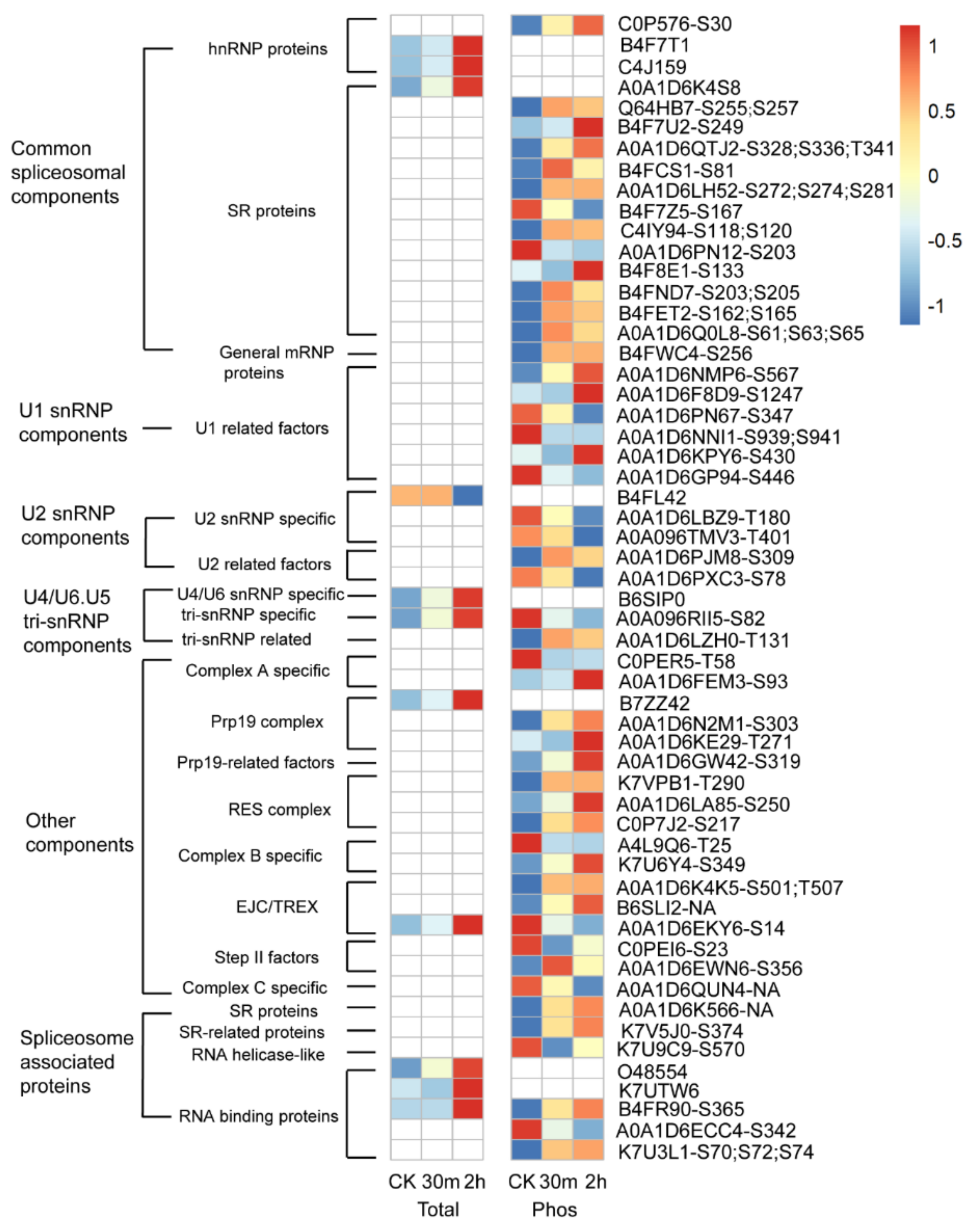
2.5. Cold-Responsive Photosynthesis and Spliceosome Proteins
2.6. Cold-Responsive Kinases and Transcription Factors
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Protein Extraction, Trypsin Digestion, and TMT Labeling
4.3. Peptide Fractionation and Phosphopeptide Enrichment
4.4. LC-MS/MS Analysis
4.5. LC-MS/MS Data Analysis and Bioinformatics Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Frascaroli, E.; Revilla, P. Genomics of cold tolerance in maize. In The Maize Genome; Springer: Cham, Switzerland, 2018; pp. 287–303. [Google Scholar]
- Farooq, M.; Aziz, T.; Wahid, A.; Lee, D.J.; Siddique, K.H.M. Chilling tolerance in maize: Agronomic and physiological approaches. Crop. Pasture Sci. 2009, 60, 501–516. [Google Scholar] [CrossRef]
- Zhou, X.; Muhammad, I.; Lan, H.; Xia, C. Recent Advances in the Analysis of Cold Tolerance in Maize. Front. Plant Sci. 2022, 13, 1040. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Malvar, R.A.; Alvarez-Iglesias, L.; Ordas, B.; Revilla, P. Dissecting the genetics of cold tolerance in a multiparental maize population. Appl. Genet. 2020, 133, 503–516. [Google Scholar] [CrossRef] [PubMed]
- Yi, Q.; Alvarez-Iglesias, L.; Malvar, R.A.; Romay, M.C.; Revilla, P. A worldwide maize panel revealed new genetic variation for cold tolerance. Appl. Genet. 2021, 134, 1083–1094. [Google Scholar] [CrossRef]
- Yadav, S.K. Cold stress tolerance mechanisms in plants. A review. Agron. Sustain. Dev. 2010, 30, 515–527. [Google Scholar] [CrossRef] [Green Version]
- Miura, K.; Furumoto, T. Cold signaling and cold response in plants. Int. J. Mol. Sci. 2013, 14, 5312–5337. [Google Scholar] [CrossRef] [Green Version]
- Guo, X.; Liu, D.; Chong, K. Cold signaling in plants: Insights into mechanisms and regulation. J. Integr. Plant Biol. 2018, 60, 745–756. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y.; Ding, Y.; Yang, S. Molecular Regulation of CBF Signaling in Cold Acclimation. Trends Plant Sci. 2018, 23, 623–637. [Google Scholar] [CrossRef]
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324. [Google Scholar] [CrossRef] [Green Version]
- Thomashow, M.F.; Stocldnger, E.J.; Jaglo-ottosen, K.R.; Gilmour, S.J.; Zarka, D.G. Function and regulation ofArabidopsis thaliana COR (cold-regulated) genes. Acta Physiol. Plant. 1997, 19, 497–504. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef]
- Ma, Y.; Dai, X.; Xu, Y.; Luo, W.; Zheng, X.; Zeng, D.; Pan, Y.; Lin, X.; Liu, H.; Zhang, D.; et al. COLD1 confers chilling tolerance in rice. Cell 2015, 160, 1209–1221. [Google Scholar] [CrossRef] [Green Version]
- Pandey, S.; Nelson, D.C.; Assmann, S.M. Two novel GPCR-type G proteins are abscisic acid receptors in Arabidopsis. Cell 2009, 136, 136–148. [Google Scholar] [CrossRef] [Green Version]
- Lamers, J.; van der Meer, T.; Testerink, C. How Plants Sense and Respond to Stressful Environments. Plant Physiol. 2020, 182, 1624–1635. [Google Scholar] [CrossRef] [Green Version]
- Burnett, A.C.; Kromdijk, J. Can we improve the chilling tolerance of maize photosynthesis through breeding? J. Exp. Bot. 2022, 73, 3138–3156. [Google Scholar] [CrossRef]
- Zeng, R.; Li, Z.; Shi, Y.; Fu, D.; Yin, P.; Cheng, J.; Jiang, C.; Yang, S. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 2021, 12, 4713. [Google Scholar] [CrossRef]
- Takac, T. The relationship of antioxidant enzymes and some physiological parameters in maize during chilling. Plant Soil Environ. 2004, 50, 27–32. [Google Scholar] [CrossRef] [Green Version]
- Waititu, J.K.; Cai, Q.; Sun, Y.; Sun, Y.; Li, C.; Zhang, C.; Liu, J.; Wang, H. Transcriptome Profiling of Maize (Zea mays L.) Leaves Reveals Key Cold-Responsive Genes, Transcription Factors, and Metabolic Pathways Regulating Cold Stress Tolerance at the Seedling Stage. Genes 2021, 12, 1638. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, J.; Cao, J.; Cai, Q.; Li, X.; Sun, Y.; Li, S.; Li, Y.; Hu, G.; Cao, S.; et al. Leaf transcriptomic response mediated by cold stress in two maize inbred lines with contrasting tolerance levels. Genomics 2021, 113, 782–794. [Google Scholar] [CrossRef]
- Li, M.; Sui, N.; Lin, L.; Yang, Z.; Zhang, Y. Transcriptomic profiling revealed genes involved in response to cold stress in maize. Funct. Plant Biol. 2019, 46, 830–844. [Google Scholar] [CrossRef]
- Frey, F.P.; Pitz, M.; Schon, C.C.; Hochholdinger, F. Transcriptomic diversity in seedling roots of European flint maize in response to cold. BMC Genom. 2020, 21, 300. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Fu, D.; Wang, X.; Zeng, R.; Zhang, X.; Tian, J.; Zhang, S.; Yang, X.; Tian, F.; Lai, J.; et al. Natural variation in the bZIP68 promoter modulates cold tolerance and was targeted during maize domestication. Plant Cell 2022, koac137. [Google Scholar] [CrossRef]
- Ding, Y.; Yang, S. Surviving and thriving: How plants perceive and respond to temperature stress. Dev. Cell 2022, 57, 947–958. [Google Scholar] [CrossRef]
- Yang, T.; Shad Ali, G.; Yang, L.; Du, L.; Reddy, A.; Poovaiah, B. Calcium/calmodulin-regulated receptor-like kinase CRLK1 interacts with MEKK1 in plants. Plant Signal. Behav. 2010, 5, 991–994. [Google Scholar] [CrossRef]
- Furuya, T.; Matsuoka, D.; Nanmori, T. Phosphorylation of Arabidopsis thaliana MEKK1 via Ca2+ signaling as a part of the cold stress response. J. Plant Res. 2013, 126, 833–840. [Google Scholar] [CrossRef]
- Zhao, C.; Wang, P.; Si, T.; Hsu, C.C.; Wang, L.; Zayed, O.; Yu, Z.; Zhu, Y.; Dong, J.; Tao, W.A.; et al. MAP Kinase Cascades Regulate the Cold Response by Modulating ICE1 Protein Stability. Dev. Cell 2017, 43, 618–629.e5. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Ding, Y.; Shi, Y.; Zhang, X.; Zhang, S.; Gong, Z.; Yang, S. MPK3-and MPK6-mediated ICE1 phosphorylation negatively regulates ICE1 stability and freezing tolerance in Arabidopsis. Dev. Cell 2017, 43, 630–642. [Google Scholar] [CrossRef]
- Li, T.; Xu, S.L.; Oses-Prieto, J.A.; Putil, S.; Xu, P.; Wang, R.J.; Li, K.H.; Maltby, D.A.; An, L.H.; Burlingame, A.L.; et al. Proteomics analysis reveals post-translational mechanisms for cold-induced metabolic changes in Arabidopsis. Mol. Plant 2011, 4, 361–374. [Google Scholar] [CrossRef] [Green Version]
- Kamal, M.M.; Ishikawa, S.; Takahashi, F.; Suzuki, K.; Kamo, M.; Umezawa, T.; Shinozaki, K.; Kawamura, Y.; Uemura, M. Large-scale phosphoproteomic study of arabidopsis membrane proteins reveals early signaling events in response to cold. Int. J. Mol. Sci. 2020, 21, 8631. [Google Scholar] [CrossRef]
- Tan, J.; Zhou, Z.; Feng, H.; Xing, J.; Niu, Y.; Deng, Z. Data-Independent Acquisition-Based Proteome and Phosphoproteome Profiling Reveals Early Protein Phosphorylation and Dephosphorylation Events in Arabidopsis Seedlings upon Cold Exposure. Int. J. Mol. Sci. 2021, 22, 12856. [Google Scholar] [CrossRef] [PubMed]
- Gan, C.S.; Chong, P.K.; Pham, T.K.; Wright, P.C. Technical, experimental, and biological variations in isobaric tags for relative and absolute quantitation (iTRAQ). J. Proteome Res. 2007, 6, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Karp, N.A.; Huber, W.; Sadowski, P.G.; Charles, P.D.; Hester, S.V.; Lilley, K.S. Addressing accuracy and precision issues in iTRAQ quantitation. Mol. Cell Proteom. 2010, 9, 1885–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, A.; Grant, C.E.; Noble, W.S.; Bailey, T.L. MoMo: Discovery of statistically significant post-translational modification motifs. Bioinformatics 2019, 35, 2774–2782. [Google Scholar] [CrossRef]
- Schwartz, D.; Gygi, S.P. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat. Biotechnol. 2005, 23, 1391–1398. [Google Scholar] [CrossRef]
- van Wijk, K.J.; Friso, G.; Walther, D.; Schulze, W.X. Meta-Analysis of Arabidopsis thaliana Phospho-Proteomics Data Reveals Compartmentalization of Phosphorylation Motifs. Plant Cell 2014, 26, 2367–2389. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Hsu, C.C.; Du, Y.; Zhu, P.; Zhao, C.; Fu, X.; Zhang, C.; Paez, J.S.; Macho, A.P.; Tao, W.A.; et al. Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. USA 2020, 117, 3270–3280. [Google Scholar] [CrossRef]
- Roberts, D.M.; Harmon, A.C. Calcium-modulated proteins: Targets of intracellular calcium signals in higher plants. Annu. Rev. Plant Biol. 1992, 43, 375–414. [Google Scholar] [CrossRef]
- Christie, P.J.; Alfenito, M.R.; Walbot, V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef]
- Matera, A.G.; Wang, Z. A day in the life of the spliceosome. Nat. Rev. Mol. Cell Biol. 2014, 15, 108–121. [Google Scholar] [CrossRef] [Green Version]
- Calixto, C.P.G.; Guo, W.; James, A.B.; Tzioutziou, N.A.; Entizne, J.C.; Panter, P.E.; Knight, H.; Nimmo, H.G.; Zhang, R.; Brown, J.W.S. Rapid and Dynamic Alternative Splicing Impacts the Arabidopsis Cold Response Transcriptome. Plant Cell 2018, 30, 1424–1444. [Google Scholar] [CrossRef] [Green Version]
- Teige, M.; Scheikl, E.; Eulgem, T.; Dóczi, R.; Ichimura, K.; Shinozaki, K.; Dangl, J.L.; Hirt, H. The MKK2 pathway mediates cold and salt stress signaling in Arabidopsis. Mol. Cell 2004, 15, 141–152. [Google Scholar] [CrossRef]
- Boudsocq, M.; Sheen, J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013, 18, 30–40. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xu, C.; Zhu, Y.; Zhang, L.; Chen, T.; Zhou, F.; Chen, H.; Lin, Y. The calcium-dependent kinase OsCPK24 functions in cold stress responses in rice. J. Integr. Plant Biol. 2018, 60, 173–188. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Park, S.; Gilmour, S.J.; Thomashow, M.F. Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 2013, 75, 364–376. [Google Scholar] [CrossRef]
- Kidokoro, S.; Yoneda, K.; Takasaki, H.; Takahashi, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Different Cold-Signaling Pathways Function in the Responses to Rapid and Gradual Decreases in Temperature. Plant Cell 2017, 29, 760–774. [Google Scholar] [CrossRef] [Green Version]
- Doherty, C.J.; Van Buskirk, H.A.; Myers, S.J.; Thomashow, M.F. Roles for Arabidopsis CAMTA Transcription Factors in Cold-Regulated Gene Expression and Freezing Tolerance. Plant Cell 2009, 21, 972–984. [Google Scholar] [CrossRef] [Green Version]
- Gangappa, S.N.; Botto, J.F. The multifaceted roles of HY5 in plant growth and development. Mol. Plant 2016, 9, 1353–1365. [Google Scholar] [CrossRef] [Green Version]
- Hardtke, C.S.; Gohda, K.; Osterlund, M.T.; Oyama, T.; Okada, K.; Deng, X.W. HY5 stability and activity in Arabidopsis is regulated by phosphorylation in its COP1 binding domain. EMBO J. 2000, 19, 4997–5006. [Google Scholar] [CrossRef]
- Savitski, M.M.; Mathieson, T.; Zinn, N.; Sweetman, G.; Doce, C.; Becher, I.; Pachl, F.; Kuster, B.; Bantscheff, M. Measuring and managing ratio compression for accurate iTRAQ/TMT quantification. J. Proteome Res. 2013, 12, 3586–3598. [Google Scholar] [CrossRef]
- Wang, X.; Shan, X.; Wu, Y.; Su, S.; Li, S.; Liu, H.; Han, J.; Xue, C.; Yuan, Y. iTRAQ-based quantitative proteomic analysis reveals new metabolic pathways responding to chilling stress in maize seedlings. J. Proteom. 2016, 146, 14–24. [Google Scholar] [CrossRef]
- Żelisko, A.; García-Lorenzo, M.; Jackowski, G.; Jansson, S.; Funk, C. AtFtsH6 is involved in the degradation of the light-harvesting complex II during high-light acclimation and senescence. Proc. Natl. Acad. Sci. USA 2005, 102, 13699–13704. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Yuan, J.; Li, Y.; Sun, H.; Ma, T.; Huai, J.; Yang, W.; Zhang, W.; Lin, R. The CDC48 complex mediates ubiquitin-dependent degradation of intra-chloroplast proteins in plants. Cell Rep. 2022, 39, 110664. [Google Scholar] [CrossRef]
- Staiger, D.; Brown, J.W. Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 2013, 25, 3640–3656. [Google Scholar] [CrossRef] [Green Version]
- Laloum, T.; Martin, G.; Duque, P. Alternative Splicing Control of Abiotic Stress Responses. Trends Plant Sci. 2018, 23, 140–150. [Google Scholar] [CrossRef] [Green Version]
- Thatcher, S.R.; Danilevskaya, O.N.; Meng, X.; Beatty, M.; Zastrow-Hayes, G.; Harris, C.; Van Allen, B.; Habben, J.; Li, B. Genome-Wide Analysis of Alternative Splicing during Development and Drought Stress in Maize. Plant Physiol. 2016, 170, 586–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Tang, J.; Bassham, D.C.; Howell, S.H. Daily temperature cycles promote alternative splicing of RNAs encoding SR45a, a splicing regulator in maize. Plant Physiol. 2021, 186, 1318–1335. [Google Scholar] [CrossRef] [PubMed]
- Stamm, S. Regulation of alternative splicing by reversible protein phosphorylation. J. Biol. Chem. 2008, 283, 1223–1227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Fu, X.D. Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 2013, 122, 191–207. [Google Scholar] [CrossRef]
- Kanno, T.; Venhuizen, P.; Wen, T.N.; Lin, W.D.; Chiou, P.; Kalyna, M.; Matzke, A.J.M.; Matzke, M. PRP4KA, a Putative Spliceosomal Protein Kinase, Is Important for Alternative Splicing and Development in Arabidopsis thaliana. Genetics 2018, 210, 1267–1285. [Google Scholar] [CrossRef] [Green Version]
- Deng, Z.; Zhang, X.; Tang, W.; Oses-Prieto, J.A.; Suzuki, N.; Gendron, J.M.; Chen, H.; Guan, S.; Chalkley, R.J.; Peterman, T.K.; et al. A proteomics study of brassinosteroid response in Arabidopsis. Mol. Cell Proteom. 2007, 6, 2058–2071. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Sato, Y.; Morishima, K. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 2016, 428, 726–731. [Google Scholar] [CrossRef] [Green Version]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Cham, Switzerland, 2016. [Google Scholar]
- Gao, C.H.; Yu, G.; Cai, P. ggVennDiagram: An Intuitive, Easy-to-Use, and Highly Customizable R Package to Generate Venn Diagram. Front. Genet. 2021, 12, 706907. [Google Scholar] [CrossRef]
- Ma, J.; Chen, T.; Wu, S.; Yang, C.; Bai, M.; Shu, K.; Li, K.; Zhang, G.; Jin, Z.; He, F.; et al. iProX: An integrated proteome resource. Nucleic Acids Res. 2019, 47, D1211–D1217. [Google Scholar] [CrossRef] [Green Version]

| Accession | Ratio (30 min) | p Value (30 min) | Ratio (2 h) | p Value (2 h) | Description |
|---|---|---|---|---|---|
| Kinases | |||||
| A0A1D6FNF9 | 0.96 | 7.27 × 10−1 | 0.76 | 8.14 × 10−3 | ABC1-like kinase |
| A0A1D6HW78 | 0.87 | 1.34 × 10−1 | 0.71 | 1.47 × 10−3 | ABC2 homolog 13 |
| A0A1D6JD09 | 1.03 | 7.53 × 10−1 | 0.74 | 2.67 × 10−3 | Calcium-dependent protein kinase |
| B4F9P5 | 1.09 | 1.95 × 10−1 | 0.76 | 1.09 × 10−3 | Protein-serine/threonine kinase |
| B4FF99 | 0.76 | 1.32 × 10−2 | 0.70 | 8.59 × 10−4 | Calcium-dependent protein kinase 7 |
| B6SVK8 | 1.02 | 8.50 × 10−1 | 0.74 | 5.73 × 10−3 | Serine/threonine-protein kinase NAK |
| P28523 | 0.91 | 3.87 × 10−1 | 0.73 | 7.04 × 10−3 | Casein kinase II subunit alpha |
| Transcription factors | |||||
| A0A1D6JVI5 | 1.02 | 8.94 × 10−1 | 2.88 | 2.38 × 10−5 | bZIP transcription factor 16 |
| B4FIJ2 | 1.15 | 8.66 × 10−2 | 1.35 | 1.42 × 10−3 | Zinc ion binding |
| K7V3U5 | 0.64 | 6.49 × 10−3 | 0.99 | 9.31 × 10−1 | WRKY DNA-binding domain protein |
| K7V9Y4 | 1.06 | 4.10 × 10−1 | 1.31 | 1.87 × 10−3 | HSF28 HSF type transcription factor |
| Accession | Ratio (30 min) | p Value (30 min) | Ratio (2 h) | p Value (2 h) | Phos-Site | Description |
|---|---|---|---|---|---|---|
| Kinases | ||||||
| A0A1D6F0U9 | 1.35 | 3.17 × 10−2 | 2.07 | 2.71 × 10−4 | S498/T501 | MAPK |
| A0A1D6G325 | 0.64 | 6.58 × 10−4 | 0.66 | 7.95 × 10−4 | S279 | Casein kinase family protein |
| A0A1D6GJU6 | 0.78 | 8.83 × 10−3 | 0.63 | 2.71 × 10−4 | S115 | MAPKKK1 |
| A0A1D6GJU6 | 0.75 | 1.77 × 10−2 | 0.63 | 9.43 × 10−4 | S200 | MAPKKK1 |
| A0A1D6ICZ3 | 0.64 | 9.04 × 10−4 | 0.63 | 5.87 × 10−4 | T52 | CDPK8 |
| A0A1D6MYW4 | 0.68 | 2.64 × 10−3 | 0.68 | 2.22 × 10−3 | S253 | CPK21 |
| A0A1D6P094 | 1.68 | 2.92 × 10−3 | 1.03 | 8.48 × 10−1 | S436 | Protein kinase |
| B4FBJ3 | 0.77 | 8.28 × 10−3 | 0.64 | 3.24 × 10−4 | S464 | Casein kinase 1 |
| O49975 | 0.51 | 1.42 × 10−4 | 0.57 | 3.52 × 10−4 | T30 | MEK1 |
| Transcription factors | ||||||
| A0A1D6EHU0 | 0.67 | 6.5 × 10−2 | 0.54 | 6.5 × 10−3 | S130 | VIP1 transcription factor |
| A0A1D6EHU0 | 0.34 | 1.6 × 10−4 | 0.53 | 2.5 × 10−3 | S31 | VIP1 transcription factor |
| A0A1D6EK20 | 0.71 | 3.6 × 10−3 | 0.74 | 4.7 × 10−3 | S930 | RNA binding family protein |
| A0A1D6EK20 | 0.58 | 2.5 × 10−4 | 0.72 | 2.7 × 10−3 | S949 | RNA binding family protein |
| A0A1D6ELA3 | 1.15 | 1.8 × 10−1 | 1.40 | 5.8 × 10−3 | S180 | G-box-binding factor 1 |
| A0A1D6H3I5 | 1.27 | 5.4 × 10−2 | 1.43 | 6.3 × 10−3 | S83 | HY5 transcription factor homolog |
| A0A1D6IJ69 | 1.56 | 6.9 × 10−4 | 1.46 | 1.3 × 10−3 | S205 | NF-Y subunit B-3 |
| A0A1D6IK52 | 0.81 | 5.2 × 10−2 | 0.70 | 3.3 × 10−3 | S172 | CAMTA 2 |
| A0A1D6JVI5 | 0.93 | 7.0 × 10−1 | 0.46 | 8.8 × 10−4 | S203 | bZIP transcription factor 16 |
| A0A1D6JVI5 | 0.95 | 6.4 × 10−1 | 0.41 | 6.9 × 10−5 | S148; S151 | bZIP transcription factor 16 |
| A0A1D6JVI5 | 1.06 | 5.4 × 10−1 | 0.38 | 3.3 × 10−3 | S290 | bZIP transcription factor 16 |
| A0A1D6K5M3 | 1.57 | 4.9 × 10−3 | 1.52 | 4.9 × 10−3 | S121 | NF-Y subunit B-2 |
| A0A1D6K5M3 | 1.47 | 3.5 × 10−3 | 1.36 | 6.9 × 10−3 | S212 | NF-Y subunit B-2 |
| A0A1D6K5M3 | 1.53 | 2.1 × 10−2 | 1.61 | 7.9 × 10−3 | S210; S213 | NF-Y subunit B-2 |
| A0A1D6MZQ6 | 1.39 | 1.1 × 10−2 | 1.45 | 4.5 × 10−3 | S124 | VIP1 transcription factor |
| A0A1D6PUS5 | 1.65 | 3.2 × 10−3 | 1.24 | 7.6 × 10−2 | S89 | Homeobox-leucine zipper protein MERISTEM L1 |
| B4F937 | 1.51 | 3.5 × 10−3 | 1.33 | 1.4 × 10−2 | S147 | G-box binding factor |
| B4FWJ9 | 0.53 | 2.7 × 10−4 | 0.47 | 1.1 × 10−4 | S139 | bZIP transcription factor |
| B4FWJ9 | 0.60 | 8.5 × 10−5 | 0.66 | 2.7 × 10−4 | S31 | bZIP transcription factor |
| C4J4L1 | 0.69 | 7.5 × 10−3 | 0.69 | 5.2 × 10−3 | T134 | ABI5-like protein 2 |
| K7TX82 | 0.78 | 1.7 × 10−2 | 0.66 | 8.9 × 10−4 | S192 | VIP1 transcription factor |
| K7VAC7 | 1.07 | 6.9 × 10−1 | 1.93 | 1.6 × 10−3 | S132 | HY5 transcription factor homolog |
| K7VQH0 | 1.12 | 3.4 × 10−1 | 1.50 | 4.7 × 10−3 | S30 | HY5 transcription factor homolog |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xing, J.; Tan, J.; Feng, H.; Zhou, Z.; Deng, M.; Luo, H.; Deng, Z. Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings. Int. J. Mol. Sci. 2022, 23, 6493. https://doi.org/10.3390/ijms23126493
Xing J, Tan J, Feng H, Zhou Z, Deng M, Luo H, Deng Z. Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings. International Journal of Molecular Sciences. 2022; 23(12):6493. https://doi.org/10.3390/ijms23126493
Chicago/Turabian StyleXing, Jiayun, Jinjuan Tan, Hanqian Feng, Zhongjing Zhou, Min Deng, Hongbing Luo, and Zhiping Deng. 2022. "Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings" International Journal of Molecular Sciences 23, no. 12: 6493. https://doi.org/10.3390/ijms23126493
APA StyleXing, J., Tan, J., Feng, H., Zhou, Z., Deng, M., Luo, H., & Deng, Z. (2022). Integrative Proteome and Phosphoproteome Profiling of Early Cold Response in Maize Seedlings. International Journal of Molecular Sciences, 23(12), 6493. https://doi.org/10.3390/ijms23126493






