Amyloid Formation in Nanoliter Droplets
Abstract
:1. Introduction
2. Results
2.1. Microfluidic System Generating Nanoliter Droplets for Observation of Amyloid Formation
2.2. Size Modulation of Nanoliter Droplets in Microfluidic System
2.3. Fluorescence Assay for Observation of Amyloid Formation in Nanoliter Droplets
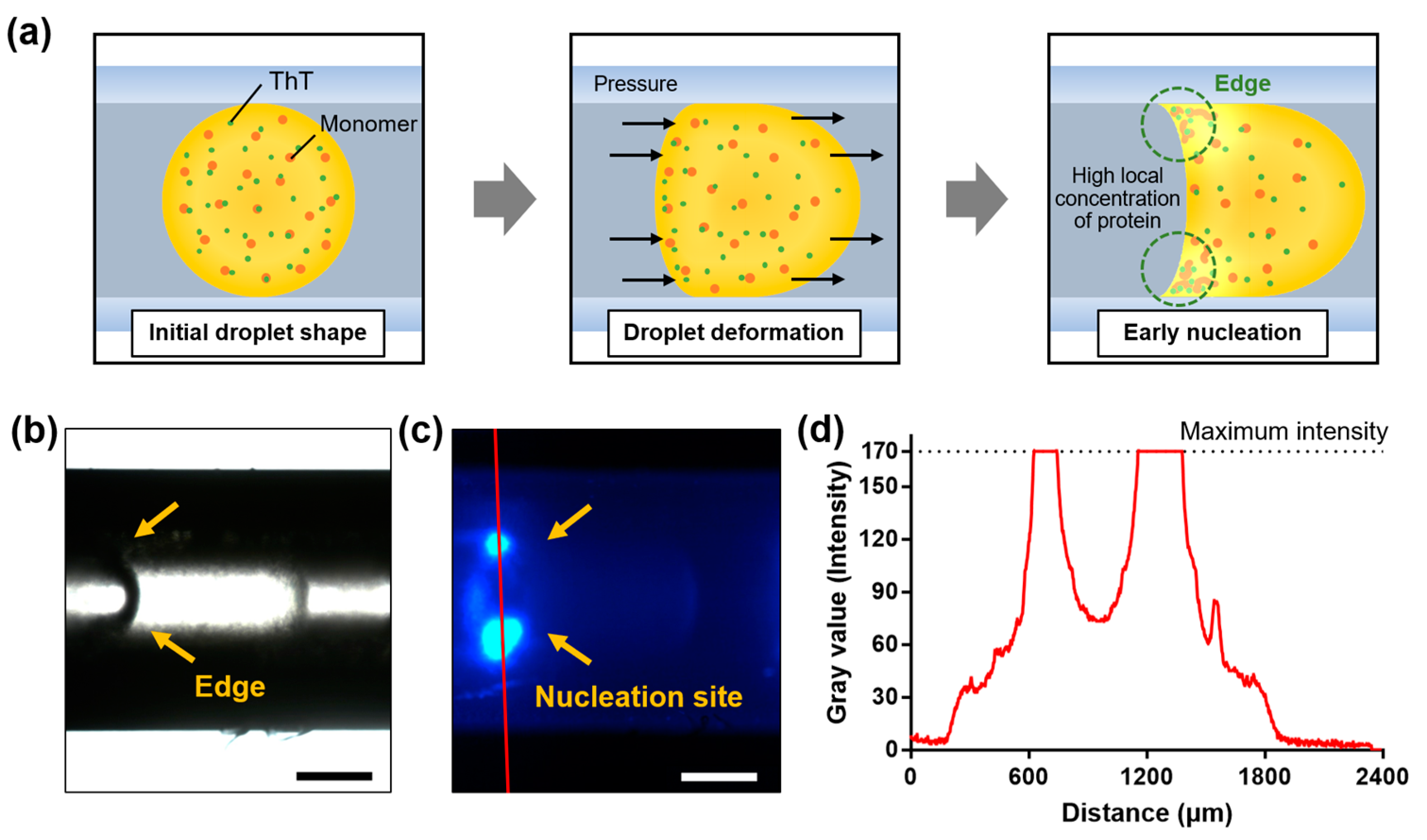
2.4. Rapid Amyloid Nucleation in Asymmetric Droplet
2.5. Polymorphism of HEWL Amyloid Fibrils Synthesized within Nanoliter Droplet
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Reagents and Materials
5.2. Preparation of HEWL Solution and HEWL Amyloid Fibrillation in Bulk Solution
5.3. Generation of Nanoliter Droplets
5.4. Fluorescence Assay for Detection of Amyloid Fibrillation in Nanoliter Droplets
5.5. AFM Imaging for Structural Analysis of HEWL Amyloid Fibrils
5.6. Statistical Analysis of AFM Images
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- McKee, A.C.; Stein, T.D.; Kiernan, P.T.; Alvarez, V.E. The neuropathology of chronic traumatic encephalopathy. Brain Pathol. 2015, 25, 350–364. [Google Scholar] [CrossRef] [Green Version]
- Caughey, B.; Lansbury, P.T., Jr. Protofibrils, pores, fibrils, and neurodegeneration: Separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003, 26, 267–298. [Google Scholar] [CrossRef] [PubMed]
- Selkoe, D.J. Folding proteins in fatal ways. Nature 2003, 426, 900–904. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, L.E.; Muffat, J.A.; Cherny, R.A.; Moir, R.D.; Ericsson, M.H.; Huang, X.; Mavros, C.; Coccia, J.A.; Faget, K.Y.; Fitch, K.A. Cytosolic β-amyloid deposition and supranuclear cataracts in lenses from people with Alzheimer’s disease. Lancet 2003, 361, 1258–1265. [Google Scholar] [CrossRef]
- Gracia, P.; Camino, J.D.; Volpicelli-Daley, L.; Cremades, N. Multiplicity of α-synuclein aggregated species and their possible roles in disease. Int. J. Mol. Sci. 2020, 21, 8043. [Google Scholar] [CrossRef]
- Adamcik, J.; Jung, J.-M.; Flakowski, J.; De Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Choi, Y.; Lee, S.W.; Kim, I.; Lee, D.; Hong, Y.; Lee, G.; Yoon, D.S. Microwave-induced formation of oligomeric amyloid aggregates. Nanotechnology 2018, 29, 345604. [Google Scholar] [CrossRef]
- Lee, G.; Lee, W.; Lee, H.; Lee, C.Y.; Eom, K.; Kwon, T. Self-assembled amyloid fibrils with controllable conformational heterogeneity. Sci. Rep. 2015, 5, 16220. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.W.; Choi, H.; Lee, G.; Choi, Y.; Lee, H.; Kim, G.; Lee, H.; Lee, W.; Park, J.; Yoon, D.S. Conformation Control of Amyloid Filaments by Repeated Thermal Perturbation. ACS Macro Lett. 2021, 10, 1549–1554. [Google Scholar] [CrossRef]
- Knowles, T.P.; White, D.A.; Abate, A.R.; Agresti, J.J.; Cohen, S.I.; Sperling, R.A.; De Genst, E.J.; Dobson, C.M.; Weitz, D.A. Observation of spatial propagation of amyloid assembly from single nuclei. Proc. Natl. Acad. Sci. USA 2011, 108, 14746–14751. [Google Scholar] [CrossRef] [Green Version]
- Courtney, M.; Chen, X.; Chan, S.; Mohamed, T.; Rao, P.P.; Ren, C.L. Droplet microfluidic system with on-demand trapping and releasing of droplet for drug screening applications. Anal. Chem. 2017, 89, 910–915. [Google Scholar] [CrossRef] [PubMed]
- Di Lorenzo, F.; Seiffert, S. Effect of Droplet Size in Acrylamide-Based Microgel Formation by Microfluidics. Macromol. React. Eng. 2016, 10, 201–205. [Google Scholar] [CrossRef]
- Biancalana, M.; Koide, S. Molecular mechanism of Thioflavin-T binding to amyloid fibrils. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2010, 1804, 1405–1412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ikenoue, T.; Lee, Y.-H.; Kardos, J.; Yagi, H.; Ikegami, T.; Naiki, H.; Goto, Y. Heat of supersaturation-limited amyloid burst directly monitored by isothermal titration calorimetry. Proc. Natl. Acad. Sci. USA 2014, 111, 6654–6659. [Google Scholar] [CrossRef] [Green Version]
- Jia, D.; Hamilton, J.; Zaman, L.M.; Goonewardene, A. The time, size, viscosity, and temperature dependence of the Brownian motion of polystyrene microspheres. Am. J. Phys. 2007, 75, 111–115. [Google Scholar] [CrossRef] [Green Version]
- Xue, C.; Lin, T.Y.; Chang, D.; Guo, Z. Thioflavin T as an amyloid dye: Fibril quantification, optimal concentration and effect on aggregation. R. Soc. Open Sci. 2017, 4, 160696. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, J.; Castro, F.; Rocha, F.; Kuhn, S. Protein crystallization in a droplet-based microfluidic device: Hydrodynamic analysis and study of the phase behaviour. Chem. Eng. Sci. 2018, 191, 232–244. [Google Scholar] [CrossRef]
- Maeki, M.; Teshima, Y.; Yoshizuka, S.; Yamaguchi, H.; Yamashita, K.; Miyazaki, M. Controlling Protein Crystal Nucleation by Droplet-Based Microfluidics. Chem.–A Eur. J. 2014, 20, 1049–1056. [Google Scholar] [CrossRef]
- Adamcik, J.; Mezzenga, R. Amyloid polymorphism in the protein folding and aggregation energy landscape. Angew. Chem. Int. Ed. 2018, 57, 8370–8382. [Google Scholar] [CrossRef]
- Zhou, J.; Ruggeri, F.S.; Zimmermann, M.R.; Meisl, G.; Longo, G.; Sekatskii, S.K.; Knowles, T.P.; Dietler, G. Effects of sedimentation, microgravity, hydrodynamic mixing and air–water interface on α-synuclein amyloid formation. Chem. Sci. 2020, 11, 3687–3693. [Google Scholar] [CrossRef] [Green Version]
- Malinowski, R.; Volpe, G.; Parkin, I.P.; Volpe, G. Dynamic control of particle deposition in evaporating droplets by an external point source of vapor. J. Phys. Chem. Lett. 2018, 9, 659–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, H.; Sanes, J.R. Neuronal cell-type classification: Challenges, opportunities and the path forward. Nat. Rev. Neurosci. 2017, 18, 530–546. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Gomez, D. Smooth muscle cell phenotypic diversity: At the crossroads of lineage tracing and single-cell transcriptomics. Arterioscler. Thromb. Vasc. Biol. 2019, 39, 1715–1723. [Google Scholar] [CrossRef]
- Choi, Y.J.; Chae, S.; Kim, J.H.; Barald, K.F.; Park, J.Y.; Lee, S.-H. Neurotoxic amyloid beta oligomeric assemblies recreated in microfluidic platform with interstitial level of slow flow. Sci. Rep. 2013, 3, 1921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, G.; Su, Z.; Reynolds, N.P.; Arosio, P.; Hamley, I.W.; Gazit, E.; Mezzenga, R. Self-assembling peptide and protein amyloids: From structure to tailored function in nanotechnology. Chem. Soc. Rev. 2017, 46, 4661–4708. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.; Sahoo, B.R.; Ozawa, D.; Kinoshita, M.; Kang, J.; Lim, M.H.; Okumura, M.; Huh, Y.H.; Moon, E.; Jang, J.H. Diverse structural conversion and aggregation pathways of Alzheimer’s amyloid-β (1–40). ACS Nano 2019, 13, 8766–8783. [Google Scholar] [CrossRef]
- Cao, A.; Hu, D.; Lai, L. Formation of amyloid fibrils from fully reduced hen egg white lysozyme. Protein Sci. 2004, 13, 319–324. [Google Scholar] [CrossRef]
- Van den Akker, C.C.; Engel, M.F.; Velikov, K.P.; Bonn, M.; Koenderink, G.H. Morphology and persistence length of amyloid fibrils are correlated to peptide molecular structure. J. Am. Chem. Soc. 2011, 133, 18030–18033. [Google Scholar] [CrossRef]
- Ray, S.; Singh, N.; Kumar, R.; Patel, K.; Pandey, S.; Datta, D.; Mahato, J.; Panigrahi, R.; Navalkar, A.; Mehra, S. α-Synuclein aggregation nucleates through liquid–liquid phase separation. Nat. Chem. 2020, 12, 705–716. [Google Scholar] [CrossRef]
- Törnquist, M.; Michaels, T.C.; Sanagavarapu, K.; Yang, X.; Meisl, G.; Cohen, S.I.; Knowles, T.P.; Linse, S. Secondary nucleation in amyloid formation. Chem. Commun. 2018, 54, 8667–8684. [Google Scholar] [CrossRef] [Green Version]
- Stein, K.C.; True, H.L. Prion strains and amyloid polymorphism influence phenotypic variation. PLoS Pathog. 2014, 10, e1004328. [Google Scholar] [CrossRef] [Green Version]
- Noji, M.; Sasahara, K.; Yamaguchi, K.; So, M.; Sakurai, K.; Kardos, J.; Naiki, H.; Goto, Y. Heating during agitation of β2-microglobulin reveals that supersaturation breakdown is required for amyloid fibril formation at neutral pH. J. Biol. Chem. 2019, 294, 15826–15835. [Google Scholar] [CrossRef] [PubMed]
- Ziaunys, M.; Sakalauskas, A.; Mikalauskaite, K.; Smirnovas, V. Polymorphism of Alpha-Synuclein Amyloid Fibrils Depends on Ionic Strength and Protein Concentration. Int. J. Mol. Sci. 2021, 22, 12382. [Google Scholar] [CrossRef] [PubMed]
- Fedunova, D.; Antosova, A.; Marek, J.; Vanik, V.; Demjen, E.; Bednarikova, Z.; Gazova, Z. Effect of 1-Ethyl-3-methylimidazolium Tetrafluoroborate and Acetate Ionic Liquids on Stability and Amyloid Aggregation of Lysozyme. Int. J. Mol. Sci. 2022, 23, 783. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Lee, S.W.; Lee, G.; Yoon, D.S. Atomic force microscopy analysis of EPPS-Driven degradation and reformation of amyloid-β aggregates. J. Alzheimer’s Dis. Rep. 2018, 2, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Lee, W.; Kim, I.; Lee, S.W.; Lee, H.; Lee, G.; Kim, S.; Lee, S.W.; Yoon, D.S. Quantifying L-ascorbic acid-driven inhibitory effect on amyloid fibrillation. Macromol. Res. 2016, 24, 868–873. [Google Scholar] [CrossRef]
- Guo, M.; Pegoraro, A.F.; Mao, A.; Zhou, E.H.; Arany, P.R.; Han, Y.; Burnette, D.T.; Jensen, M.H.; Kasza, K.E.; Moore, J.R. Cell volume change through water efflux impacts cell stiffness and stem cell fate. Proc. Natl. Acad. Sci. USA 2017, 114, E8618–E8627. [Google Scholar] [CrossRef] [Green Version]
- Müller, B.; Lang, S.; Dominietto, M.; Rudin, M.; Schulz, G.; Deyhle, H.; Germann, M.; Pfeiffer, F.; David, C.; Weitkamp, T. High-resolution tomographic imaging of microvessels. In Developments in X-ray Tomography VI; International Society for Optics and Photonics: San Diego, CA, USA, 2008; p. 70780B. [Google Scholar]
- Sharma, K.; Roth, B.J. A multiscale mechanical bidomain model of cardiac tissue with complex fiber geometry. In Proceedings of the 5th International Conference on Computational and Mathematical Biomedical Engineering, Pittsburgh, PA, USA, 10–12 April 2017. [Google Scholar]
- Merlini, G.; Bellotti, V. Molecular mechanisms of amyloidosis. N. Engl. J. Med. 2003, 349, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Andreasen, M.; Meisl, G.; Taylor, J.D.; Michaels, T.C.; Levin, A.; Otzen, D.E.; Chapman, M.R.; Dobson, C.M.; Matthews, S.J.; Knowles, T.P. Physical determinants of amyloid assembly in biofilm formation. MBio 2019, 10, e02279-18. [Google Scholar] [CrossRef] [Green Version]
- Lara, C.; Adamcik, J.; Jordens, S.; Mezzenga, R. General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 2011, 12, 1868–1875. [Google Scholar] [CrossRef]
- Lamour, G.; Kirkegaard, J.B.; Li, H.; Knowles, T.P.; Gsponer, J. Easyworm: An open-source software tool to determine the mechanical properties of worm-like chains. Source Code Biol. Med. 2014, 9, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheong, D.Y.; Lee, W.; Park, I.; Park, J.; Lee, G. Amyloid Formation in Nanoliter Droplets. Int. J. Mol. Sci. 2022, 23, 5480. https://doi.org/10.3390/ijms23105480
Cheong DY, Lee W, Park I, Park J, Lee G. Amyloid Formation in Nanoliter Droplets. International Journal of Molecular Sciences. 2022; 23(10):5480. https://doi.org/10.3390/ijms23105480
Chicago/Turabian StyleCheong, Da Yeon, Wonseok Lee, Insu Park, Jinsung Park, and Gyudo Lee. 2022. "Amyloid Formation in Nanoliter Droplets" International Journal of Molecular Sciences 23, no. 10: 5480. https://doi.org/10.3390/ijms23105480
APA StyleCheong, D. Y., Lee, W., Park, I., Park, J., & Lee, G. (2022). Amyloid Formation in Nanoliter Droplets. International Journal of Molecular Sciences, 23(10), 5480. https://doi.org/10.3390/ijms23105480








