m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages
Abstract
:1. Introduction
2. Results
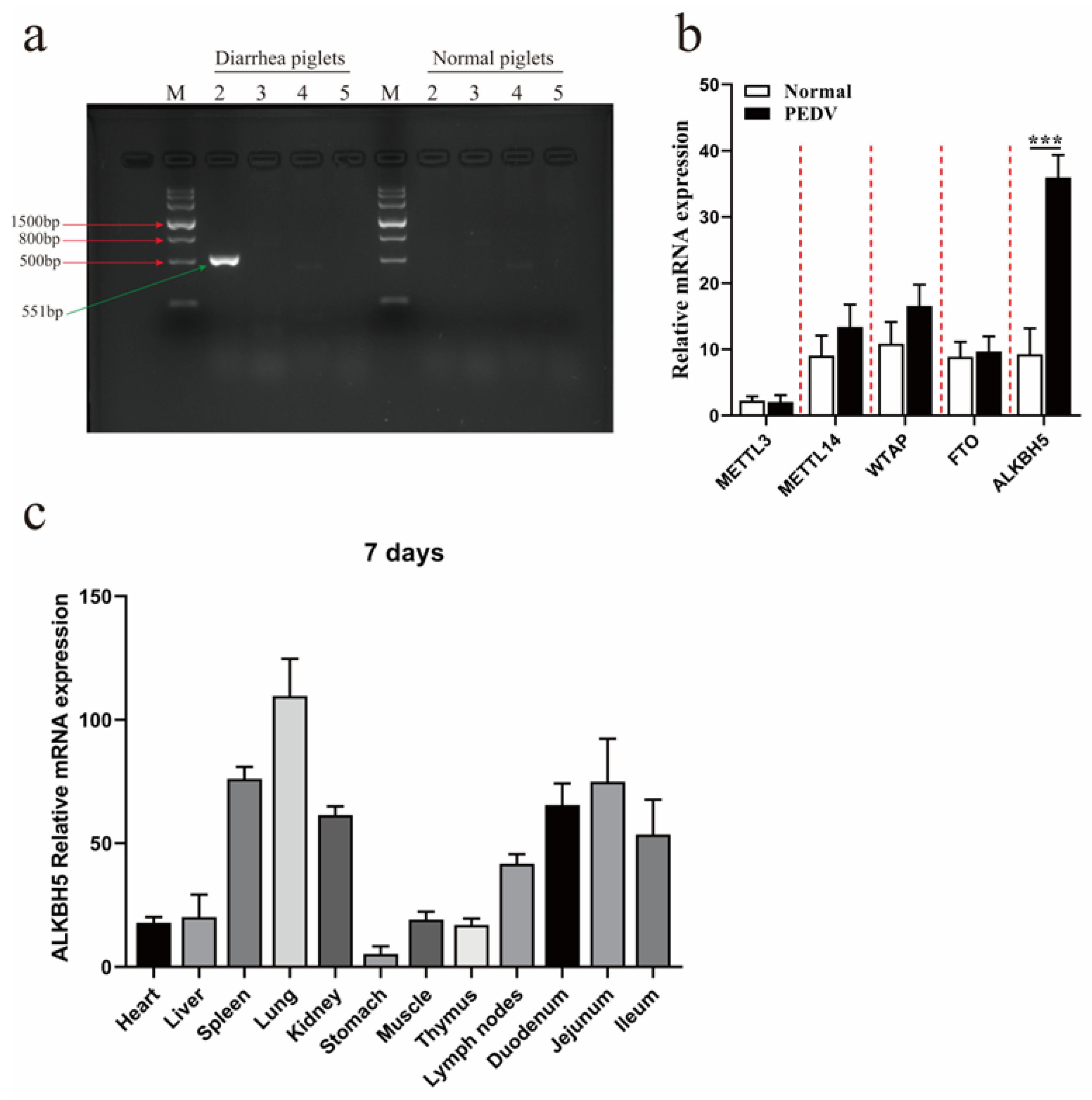
2.1. PEDV Infection Promotes the Expression of ALKBH5 in the Lung Tissues of Piglets
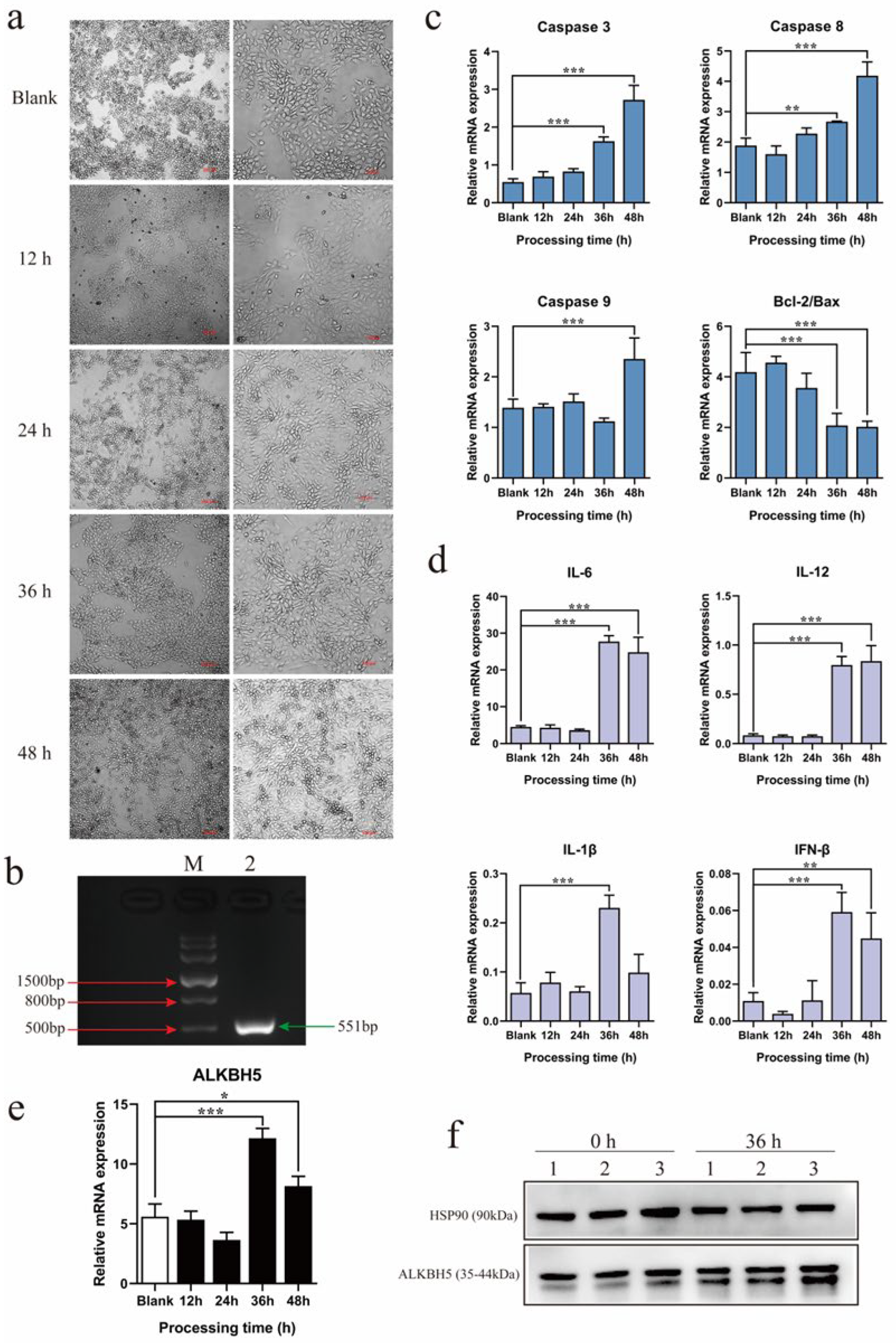
2.2. PEDV Infection Up-Regulates ALKBH5 Expression in 3D4/21 Alveolar Macrophage Cell Line
2.3. Disruption of ALKBH5 Expression Increases PEDV Infection Capacity
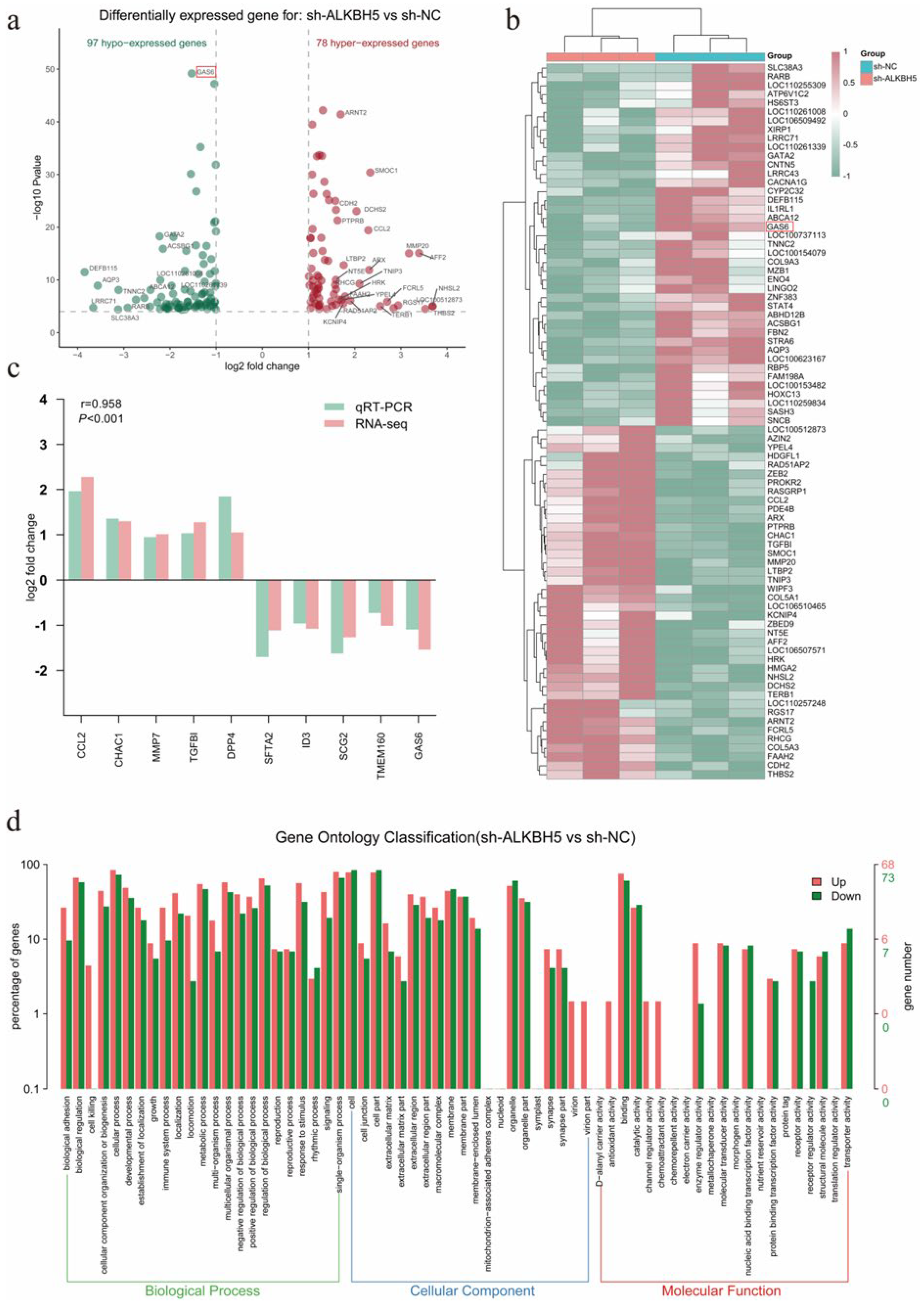
2.4. Transcriptome Analysis Identified GAS6 as a Putative Target Gene of ALKBH5
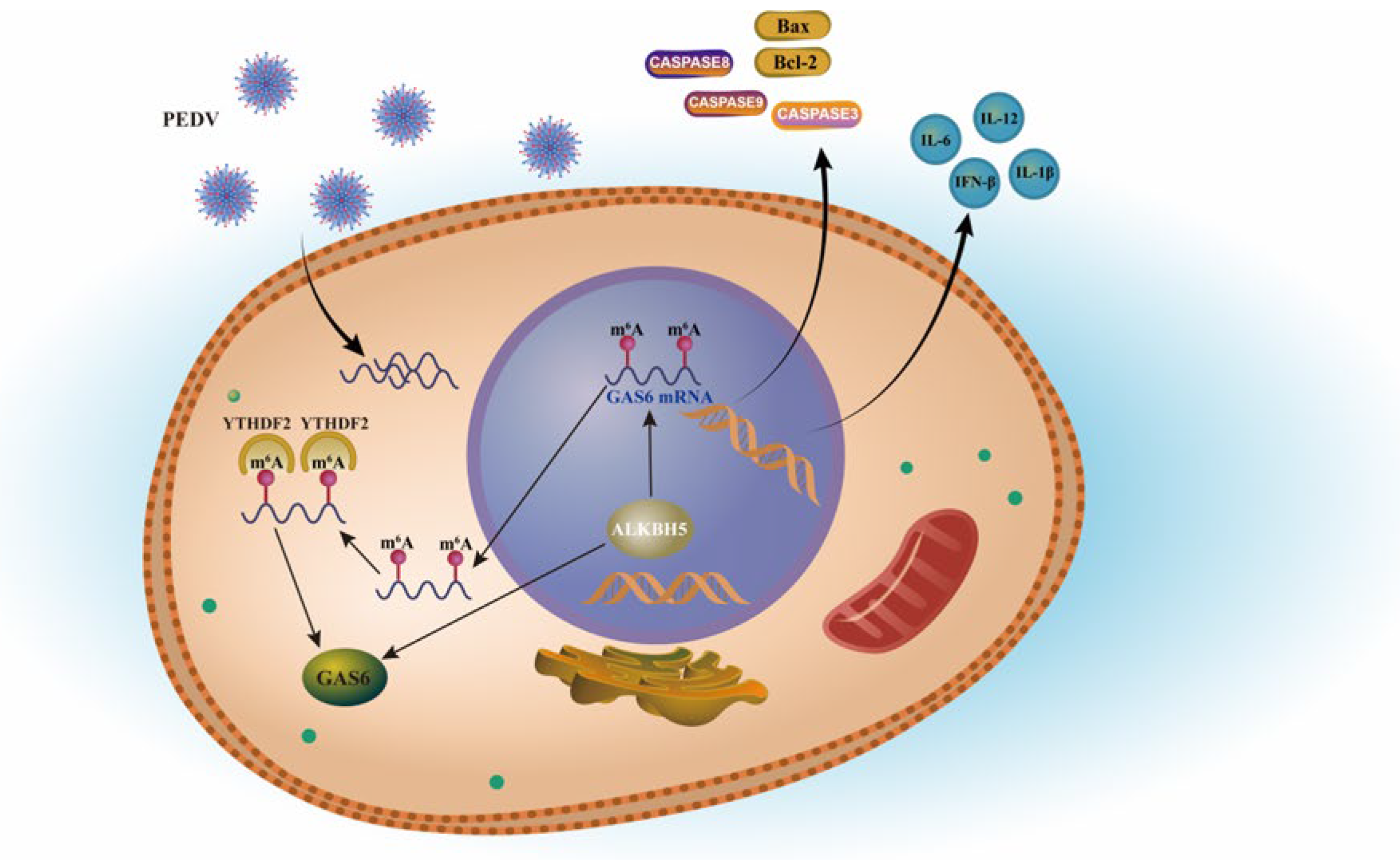
2.5. ALKBH5 Depends on YTHDF2 to Affect the Expression of Target Gene GAS6
3. Discussion
4. Materials and Methods
4.1. Experimental Animals and Sample Collection
4.2. Proliferation and Isolation of PEDV
4.3. Virus Titer Determination
4.4. PEDV Infection of 3D4/21 Cells
4.5. Construction of 3D4/21 Cell Line with ALKBH5 Gene Interference
4.6. RNA Extraction and cDNA Synthesis
4.7. RT-PCR
4.8. qPCR
4.9. Detection of Copy Number of PEDV-M Gene
4.10. Western Blotting
4.11. RNA Sequencing
4.12. Methylated RNA Immunoprecipitation Coupled with qPCR (MeRIP-qPCR)
4.13. mRNA Stability Analysis
4.14. Statistical Analyses
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Song, D.; Park, B. Porcine epidemic diarrhoea virus: A comprehensive review of molecular epidemiology, diagnosis, and vaccines. Virus Genes 2012, 44, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Vlasova, A.N.; Marthaler, D.; Wang, Q.H.; Culhane, M.R.; Rossow, K.D.; Rovira, A.; Collins, J.; Saif, L.J. Distinct characteristics and complex evolution of PEDV strains, North America, May 2013–February 2014. Emerg. Infect. Dis. 2014, 20, 1620–1628. [Google Scholar] [CrossRef] [Green Version]
- Choudhury, B.; Dastjerdi, A.; Doyle, N.; Frossard, J.P.; Steinbach, F. From the field to the lab—An European view on the global spread of PEDV. Virus Res. 2016, 226, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Wu, Q.X.; Huang, L.L.; Yuan, C.; Wang, J.L.; Yang, Q. An alternative pathway of enteric PEDV dissemination from nasal cavity to intestinal mucosa in swine. Nat. Commun. 2018, 9, 3811. [Google Scholar] [CrossRef] [PubMed]
- Jung, K.; Saif, L.J. Porcine epidemic diarrhea virus infection: Etiology, epidemiology, pathogenesis and immunoprophylaxis. Vet. J. 2015, 204, 134–143. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.F.; Luo, S.X.; Gu, J.; Li, Z.Q.; Li, K.; Yuan, W.F.; Ye, Y.; Li, H.; Ding, Z.; Song, D.P.; et al. Prevalence and phylogenetic analysis of porcine diarrhea associated viruses in southern China from 2012 to 2018. BMC Vet. Res. 2019, 15, 470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuda, T.; Murakami, S.; Takahashi, O.; Miyazaki, A.; Ohashi, S.; Yamasato, H.; Suzuki, T. New porcine epidemic diarrhoea virus variant with a large deletion in the spike gene identified in domestic pigs. Arch. Virol. 2015, 160, 2565–2568. [Google Scholar] [CrossRef]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA modifications in gene expression regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [Green Version]
- Roignant, J.Y.; Soller, M. m6A in mRNA: An ancient mechanism for fine-tuning gene expression. Trends Genet. 2017, 33, 380–390. [Google Scholar] [CrossRef]
- Imam, H.; Kim, G.W.; Siddiqui, A. Epitranscriptomic(N6-methyladenosine) modification of viral RNA and virus-host interactions. Front. Cell. Infect. Microbiol. 2020, 10, 584283. [Google Scholar] [CrossRef]
- Du, K.Z.; Zhang, L.B.; Lee, T.; Sun, T. m6A RNA methylation controls neural development and is involved in human diseases. Mol. Neurobiol. 2019, 56, 1596–1606. [Google Scholar] [CrossRef] [PubMed]
- Patil, D.P.; Chen, C.K.; Pickering, B.F.; Chow, A.; Jackson, C.; Guttman, M.; Jaffrey, S.R. m(6)A RNA methylation promotes XIST-mediated transcriptional repression. Nature 2016, 537, 369–373. [Google Scholar] [CrossRef]
- Rana, A.K.; Ankri, S. Reviving the RNA world: An insight into the appearance of RNA methyltransferases. Front. Genet. 2016, 7, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, H.; Zhao, Y.; He, J.Q.; Zhang, Y.; Xi, H.R.; Liu, M.F.; Ma, J.B.; Wu, L.G. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat. Commun. 2016, 7, 12626. [Google Scholar] [CrossRef] [PubMed]
- Xiao, W.; Adhikari, S.; Dahal, U.; Chen, Y.S.; Hao, Y.J.; Sun, B.F.; Sun, H.Y.; Li, A.; Ping, X.L.; Lai, W.Y.; et al. Nuclear m(6)A reader YTHDC1 regulates mRNA splicing. Mol. Cell 2016, 61, 507–519. [Google Scholar] [CrossRef] [Green Version]
- Lichinchi, G.; Gao, S.; Saletore, Y.; Gonzalez, G.M.; Bansal, V.; Wang, Y.S.; Mason, C.E.; Rana, T.M. Dynamics of the human and viral m(6)A RNA methylomes during HIV-1 infection of T cells. Nat. Microbiol. 2016, 1, 16011. [Google Scholar] [CrossRef]
- Courtney, D.G.; Kennedy, E.M.; Dumm, R.E.; Bogerd, H.P.; Tsai, K.; Heaton, N.S.; Cullen, B.R. Epitranscriptomic enhancement of influenza a virus gene expression and replication. Cell Host Microbe 2017, 22, 377–386. [Google Scholar] [CrossRef] [Green Version]
- Tsai, K.; Courtney, D.G.; Cullen, B.R. Addition of m6A to SV40 late mRNAs enhances viral structural gene expression and replication. PLoS Pathog. 2018, 14, e1006919. [Google Scholar] [CrossRef]
- Lichinchi, G.; Zhao, B.S.; Wu, Y.G.; Lu, Z.K.; Qin, Y.; He, C.; Rana, T.M. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 2016, 20, 666–673. [Google Scholar] [CrossRef] [Green Version]
- Chen, J.; Jin, L.; Wang, Z.; Wang, L.; Chen, Q.; Cui, Y.; Liu, G. N6-methyladenosine regulates PEDV replication and host gene expression. Virology 2020, 548, 59–72. [Google Scholar] [CrossRef]
- Zhang, L.Z.; Richard, A.S.; Jackson, C.B.; Ojha, A.; Choe, H. Phosphatidylethanolamine and phosphatidylserine synergize to enhance GAS6/AXL-mediated virus infection and efferocytosis. J. Virol. 2020, 95, e02079-20. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Zeng, P.; Li, Y.H.; Zhang, Z.D.; Cui, Q.H. SRAMP: Prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 2016, 44, e91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xuan, J.J.; Sun, W.J.; Lin, P.H.; Zhou, K.R.; Liu, S.; Zheng, L.L.; Qu, L.H.; Yang, J.H. RMBase v2.0: Deciphering the map of RNA modifications from epitranscriptome sequencing data. Nucleic Acids Res. 2018, 46, D327–D334. [Google Scholar] [CrossRef] [PubMed]
- Alonso, C.; Goede, D.P.; Morrison, R.B.; Davies, P.R.; Rovira, A.; Marthaler, D.G.; Torremorell, M. Evidence of infectivity of airborne porcine epidemic diarrhea virus and detection of airborne viral RNA at long distances from infected herds. Vet. Res. 2014, 45, 73. [Google Scholar] [CrossRef] [PubMed]
- Park, J.E.; Shin, H.J. Porcine epidemic diarrhea virus infects and replicates in porcine alveolar macrophages. Virus Res. 2014, 191, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Joshi, N.; Walter, J.M.; Misharin, A.V. Alveolar macrophages. Cell. Immunol. 2018, 330, 86–90. [Google Scholar] [CrossRef] [PubMed]
- Rubio, R.M.; Depledge, D.P.; Bianco, C.; Thompson, L.; Mohr, I. RNA m6A modification enzymes shape innate responses to DNA by regulating interferon β. Gene. Dev. 2018, 32, 1472–1484. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Zhang, X.L.; Hu, J.J.; Qu, R.H.; Yu, Z.B.; Xu, H.; Chen, H.F.; Yan, L.C.; Ding, C.B.; Zou, Q.; et al. m6A demethylase ALKBH5 controls CD4+ T cell pathogenicity and promotes autoimmunity. Sci. Adv. 2021, 7, eabg0470. [Google Scholar] [CrossRef]
- Zheng, Q.L.; Hou, J.; Zhou, Y.; Li, Z.Y.; Cao, X.T. The RNA helicase DDX46 inhibits innate immunity by entrapping m6A-demethylated antiviral transcripts in the nucleus. Nat. Immunol. 2017, 18, 1094–1103. [Google Scholar] [CrossRef]
- Hafizi, S.; Gustafsson, A.; Stenhoff, J.; Dahlbäck, B. The Ran binding protein RanBPM interacts with Axl and Sky receptor tyrosine kinases. Int. J. Biochem. Cell B. 2005, 37, 2344–2356. [Google Scholar] [CrossRef]
- Lemke, G.; Burstyn-Cohen, T. TAM receptors and the clearance of apoptotic cells. Ann. N. Y. Acad. Sci. 2010, 1209, 23–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rothlin, C.V.; Lemke, G. TAM receptor signaling and autoimmune disease. Curr. Opin. Immunol. 2010, 22, 740–746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Lim, E.J.; Yoon, Y.S.; Ahn, Y.H.; Park, E.M.; Kim, H.S.; Kang, J.L. Liver X receptor and STAT1 cooperate downstream of Gas6/Mer to induce anti-inflammatory arginase 2 expression in macrophages. Sci. Rep. 2016, 6, 29673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cohen, P.L.; Shao, W.H. Gas6/TAM receptors in systemic lupus erythematosus. Dis. Markers. 2019, 2019, 7838195. [Google Scholar] [CrossRef]
- Bellan, M.; Cittone, M.G.; Tonello, S.; Rigamonti, C.; Castello, L.M.; Gavelli, F.; Pirisi, M.; Sainaghi, P.P. Gas6/TAM system: A key modulator of the interplay between inflammation and fibrosis. Int. J. Mol. Sci. 2019, 20, 5070. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibata, T.; Makino, A.; Ogata, R.; Nakamura, S.; Ito, T.; Nagata, K.; Terauchi, Y.; Oishi, T.; Fujieda, M.; Takahashi, Y.; et al. Respiratory syncytial virus infection exacerbates pneumococcal pneumonia via Gas6/Axl-mediated macrophage polarization. J. Clin. Investig. 2020, 130, 3021–3037. [Google Scholar] [CrossRef]
- Nidetz, N.F.; Gallagher, T.M.; Wiethoff, C.M. Inhibition of type I interferon responses by adenovirus serotype-dependent Gas6 binding. Virology 2018, 515, 150–157. [Google Scholar] [CrossRef]
- Moller-Tank, S.; Maury, W. Phosphatidylserine receptors: Enhancers of enveloped virus entry and infection. Virology 2014, 468, 565–580. [Google Scholar] [CrossRef] [Green Version]
- Silva-Filho, J.L.; de Oliveira, L.G.; Monteiro, L.; Parise, P.L.; Zanluqui, N.G.; Polonio, C.M.; de Freitas, C.L.; Toledo-Teixeira, D.A.; de Souza, W.M.; Bittencourt, N.; et al. Gas6 drives Zika virus-induced neurological complications in humans and congenital syndrome in immunocompetent mice. Brain Behav. Immun. 2021, 97, 260–274. [Google Scholar] [CrossRef]
- Zhao, Y.C.; Shi, Y.F.; Shen, H.F.; Xie, W.Z. m6A-binding proteins: The emerging crucial performers in epigenetics. J. Hematol. Oncol. 2020, 13, 35. [Google Scholar] [CrossRef] [Green Version]
- Wu, R.F.; Liu, Y.H.; Zhao, Y.L.; Bi, Z.; Yao, Y.X.; Liu, Q.; Wang, F.Q.; Wang, Y.Z.; Wang, X.X. m6A methylation controls pluripotency of porcine induced pluripotent stem cells by targeting SOCS3/JAK2/STAT3 pathway in a YTHDF1/YTHDF2-orchestrated manner. Cell Death Dis. 2019, 10, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, X.Y.; Cao, Y.; Wu, S.L.; Wu, Z.C.; Bao, W.B. miR-129a-3p inhibits PEDV replication by targeting the EDA-mediated NF-κB pathway in IPEC-J2 cells. Int. J. Mol. Sci. 2021, 22, 8133. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.J.; Ren, Z.S.; Zhang, S.; Wang, H.F.; Wu, S.L.; Bao, W.B. Analysis of intestinal mucosa integrity and GLP-2 gene functions upon porcine epidemic diarrhea virus infection in pigs. Animals 2021, 11, 644. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, X.; Lu, Z.K.; Gomez, A.; Hon, G.C.; Yue, Y.N.; Han, D.L.; Fu, Y.; Parisien, M.; Dai, Q.; Jia, G.F.; et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014, 505, 117–120. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, J.; Xu, C.; Wu, S.; Wu, Z.; Wu, S.; Sun, M.; Bao, W. m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages. Int. J. Mol. Sci. 2022, 23, 6191. https://doi.org/10.3390/ijms23116191
Jin J, Xu C, Wu S, Wu Z, Wu S, Sun M, Bao W. m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages. International Journal of Molecular Sciences. 2022; 23(11):6191. https://doi.org/10.3390/ijms23116191
Chicago/Turabian StyleJin, Jian, Chao Xu, Sen Wu, Zhengchang Wu, Shenglong Wu, Mingan Sun, and Wenbin Bao. 2022. "m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages" International Journal of Molecular Sciences 23, no. 11: 6191. https://doi.org/10.3390/ijms23116191
APA StyleJin, J., Xu, C., Wu, S., Wu, Z., Wu, S., Sun, M., & Bao, W. (2022). m6A Demethylase ALKBH5 Restrains PEDV Infection by Regulating GAS6 Expression in Porcine Alveolar Macrophages. International Journal of Molecular Sciences, 23(11), 6191. https://doi.org/10.3390/ijms23116191






