Association of DNA Methylation of the NLRP3 Gene with Changes in Cortical Thickness in Major Depressive Disorder
Abstract
:1. Introduction
2. Results
2.1. Differential Methylation Analysis
2.2. Cortical Thickness Analysis
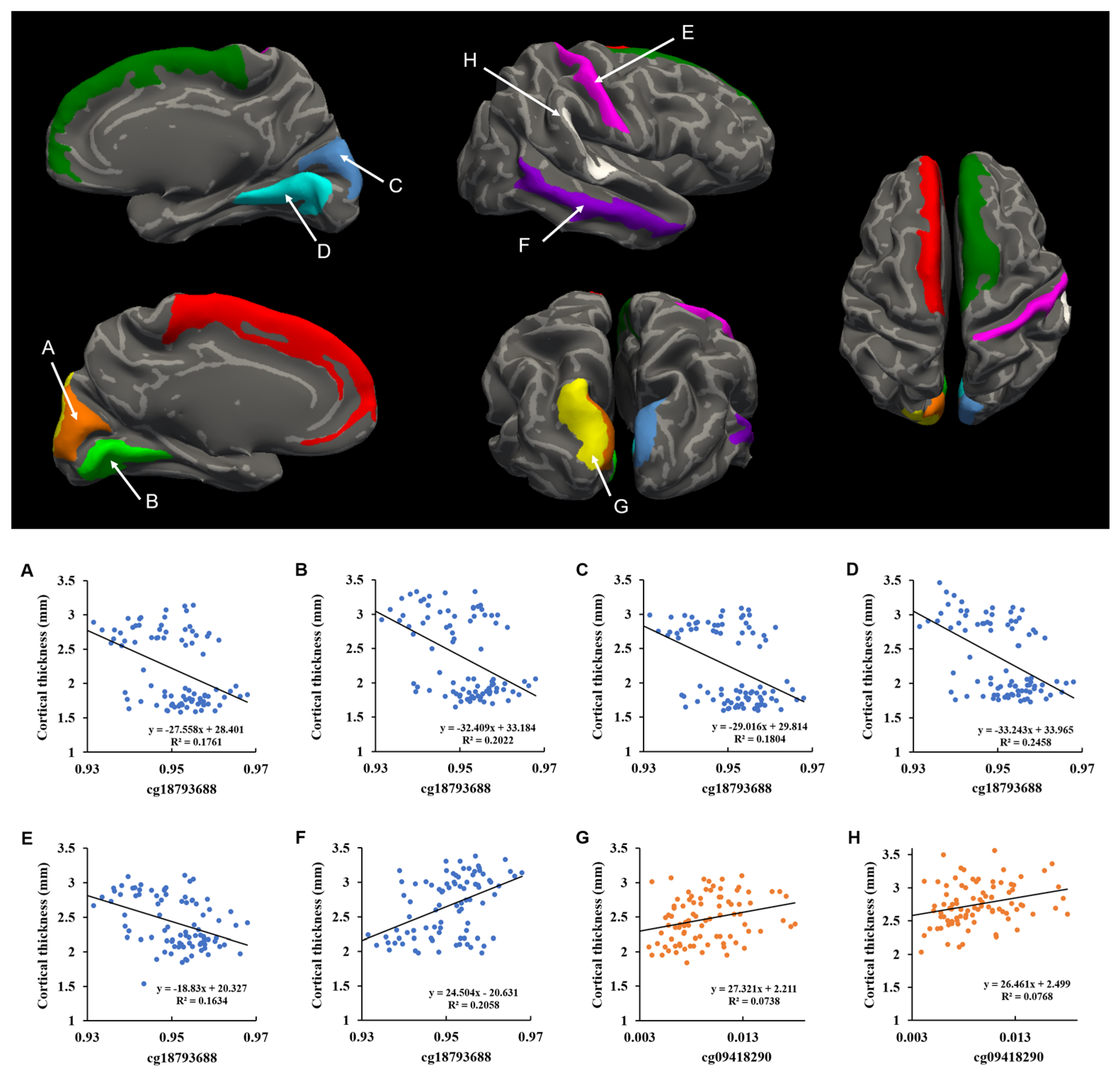
2.3. Correlation between DNA Methylation and Cortical Thickness
3. Discussion
4. Materials and Methods
4.1. Participants
4.2. Methylomic Profiling of NLRP3 Gene
4.3. Genome-Wide DNA Methylation Analysis of the NRLP3 Gene
4.4. MRI Data Acquisition
4.5. Neuroimage Processing
4.6. Statistical Methods for Neuroimaging–DNA Methylation Correlation Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Otte, C.; Gold, S.M.; Penninx, B.W.; Pariante, C.M.; Etkin, A.; Fava, M.; Mohr, D.C.; Schatzberg, A.F. Major depressive disorder. Nat. Rev. Dis. Primers 2016, 2, 16065. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, K.M.; Ham, B.J. How Inflammation Affects the Brain in Depression: A Review of Functional and Structural MRI Studies. J. Clin. Neurol. 2021, 17, 503–515. [Google Scholar] [CrossRef] [PubMed]
- Raison, C.L.; Capuron, L.; Miller, A.H. Cytokines sing the blues: Inflammation and the pathogenesis of depression. Trends Immunol. 2006, 27, 24–31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldsmith, D.R.; Rapaport, M.H.; Miller, B.J. A meta-analysis of blood cytokine network alterations in psychiatric patients: Comparisons between schizophrenia, bipolar disorder and depression. Mol. Psychiatry 2016, 21, 1696–1709. [Google Scholar] [CrossRef] [PubMed]
- Köhler, C.A.; Freitas, T.H.; Maes, M.; de Andrade, N.Q.; Liu, C.S.; Fernandes, B.S.; Stubbs, B.; Solmi, M.; Veronese, N.; Herrmann, N.; et al. Peripheral cytokine and chemokine alterations in depression: A meta-analysis of 82 studies. Acta Psychiatr. Scand. 2017, 135, 373–387. [Google Scholar] [CrossRef]
- Osimo, E.F.; Pillinger, T.; Rodriguez, I.M.; Khandaker, G.M.; Pariante, C.M.; Howes, O.D. Inflammatory markers in depression: A meta-analysis of mean differences and variability in 5166 patients and 5083 controls. Brain Behav. Immun. 2020, 87, 901–909. [Google Scholar] [CrossRef]
- Lohoff, F.W. Overview of the genetics of major depressive disorder. Curr. Psychiatry Rep. 2010, 12, 539–546. [Google Scholar] [CrossRef] [Green Version]
- Agid, O.; Kohn, Y.; Lerer, B. Environmental stress and psychiatric illness. Biomed. Pharm. 2000, 54, 135–141. [Google Scholar] [CrossRef]
- Berk, M.; Williams, L.J.; Jacka, F.N.; O’Neil, A.; Pasco, J.A.; Moylan, S.; Allen, N.B.; Stuart, A.L.; Hayley, A.C.; Byrne, M.L.; et al. So depression is an inflammatory disease, but where does the inflammation come from? BMC Med. 2013, 11, 200. [Google Scholar] [CrossRef] [Green Version]
- Barbu, M.C.; Shen, X.; Walker, R.M.; Howard, D.M.; Evans, K.L.; Whalley, H.C.; Porteous, D.J.; Morris, S.W.; Deary, I.J.; Zeng, Y.; et al. Epigenetic prediction of major depressive disorder. Mol. Psychiatry 2021, 26, 5112–5123. [Google Scholar] [CrossRef]
- Crawford, B.; Craig, Z.; Mansell, G.; White, I.; Smith, A.; Spaull, S.; Imm, J.; Hannon, E.; Wood, A.; Yaghootkar, H.; et al. DNA methylation and inflammation marker profiles associated with a history of depression. Hum. Mol. Genet. 2018, 27, 2840–2850. [Google Scholar] [CrossRef] [PubMed]
- Clark, S.L.; Hattab, M.W.; Chan, R.F.; Shabalin, A.A.; Han, L.K.M.; Zhao, M.; Smit, J.H.; Jansen, R.; Milaneschi, Y.; Xie, L.Y.; et al. A methylation study of long-term depression risk. Mol. Psychiatry 2020, 25, 1334–1343. [Google Scholar] [CrossRef] [PubMed]
- Wray, N.R.; Ripke, S.; Mattheisen, M.; Trzaskowski, M.; Byrne, E.M.; Abdellaoui, A.; Adams, M.J.; Agerbo, E.; Air, T.M.; Andlauer, T.M.F.; et al. Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nat. Genet. 2018, 50, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.-K.; Won, E. The influence of stress on neuroinflammation and alterations in brain structure and function in major depressive disorder. Behav. Brain Res. 2017, 329, 6–11. [Google Scholar] [CrossRef] [PubMed]
- Kaufmann, F.N.; Costa, A.P.; Ghisleni, G.; Diaz, A.P.; Rodrigues, A.L.S.; Peluffo, H.; Kaster, M.P. NLRP3 inflammasome-driven pathways in depression: Clinical and preclinical findings. Brain Behav. Immun. 2017, 64, 367–383. [Google Scholar] [CrossRef]
- Iwata, M.; Ota, K.T.; Li, X.Y.; Sakaue, F.; Li, N.; Dutheil, S.; Banasr, M.; Duric, V.; Yamanashi, T.; Kaneko, K.; et al. Psychological Stress Activates the Inflammasome via Release of Adenosine Triphosphate and Stimulation of the Purinergic Type 2X7 Receptor. Biol. Psychiatry 2016, 80, 12–22. [Google Scholar] [CrossRef]
- Pan, Y.; Chen, X.Y.; Zhang, Q.Y.; Kong, L.D. Microglial NLRP3 inflammasome activation mediates IL-1β-related inflammation in prefrontal cortex of depressive rats. Brain Behav. Immun. 2014, 41, 90–100. [Google Scholar] [CrossRef]
- Alcocer-Gómez, E.; de Miguel, M.; Casas-Barquero, N.; Núñez-Vasco, J.; Sánchez-Alcazar, J.A.; Fernández-Rodríguez, A.; Cordero, M.D. NLRP3 inflammasome is activated in mononuclear blood cells from patients with major depressive disorder. Brain Behav. Immun. 2014, 36, 111–117. [Google Scholar] [CrossRef]
- Taene, A.; Khalili-Tanha, G.; Esmaeili, A.; Mobasheri, L.; Kooshkaki, O.; Jafari, S.; Shokouhifar, A.; Sarab, G.A. The Association of Major Depressive Disorder with Activation of NLRP3 Inflammasome, Lipid Peroxidation, and Total Antioxidant Capacity. J. Mol. Neurosci. 2020, 70, 65–70. [Google Scholar] [CrossRef]
- Rubartelli, A. Redox control of NLRP3 inflammasome activation in health and disease. J. Leukoc. Biol. 2012, 92, 951–958. [Google Scholar] [CrossRef]
- Swanson, K.V.; Deng, M.; Ting, J.P. The NLRP3 inflammasome: Molecular activation and regulation to therapeutics. Nat. Rev. Immunol. 2019, 19, 477–489. [Google Scholar] [CrossRef]
- Wang, D.; Wang, H.; Gao, H.; Zhang, H.; Zhang, H.; Wang, Q.; Sun, Z. P2X7 receptor mediates NLRP3 inflammasome activation in depression and diabetes. Cell Biosci. 2020, 10, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleshner, M.; Frank, M.; Maier, S.F. Danger Signals and Inflammasomes: Stress-Evoked Sterile Inflammation in Mood Disorders. Neuropsychopharmacology 2017, 42, 36–45. [Google Scholar] [CrossRef] [Green Version]
- Franklin, T.C.; Xu, C.; Duman, R.S. Depression and sterile inflammation: Essential role of danger associated molecular patterns. Brain Behav. Immun. 2018, 72, 2–13. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, A.; Jones, D.N.C. Emerging role of the P2X7-NLRP3-IL1β pathway in mood disorders. Psychoneuroendocrinology 2018, 98, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.Y.; Nuñez, G. Sterile inflammation: Sensing and reacting to damage. Nat. Rev. Immunol. 2010, 10, 826–837. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schmaal, L.; Hibar, D.P.; Sämann, P.G.; Hall, G.B.; Baune, B.T.; Jahanshad, N.; Cheung, J.W.; van Erp, T.G.M.; Bos, D.; Ikram, M.A.; et al. Cortical abnormalities in adults and adolescents with major depression based on brain scans from 20 cohorts worldwide in the ENIGMA Major Depressive Disorder Working Group. Mol. Psychiatry 2017, 22, 900–909. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Zhao, Y.; Chen, Z.; Long, J.; Dai, J.; Huang, X.; Lui, S.; Radua, J.; Vieta, E.; Kemp, G.J.; et al. Meta-analysis of cortical thickness abnormalities in medication-free patients with major depressive disorder. Neuropsychopharmacology 2020, 45, 703–712. [Google Scholar] [CrossRef]
- Suh, J.S.; Schneider, M.A.; Minuzzi, L.; MacQueen, G.M.; Strother, S.C.; Kennedy, S.H.; Frey, B.N. Cortical thickness in major depressive disorder: A systematic review and meta-analysis. Prog. Neuropsychopharmacol. Biol. Psychiatry 2019, 88, 287–302. [Google Scholar] [CrossRef]
- Tu, P.C.; Chen, L.F.; Hsieh, J.C.; Bai, Y.M.; Li, C.T.; Su, T.P. Regional cortical thinning in patients with major depressive disorder: A surface-based morphometry study. Psychiatry Res. 2012, 202, 206–213. [Google Scholar] [CrossRef]
- Jung, J.; Kang, J.; Won, E.; Nam, K.; Lee, M.S.; Tae, W.S.; Ham, B.J. Impact of lingual gyrus volume on antidepressant response and neurocognitive functions in Major Depressive Disorder: A voxel-based morphometry study. J. Affect. Disord. 2014, 169, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Qiu, L.; Lui, S.; Kuang, W.; Huang, X.; Li, J.; Li, J.; Zhang, J.; Chen, H.; Sweeney, J.A.; Gong, Q. Regional increases of cortical thickness in untreated, first-episode major depressive disorder. Transl. Psychiatry 2014, 4, e378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kandilarova, S.; Stoyanov, D.; Sirakov, N.; Maes, M.; Specht, K. Reduced grey matter volume in frontal and temporal areas in depression: Contributions from voxel-based morphometry study. Acta Neuropsychiatr. 2019, 31, 252–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andreazza, A.C.; Shao, L.; Wang, J.F.; Young, L.T. Mitochondrial complex I activity and oxidative damage to mitochondrial proteins in the prefrontal cortex of patients with bipolar disorder. Arch. Gen. Psychiatry 2010, 67, 360–368. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.T.; Beal, M.F. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature 2006, 443, 787–795. [Google Scholar] [CrossRef]
- Steiner, J.; Walter, M.; Gos, T.; Guillemin, G.J.; Bernstein, H.G.; Sarnyai, Z.; Mawrin, C.; Brisch, R.; Bielau, H.; Meyer zu Schwabedissen, L.; et al. Severe depression is associated with increased microglial quinolinic acid in subregions of the anterior cingulate gyrus: Evidence for an immune-modulated glutamatergic neurotransmission? J. Neuroinflammation 2011, 8, 94. [Google Scholar] [CrossRef] [Green Version]
- Alcocer-Gómez, E.; Casas-Barquero, N.; Williams, M.R.; Romero-Guillena, S.L.; Cañadas-Lozano, D.; Bullón, P.; Sánchez-Alcazar, J.A.; Navarro-Pando, J.M.; Cordero, M.D. Antidepressants induce autophagy dependent-NLRP3-inflammasome inhibition in Major depressive disorder. Pharm. Res. 2017, 121, 114–121. [Google Scholar] [CrossRef]
- Dantzer, R. Depression and inflammation: An intricate relationship. Biol. Psychiatry 2012, 71, 4–5. [Google Scholar] [CrossRef]
- Dowlati, Y.; Herrmann, N.; Swardfager, W.; Liu, H.; Sham, L.; Reim, E.K.; Lanctôt, K.L. A meta-analysis of cytokines in major depression. Biol. Psychiatry 2010, 67, 446–457. [Google Scholar] [CrossRef]
- Jeon, S.W.; Kim, Y.K. Neuroinflammation and cytokine abnormality in major depression: Cause or consequence in that illness? World J. Psychiatry 2016, 6, 283–293. [Google Scholar] [CrossRef]
- Wohleb, E.S.; Franklin, T.; Iwata, M.; Duman, R.S. Integrating neuroimmune systems in the neurobiology of depression. Nat. Rev. Neurosci. 2016, 17, 497–511. [Google Scholar] [CrossRef] [PubMed]
- Sestan-Pesa, M.; Horvath, T.L. Metabolism and Mental Illness. Trends Mol. Med. 2016, 22, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Malynn, S.; Campos-Torres, A.; Moynagh, P.; Haase, J. The pro-inflammatory cytokine TNF-α regulates the activity and expression of the serotonin transporter (SERT) in astrocytes. Neurochem. Res. 2013, 38, 694–704. [Google Scholar] [CrossRef] [PubMed]
- Troubat, R.; Barone, P.; Leman, S.; Desmidt, T.; Cressant, A.; Atanasova, B.; Brizard, B.; El Hage, W.; Surget, A.; Belzung, C. Neuroinflammation and depression: A review. Eur. J. Neurosci. 2021, 53, 151–171. [Google Scholar] [CrossRef] [PubMed]
- Green, C.; Shen, X.; Stevenson, A.J.; Conole, E.L.S.; Harris, M.A.; Barbu, M.C.; Hawkins, E.L.; Adams, M.J.; Hillary, R.F.; Lawrie, S.M.; et al. Structural brain correlates of serum and epigenetic markers of inflammation in major depressive disorder. Brain Behav. Immun. 2021, 92, 39–48. [Google Scholar] [CrossRef]
- Opel, N.; Cearns, M.; Clark, S.; Toben, C.; Grotegerd, D.; Heindel, W.; Kugel, H.; Teuber, A.; Minnerup, H.; Berger, K.; et al. Large-scale evidence for an association between low-grade peripheral inflammation and brain structural alterations in major depression in the BiDirect study. J. Psychiatry Neurosci. JPN 2019, 44, 423–431. [Google Scholar] [CrossRef] [Green Version]
- Han, K.M.; Tae, W.S.; Kim, A.; Kang, Y.; Kang, W.; Kang, J.; Kim, Y.K.; Kim, B.; Seong, J.Y.; Ham, B.J. Serum FAM19A5 levels: A novel biomarker for neuroinflammation and neurodegeneration in major depressive disorder. Brain Behav. Immun. 2020, 87, 852–859. [Google Scholar] [CrossRef]
- Vidal-Pineiro, D.; Parker, N.; Shin, J.; French, L.; Grydeland, H.; Jackowski, A.P.; Mowinckel, A.M.; Patel, Y.; Pausova, Z.; Salum, G.; et al. Cellular correlates of cortical thinning throughout the lifespan. Sci. Rep. 2020, 10, 21803. [Google Scholar] [CrossRef]
- Zhou, J.; Li, M.; Wang, X.; He, Y.; Xia, Y.; Sweeney, J.A.; Kopp, R.F.; Liu, C.; Chen, C. Drug Response-Related DNA Methylation Changes in Schizophrenia, Bipolar Disorder, and Major Depressive Disorder. Front. Neurosci. 2021, 15, 674273. [Google Scholar] [CrossRef]
- Wheater, E.N.W.; Stoye, D.Q.; Cox, S.R.; Wardlaw, J.M.; Drake, A.J.; Bastin, M.E.; Boardman, J.P. DNA methylation and brain structure and function across the life course: A systematic review. Neurosci. Biobehav. Rev. 2020, 113, 133–156. [Google Scholar] [CrossRef]
- Barnes, J.; Mondelli, V.; Pariante, C.M. Genetic Contributions of Inflammation to Depression. Neuropsychopharmacology 2017, 42, 81–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stojanov, D.; Korf, J.; de Jonge, P.; Popov, G. The possibility of evidence-based psychiatry: Depression as a case. Clin. Epigenetics 2011, 2, 7–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamilton, M. A rating scale for depression. J. Neurol. Neurosurg. Psychiatry 1960, 23, 56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oldfield, R.C. The assessment and analysis of handedness: The Edinburgh inventory. Neuropsychologia 1971, 9, 97–113. [Google Scholar] [CrossRef]
- Tian, Y.; Morris, T.J.; Webster, A.P.; Yang, Z.; Beck, S.; Feber, A.; Teschendorff, A.E. ChAMP: Updated methylation analysis pipeline for Illumina BeadChips. Bioinformatics 2017, 33, 3982–3984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.P.; Triche, T.J., Jr.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Dale, A.M.; Fischl, B.; Sereno, M.I. Cortical surface-based analysis. I. Segmentation and surface reconstruction. Neuroimage 1999, 9, 179–194. [Google Scholar] [CrossRef]
- Fischl, B.; Liu, A.; Dale, A.M. Automated manifold surgery: Constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans. Med. Imaging 2001, 20, 70–80. [Google Scholar] [CrossRef] [Green Version]
- Fischl, B.; Salat, D.H.; Busa, E.; Albert, M.; Dieterich, M.; Haselgrove, C.; van der Kouwe, A.; Killiany, R.; Kennedy, D.; Klaveness, S.; et al. Whole brain segmentation: Automated labeling of neuroanatomical structures in the human brain. Neuron 2002, 33, 341–355. [Google Scholar] [CrossRef] [Green Version]
- Fischl, B.; Sereno, M.I.; Dale, A.M. Cortical surface-based analysis. II: Inflation, flattening, and a surface-based coordinate system. Neuroimage 1999, 9, 195–207. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischl, B.; van der Kouwe, A.; Destrieux, C.; Halgren, E.; Ségonne, F.; Salat, D.H.; Busa, E.; Seidman, L.J.; Goldstein, J.; Kennedy, D.; et al. Automatically parcellating the human cerebral cortex. Cereb. Cortex 2004, 14, 11–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ségonne, F.; Pacheco, J.; Fischl, B. Geometrically accurate topology-correction of cortical surfaces using nonseparating loops. IEEE Trans. Med. Imaging 2007, 26, 518–529. [Google Scholar] [CrossRef]
- Han, K.M.; Choi, S.; Jung, J.; Na, K.S.; Yoon, H.K.; Lee, M.S.; Ham, B.J. Cortical thickness, cortical and subcortical volume, and white matter integrity in patients with their first episode of major depression. J. Affect. Disord. 2014, 155, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Han, K.M.; Won, E.; Sim, Y.; Kang, J.; Han, C.; Kim, Y.K.; Kim, S.H.; Joe, S.H.; Lee, M.S.; Tae, W.S.; et al. Influence of FKBP5 polymorphism and DNA methylation on structural changes of the brain in major depressive disorder. Sci. Rep. 2017, 7, 42621. [Google Scholar] [CrossRef] [Green Version]
- Lim, J.E.; Kim, S.; Seo, S.; Kang, W.; Kim, A.; Kang, Y.; Choi, K.W.; Tae, W.S.; Ham, B.J.; Han, K.M. Association of Prefrontal Cortex Thinning with High Impulsivity in Healthy Adults. Psychiatry Investig. 2021, 18, 570–579. [Google Scholar] [CrossRef]
- Destrieux, C.; Fischl, B.; Dale, A.; Halgren, E. Automatic parcellation of human cortical gyri and sulci using standard anatomical nomenclature. Neuroimage 2010, 53, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Tae, W.S.; Kim, S.S.; Lee, K.U.; Nam, E.-C.; Kim, K.W. Validation of hippocampal volumes measured using a manual method and two automated methods (FreeSurfer and IBASPM) in chronic major depressive disorder. Neuroradiology 2008, 50, 569. [Google Scholar] [CrossRef]
| Characteristics | MDD (n = 88) | HC (n = 74) | p-Value (t, χ2) |
|---|---|---|---|
| Age | 44.38 ± 13.76 | 30.46 ± 12.06 | <0.001 (t = 6.858) |
| Sex (F/M) | 62/26 | 43/31 | 0.101 (χ2 = 2.687) |
| Education level | |||
| Elementary and middle school | 26 | 3 | <0.001 (χ2 = 17.792) |
| High school or college/university | 57 | 66 | |
| Above graduate school | 4 | 5 | |
| HDRS-17 score | 15.47 ± 6.83 | 1.15 ± 1.90 | <0.001 (t = 18.807) |
| Duration of illness (months) | 28.95 ± 45.16 | NA | NA |
| Drug-naïve/Medicated patients | 32/56 | NA | NA |
| Remitted/Non-remitted patients | 13/75 | NA | NA |
| Medication, n | |||
| SSRI | 22 | NA | NA |
| SNRI | 13 | ||
| NDRI | 3 | ||
| NaSSA | 4 | ||
| Other AD | 3 | ||
| Combination of ADs | 11 | ||
| AP | 12 | ||
| Combination of APs | 3 |
| CpG Site | Puncorr | Pcorr | Δβ | Chromosome | Position | Gene | Genomic Feature |
|---|---|---|---|---|---|---|---|
| cg06710101 | 0.002 | 0.015 | 0.007 | 1 | 247587253 | NLRP3 | Body |
| cg09418290 | 0.004 | 0.027 | 0.001 | 1 | 247579319 | NLRP3 | TSS200 |
| cg05615449 | 0.009 | 0.049 | −0.028 | 1 | 247601478 | NLRP3 | Body |
| cg18126557 | 0.023 | 0.091 | 0.012 | 1 | 247611842 | NLRP3 | 3′UTR |
| cg18793688 | 0.038 | 0.126 | −0.002 | 1 | 247588074 | NLRP3 | Body |
| Cortical Regions | MDD | HC | F | Puncorr | Pcorr | ||
|---|---|---|---|---|---|---|---|
| Mean | SD | Mean | SD | ||||
| MDD < HC | |||||||
| L Ventral posterior cingulate gyrus | 2.15 | 0.44 | 2.45 | 0.37 | 21.116 | 8.87 × 10−6 | 0.001 |
| L Middle occipital gyrus | 2.38 | 0.27 | 2.55 | 0.22 | 13.491 | 3.29 × 10−4 | 0.025 |
| L Lateral occipitotemporal gyrus | 2.40 | 0.48 | 2.77 | 0.30 | 33.403 | 3.95 × 10−8 | 3.01 × 10−6 |
| L Superior parietal lobule | 2.27 | 0.22 | 2.45 | 0.19 | 20.706 | 1.07 × 10−5 | 0.001 |
| L Planum polare | 2.96 | 0.50 | 3.33 | 0.39 | 25.519 | 1.21 × 10−6 | 9.20 × 10−5 |
| L Middle temporal gyrus | 2.67 | 0.45 | 2.96 | 0.35 | 20.095 | 1.42 × 10−5 | 0.001 |
| R Ventral posterior cingulate gyrus | 2.21 | 0.44 | 2.69 | 0.33 | 48.054 | 1.03 × 10−10 | 7.85 × 10−9 |
| R Short insular gyrus | 2.98 | 0.40 | 3.26 | 0.60 | 12.822 | 4.57 × 10−4 | 0.035 |
| R Middle occipital gyrus | 2.45 | 0.27 | 2.60 | 0.20 | 13.169 | 3.85 × 10−4 | 0.029 |
| R Lateral occipitotemporal gyrus | 2.44 | 0.41 | 2.81 | 0.28 | 35.612 | 1.56 × 10−8 | 1.19 × 10−6 |
| R Superior parietal lobule | 2.28 | 0.23 | 2.43 | 0.17 | 16.383 | 8.13 × 10−5 | 0.006 |
| R Precentral gyrus | 2.66 | 0.23 | 2.80 | 0.23 | 12.840 | 4.53 × 10−4 | 0.034 |
| R Subcallosal gyrus | 2.37 | 0.42 | 2.70 | 0.50 | 14.193 | 2.33 × 10−4 | 0.018 |
| R Planum polare | 2.87 | 0.44 | 3.16 | 0.38 | 15.408 | 1.30 × 10−4 | 0.010 |
| R Middle temporal gyrus | 2.67 | 0.43 | 2.92 | 0.30 | 17.928 | 3.91 × 10−5 | 0.003 |
| MDD > HC | |||||||
| L Paracentral lobule | 2.52 | 0.23 | 2.44 | 0.18 | 15.162 | 1.46 × 10−4 | 0.011 |
| L Posterior mid-cingulate gyrus | 2.68 | 0.19 | 2.56 | 0.23 | 23.786 | 2.63 × 10−6 | 2.00 × 10−4 |
| L Cuneus | 2.20 | 0.52 | 1.94 | 0.39 | 22.871 | 3.98 × 10−6 | 3.02 × 10−4 |
| L Superior occipital gyrus | 2.47 | 0.33 | 2.12 | 0.26 | 61.294 | 7.03 × 10−13 | 5.34 × 10−11 |
| L Lingual gyrus | 2.37 | 0.57 | 2.03 | 0.30 | 30.837 | 1.18 × 10−7 | 8.98 × 10−6 |
| L Postcentral gyrus | 2.43 | 0.34 | 2.22 | 0.19 | 33.786 | 3.36 × 10−8 | 2.56 × 10−6 |
| L Precuneus | 2.64 | 0.21 | 2.55 | 0.19 | 22.229 | 5.33 × 10−6 | 4.05 × 10−4 |
| L Lateral superior temporal gyrus | 3.18 | 0.30 | 3.00 | 0.30 | 22.420 | 4.88 × 10−6 | 3.71 × 10−4 |
| R Frontomarginal gyrus | 2.47 | 0.26 | 2.36 | 0.23 | 12.997 | 4.19 × 10−4 | 0.032 |
| R Posterior mid-cingulate gyrus | 2.71 | 0.19 | 2.59 | 0.19 | 24.728 | 1.72 × 10−6 | 1.31 × 10−4 |
| R Cuneus | 2.22 | 0.54 | 1.95 | 0.40 | 23.243 | 3.36 × 10−6 | 2.56 × 10−4 |
| R Superior occipital gyrus | 2.47 | 0.33 | 2.18 | 0.22 | 55.742 | 5.47 × 10−12 | 4.16 × 10−10 |
| R Lingual gyrus | 2.35 | 0.53 | 2.10 | 0.29 | 26.666 | 7.28 × 10−7 | 5.53 × 10−5 |
| R Postcentral gyrus | 2.42 | 0.37 | 2.22 | 0.20 | 27.926 | 4.18 × 10−7 | 3.18 × 10−5 |
| R Lateral superior temporal gyrus | 3.14 | 0.24 | 2.97 | 0.36 | 18.004 | 3.77 × 10−5 | 0.003 |
| R Planum temporale | 2.75 | 0.32 | 2.62 | 0.22 | 13.815 | 2.81 × 10−4 | 0.021 |
| Cortical Regions | cg18793688 | cg09418290 | ||||
|---|---|---|---|---|---|---|
| r | Puncorr | Pcorr | r | Puncorr | Pcorr | |
| L Cuneus * | −0.364 | 8.46 × 10−4 | 0.038 | 0.270 | 0.015 | 0.118 |
| L Superior frontal gyrus | −0.359 | 0.001 | 0.038 | 0.272 | 0.014 | 0.118 |
| L Superior occipital gyrus * | −0.276 | 0.013 | 0.118 | 0.369 | 6.87 × 10−4 | 0.037 |
| L Lingual gyrus * | −0.383 | 4.19 × 10−4 | 0.037 | 0.273 | 0.014 | 0.118 |
| R Cuneus * | −0.378 | 5.10 × 10−4 | 0.037 | 0.261 | 0.019 | 0.140 |
| R Superior frontal gyrus | −0.388 | 3.42 × 10−4 | 0.037 | 0.238 | 0.033 | 0.191 |
| R Lingual gyrus * | −0.447 | 2.94 × 10−5 | 0.011 | 0.289 | 0.009 | 0.103 |
| R Postcentral gyrus * | −0.394 | 2.72 × 10−4 | 0.037 | 0.282 | 0.011 | 0.110 |
| R Planum temporale * | −0.332 | 0.002 | 0.052 | 0.373 | 6.08 × 10−4 | 0.037 |
| R Middle temporal gyrus † | 0.362 | 9.07 × 10−4 | 0.038 | −0.254 | 0.022 | 0.151 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, K.-M.; Choi, K.W.; Kim, A.; Kang, W.; Kang, Y.; Tae, W.-S.; Han, M.-R.; Ham, B.-J. Association of DNA Methylation of the NLRP3 Gene with Changes in Cortical Thickness in Major Depressive Disorder. Int. J. Mol. Sci. 2022, 23, 5768. https://doi.org/10.3390/ijms23105768
Han K-M, Choi KW, Kim A, Kang W, Kang Y, Tae W-S, Han M-R, Ham B-J. Association of DNA Methylation of the NLRP3 Gene with Changes in Cortical Thickness in Major Depressive Disorder. International Journal of Molecular Sciences. 2022; 23(10):5768. https://doi.org/10.3390/ijms23105768
Chicago/Turabian StyleHan, Kyu-Man, Kwan Woo Choi, Aram Kim, Wooyoung Kang, Youbin Kang, Woo-Suk Tae, Mi-Ryung Han, and Byung-Joo Ham. 2022. "Association of DNA Methylation of the NLRP3 Gene with Changes in Cortical Thickness in Major Depressive Disorder" International Journal of Molecular Sciences 23, no. 10: 5768. https://doi.org/10.3390/ijms23105768
APA StyleHan, K.-M., Choi, K. W., Kim, A., Kang, W., Kang, Y., Tae, W.-S., Han, M.-R., & Ham, B.-J. (2022). Association of DNA Methylation of the NLRP3 Gene with Changes in Cortical Thickness in Major Depressive Disorder. International Journal of Molecular Sciences, 23(10), 5768. https://doi.org/10.3390/ijms23105768






