Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary
Abstract
1. Introduction
2. Results
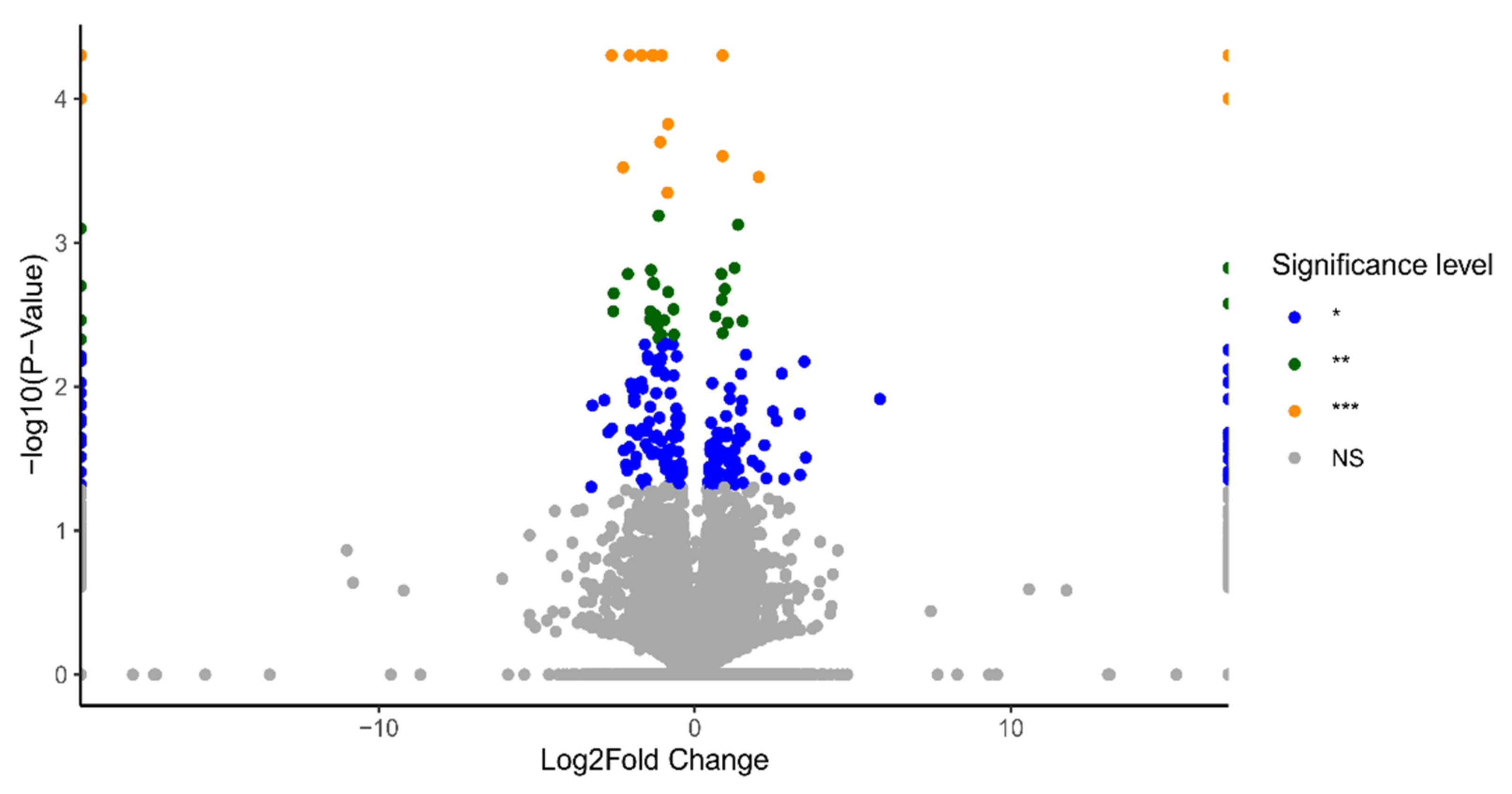
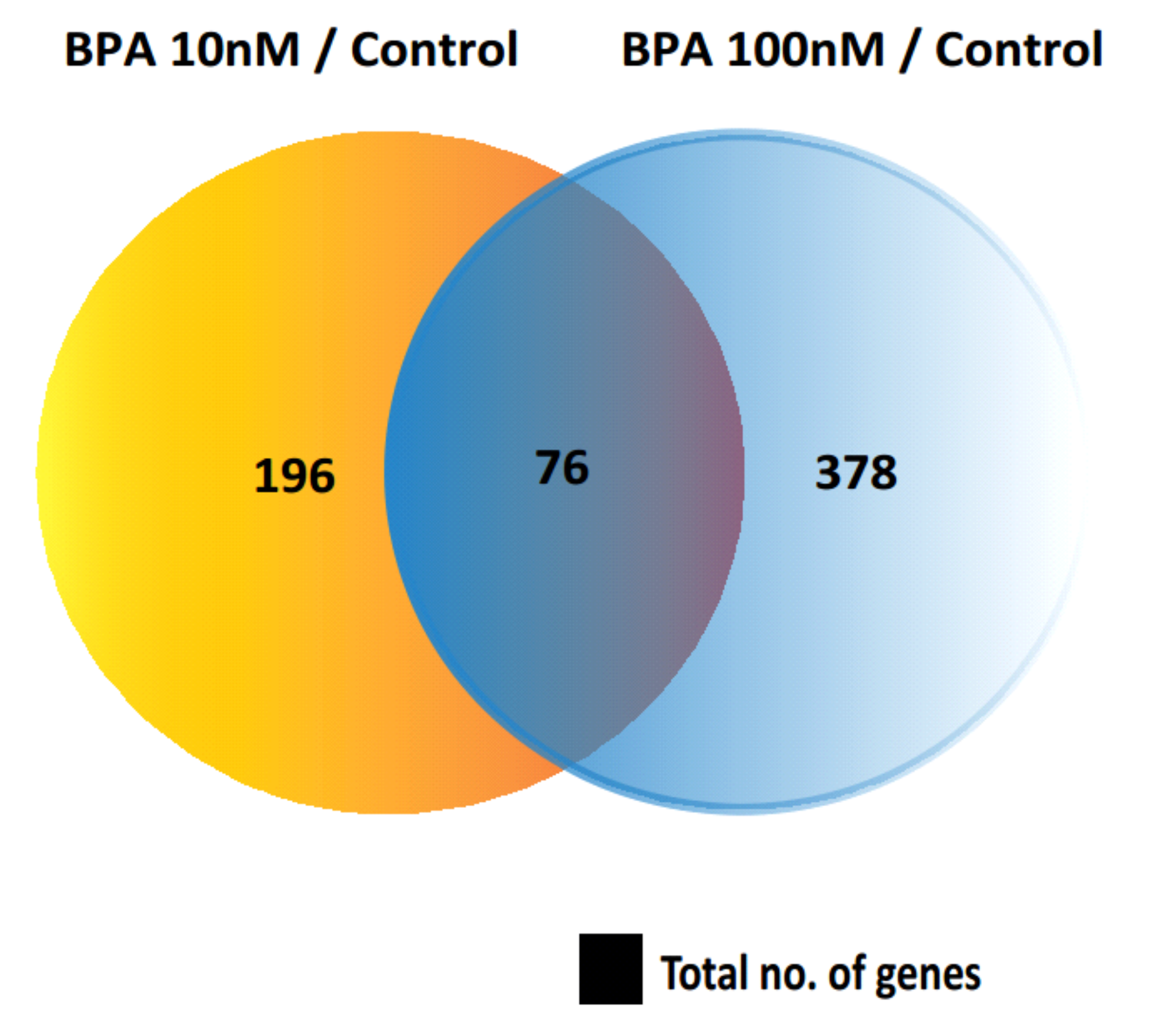
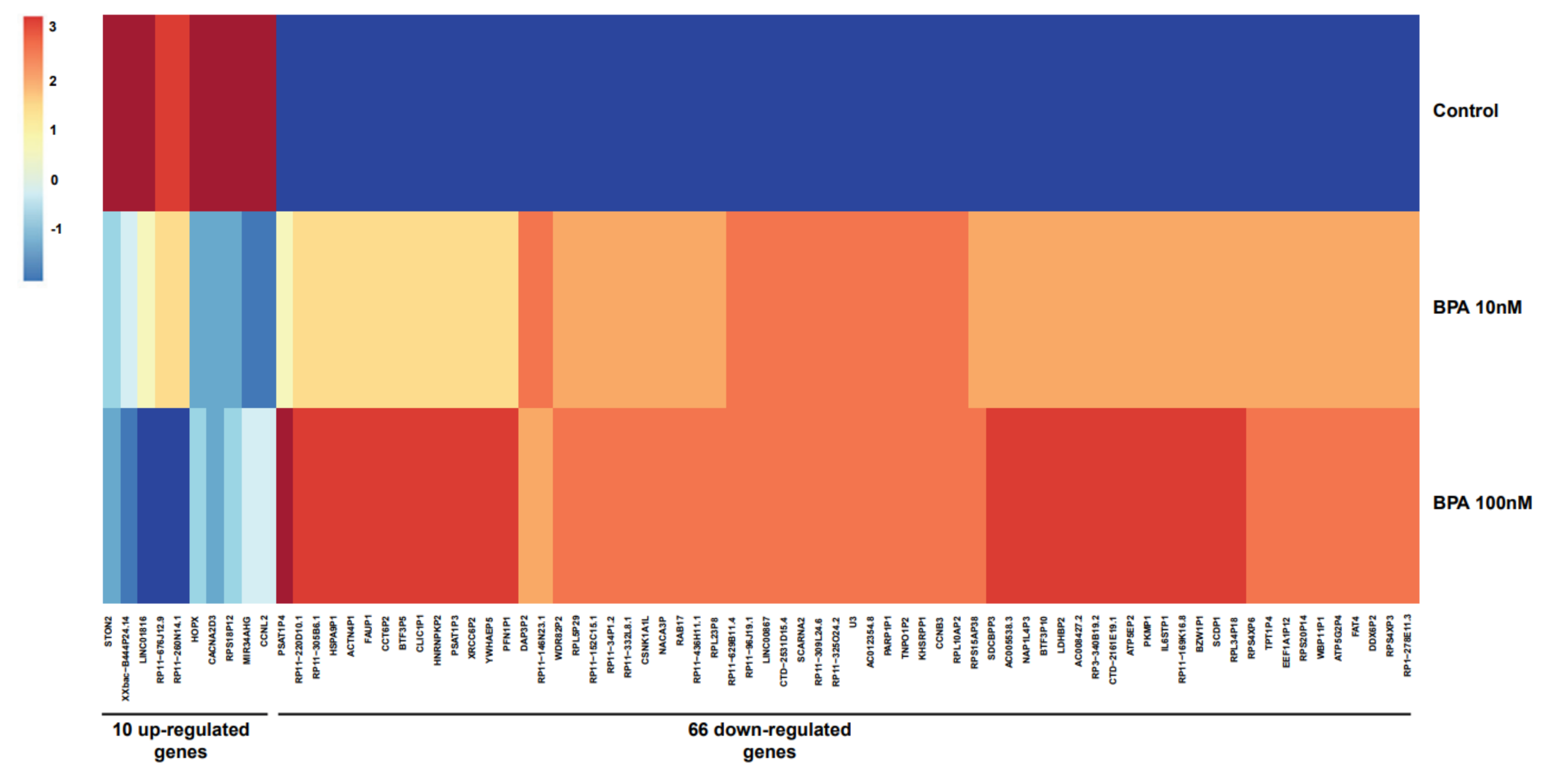
2.1. Identification of Differentially Expressed Genes (DEGs)
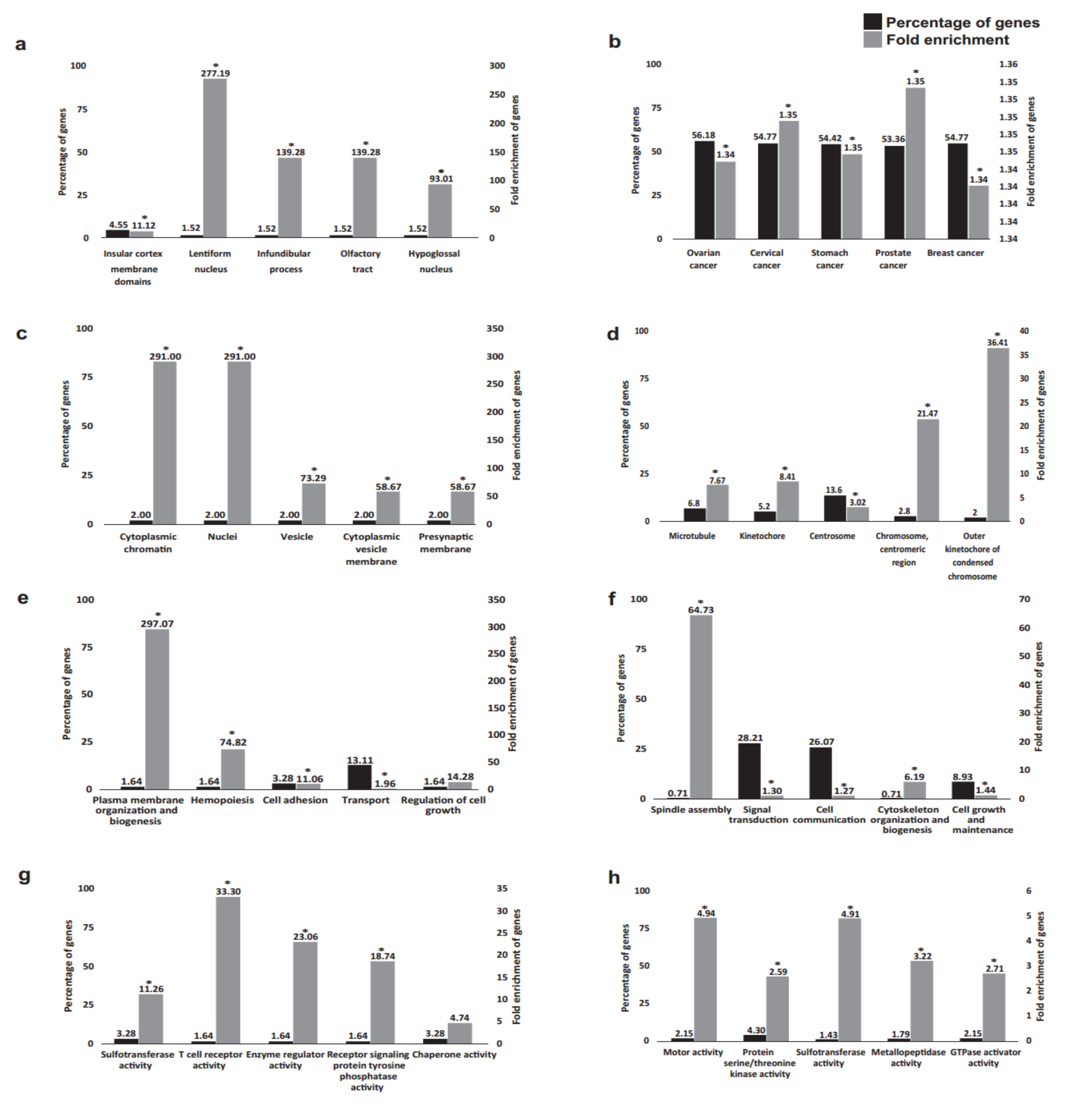
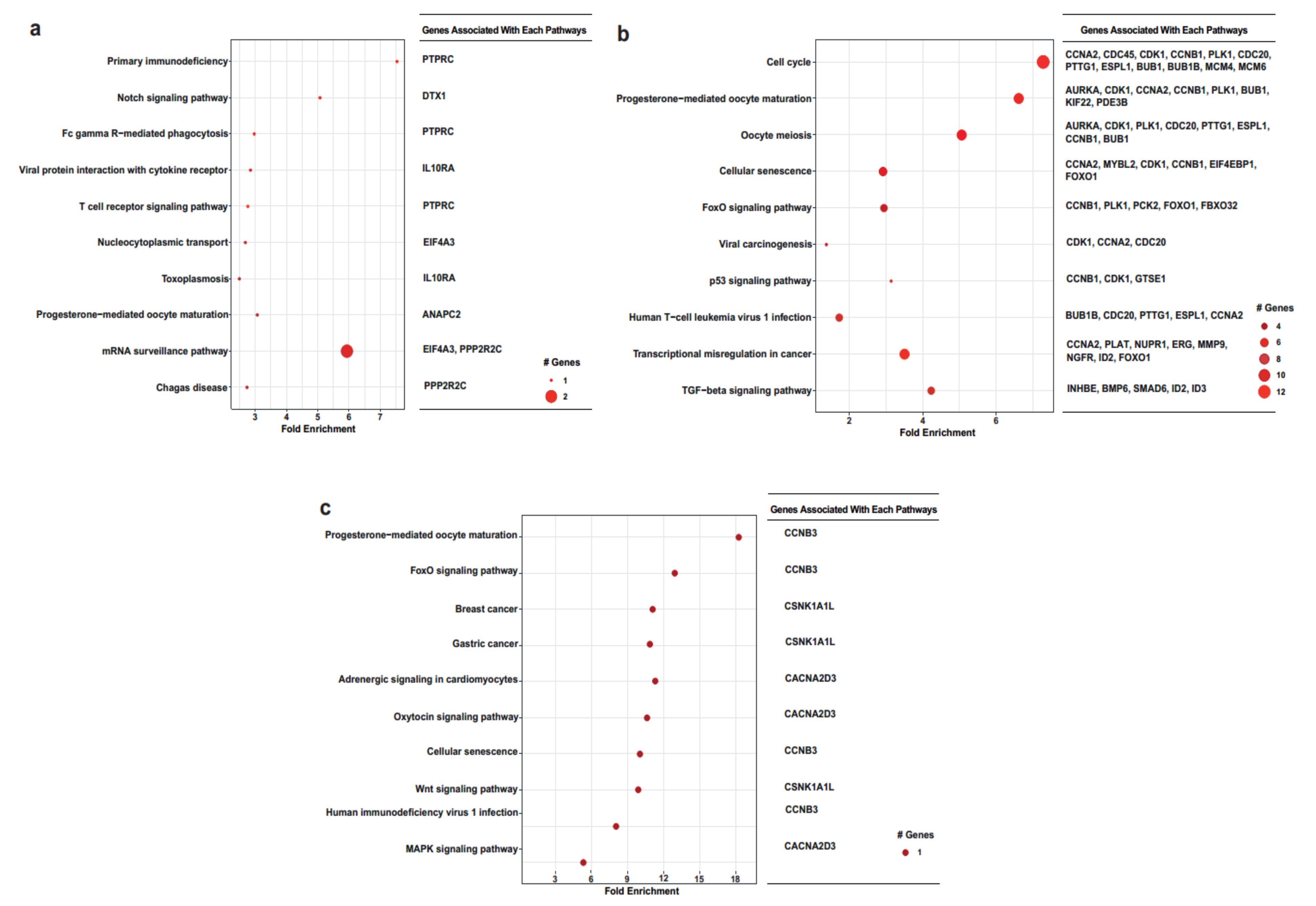
2.2. Functional Annotation Analysis of the DEGs
3. Discussion
4. Materials and Methods
4.1. Cell Culture
4.2. RNA Extraction
4.3. RNA-Sequencing (RNA-Seq), Data Generation
4.4. Statistical RNA-Sequencing Analysis
4.5. Functional Annotation
4.5.1. KEGG Pathway Database
4.5.2. Comparative Toxicogenomics Database (CTD)
4.5.3. Functional Analysis
4.5.4. The Gene Ontology Consortium
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Montes-Grajales, D.; Fennix-Agudelo, M.; Miranda-Castro, W. Occurrence of personal care products as emerging chemicals of concern in water resources: A review. Sci. Total Environ. 2017, 595, 601–614. [Google Scholar] [CrossRef] [PubMed]
- Endocrine-Disrupting Chemicals|Endocrine Society. Available online: https://www.endocrine.org/topics/edc (accessed on 7 March 2022).
- Lauretta, R.; Sansone, A.; Sansone, M.; Romanelli, F.; Appetecchia, M. Endocrine Disrupting Chemicals: Effects on Endocrine Glands. Front. Endocrinol. 2019, 10, 178. [Google Scholar] [CrossRef]
- Rubin, B.S. Bisphenol A: An endocrine disruptor with widespread exposure and multiple effects. J. Steroid Biochem. Mol. Biol. 2011, 127, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Jones, L.; Regan, F. Endocrine Disrupting Chemicals. In Encyclopedia of Analytical Science, 3rd ed.; Worsfold, P., Poole, C., Townshend, A., Miró, M., Eds.; Academic Press: Cambridge, MA, USA, 2019; pp. 31–38. ISBN 9780081019849. Available online: https://www.sciencedirect.com/science/article/pii/B9780124095472145123 (accessed on 27 March 2022). [CrossRef]
- Global Bisphenol A Market Report 2018: Analysis 2013–2017 & Forecasts 2018–2023. Available online: https://www.prnewswire.com/news-releases/global-bisphenol-a-market-report-2018-analysis-2013-2017--forecasts-2018-2023-300757673.html (accessed on 7 March 2022).
- Wang, Z.; Liu, H.; Liu, S. Low-Dose Bisphenol A Exposure: A Seemingly Instigating Carcinogenic Effect on Breast Cancer. Adv. Sci. 2017, 4, 1600248. [Google Scholar] [CrossRef] [PubMed]
- Alavian-Ghavanini, A.; Lin, P.I.; Lind, P.M.; Risén Rimfors, S.; Halin Lejonklou, M.; Dunder, L.; Tang, M.; Lindh, C.; Bornehag, C.-G.; Rüegg, J. Prenatal Bisphenol A Exposure is Linked to Epigenetic Changes in Glutamate Receptor Subunit Gene Grin2b in Female Rats and Humans. Sci. Rep. 2018, 8, 11315. [Google Scholar] [CrossRef]
- EFSA Panel on Food Contact Materials, Enzymes, Flavourings and Processing Aids (CEF). Scientific Opinion on the risks to public health related to the presence of bisphenol A (BPA) in foodstuffs. EFSA J. 2015, 13, 3978. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Colborn, T.; Hayes, T.B.; Heindel, J.J.; Jacobs, D.R., Jr.; Lee, D.H.; Myers, J.P.; Shioda, T.; Soto, A.M.; vom Saal, F.S.; et al. Regulatory decisions on endocrine disrupting chemicals should be based on the principles of endocrinology. Reprod. Toxicol. 2013, 38, 1–15. [Google Scholar] [CrossRef]
- Kortenkamp, A.; Scholze, M.; Ermler, S. Mind the gap: Can we explain declining male reproductive health with known antiandrogens? Reproduction 2014, 147, 515. [Google Scholar] [CrossRef][Green Version]
- Mendonca, K.; Hauser, R.; Calafat, A.M.; Arbuckle, T.E.; Duty, S.M. Bisphenol A concentrations in maternal breast milk and infant urine. Int. Arch. Occup. Environ. Health 2014, 87, 13. [Google Scholar] [CrossRef]
- Ottawa, C. Toxicological and Health Aspects of Bisphenol A Report of Joint FAO/WHO Expert Meeting and Report of Stakeholder Meeting on Bisphenol A Food and Agriculture Organization of the United Nations. Available online: www.who.int (accessed on 9 December 2020).
- Prusinski, L.; Al-Hendy, A.; Yang, Q. Developmental exposure to endocrine disrupting chemicals alters the epigenome: Identification of reprogrammed targets. Gynecol. Obstet. Res. Open J. 2016, 3, 1–6. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Maffini, M.V.; Sonnenschein, C.; Rubin, B.S.; Soto, A.M. Bisphenol-a and the great divide: A review of controversies in the field of endocrine disruption. Endocr. Rev. 2009, 30, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Rochester, J.R. Bisphenol A and human health: A review of the literature. Reprod. Toxicol. 2013, 42, 132–155. [Google Scholar] [CrossRef] [PubMed]
- Fuentes, N.; Silveyra, P. Estrogen receptor signaling mechanisms. Adv. Protein Chem. Struct. Biol. 2019, 116, 135. [Google Scholar] [CrossRef]
- Kim, M.-J.; Kim, T.-H.; Lee, H.-H. G-protein Coupled Estrogen Receptor (GPER/GPR30) and Women’s Health. J. Menopausal Med. 2015, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Silva, C.D.; Villegas-Pineda, J.C.; Pereira-Suárez, A.L. Expression and Role of the G Protein-Coupled Estrogen Receptor (GPR30/GPER) in the Development and Immune Response in Female Reproductive Cancers. Front. Endocrinol. 2020, 11, 544. [Google Scholar] [CrossRef]
- Hoffmann, M.; Rak, A.; Ptak, A. Bisphenol A and its derivatives decrease expression of chemerin, which reverses its stimulatory action in ovarian cancer cells. Toxicol. Lett. 2018, 291, 61–69. [Google Scholar] [CrossRef]
- Lin, H.; Li, H.; Lu, G.; Chen, Z.; Sun, W.; Shi, Y.; Fu, Z.; Huang, B.; Zhu, X.; Lu, W.; et al. Low dose of bisphenol a modulates ovarian cancer gene expression profile and promotes epithelial to mesenchymal transition via canonical wnt pathway. Toxicol. Sci. 2018, 164, 527–538. [Google Scholar] [CrossRef]
- Can, A.; Semiz, O.; Cinar, O. Bisphenol-A induces cell cycle delay and alters centrosome and spindle microtubular organization in oocytes during meiosis. Mol. Hum. Reprod. 2005, 11, 389–396. [Google Scholar] [CrossRef]
- Qiu, J.; Sun, Y.; Sun, W.; Wang, Y.; Fan, T.; Yu, J. Neonatal exposure to bisphenol A advances pubertal development in female rats. Mol. Reprod. Dev. 2020, 87, 503–511. [Google Scholar] [CrossRef]
- Machtinger, R.; Combelles, C.M.; Missmer, S.A.; Correia, K.F.; Williams, P.; Hauser, R.; Racowsky, C. Bisphenol-A and human oocyte maturation in vitro. Hum. Reprod. 2013, 28, 2735–2745. [Google Scholar] [CrossRef]
- Lin, M.; Hua, R.; Ma, J.; Zhou, Y.; Li, P.; Xu, X.; Yu, Z.; Quan, S. Bisphenol A promotes autophagy in ovarian granulosa cells by inducing AMPK/mTOR/ULK1 signalling pathway. Environ. Int. 2021, 147, 106298. [Google Scholar] [CrossRef] [PubMed]
- Molina, A.M.; Abril, N.; Lora, A.J.; Huertas-Abril, P.V.; Ayala, N.; Blanco, C.; Moyano, M.R. Proteomic profile of the effects of low-dose bisphenol A on zebrafish ovaries. Food Chem. Toxicol. 2021, 156, 112435. [Google Scholar] [CrossRef] [PubMed]
- Hung, P.H.; Van Winkle, L.S.; Williams, C.J.; Hunt, P.A.; VandeVoort, C.A. Prenatal Bisphenol A Exposure Alters Epithelial Cell Composition in the Rhesus Macaque Fetal Oviduct. Toxicol. Sci. 2019, 167, 450–457. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, R.A.M.; ElGhamrawy, T.A.; Salama, E.E.A. Effect of prenatal exposure to bisphenol a on the vagina of albino rats: Immunohistochemical and ultrastructural study. Folia Morphol. 2014, 73, 399–408. [Google Scholar] [CrossRef]
- Newbold, R.R.; Jefferson, W.N.; Padilla-Banks, E. Prenatal Exposure to Bisphenol A at environmentally relevant doses adversely affects the murine female reproductive tract later in life. Environ. Health Perspect. 2009, 117, 879–885. [Google Scholar] [CrossRef]
- Namat, A.; Xia, W.; Xiong, C.; Xu, S.; Wu, C.; Wang, A.; Li, Y.; Wu, Y.; Li, J. Association of BPA exposure during pregnancy with risk of preterm birth and changes in gestational age: A meta-analysis and systematic review. Ecotoxicol. Environ. Saf. 2021, 220, 112400. [Google Scholar] [CrossRef]
- De Aguiar Greca, S.C.; Kyrou, I.; Pink, R.; Randeva, H.; Grammatopoulos, D.; Silva, E.; Karteris, E. Involvement of the Endocrine-Disrupting Chemical Bisphenol A (BPA) in Human Placentation. J. Clin. Med. 2020, 9, 405. [Google Scholar] [CrossRef]
- Mei, L.; Chen, H.; Chen, F.; Feng, D.; Fang, F. Maintenance chemotherapy for ovarian cancer. Cochrane Database Syst. Rev. 2010, Volume 9, Page. [Google Scholar] [CrossRef]
- Dumitrascu, M.C.; Mares, C.; Petca, R.C.; Sandru, F.; Popescu, R.I.; Mehedintu, C.; Petca, A. Carcinogenic effects of bisphenol A in breast and ovarian cancers. Oncol. Lett. 2020, 20, 282. [Google Scholar] [CrossRef]
- Rodríguez, H.A.; Santambrosio, N.; Santamaría, C.G.; Muñoz-de-Toro, M.; Luque, E.H. Neonatal exposure to bisphenol A reduces the pool of primordial follicles in the rat ovary. Reprod. Toxicol. 2010, 30, 550–557. [Google Scholar] [CrossRef]
- Markey, C.M.; Coombs, M.A.; Sonnenschein, C.; Soto, A.M. Mammalian development in a changing environment: Exposure to endocrine disruptors reveals the developmental plasticity of steroid-hormone target organs. Evol. Dev. 2003, 5, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Zahra, A.; Dong, Q.; Hall, M.; Jeyaneethi, J.; Silva, E.; Karteris, E.; Sisu, C. Identification of Potential Bisphenol A (BPA) Exposure Biomarkers in Ovarian Cancer. J. Clin. Med. 2021, 10, 1979. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Feng, D.; Ding, H.; Zheng, X.; Ma, Z.; Yang, B.; Xie, M. Effects of bisphenol A exposure on DNA integrity and protamination of mouse spermatozoa. Andrology 2020, 8, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Lombó, M.; Fernández-Díez, C.; González-Rojo, S.; Herráez, M.P. Genetic and epigenetic alterations induced by bisphenol A exposure during different periods of spermatogenesis: From spermatozoa to the progeny. Sci. Rep. 2019, 9, 18029. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, S.; Keating, A.F. Bisphenol A-Induced Ovotoxicity Involves DNA Damage Induction to Which the Ovary Mounts a Protective Response Indicated by Increased Expression of Proteins Involved in DNA Repair and Xenobiotic Biotransformation. Toxicol. Sci. 2016, 152, 169–180. [Google Scholar] [CrossRef][Green Version]
- Musacchio, A.; Salmon, E.D. The spindle-assembly checkpoint in space and time. Nat. Rev. Mol. Cell Biol. 2007, 8, 379–393. [Google Scholar] [CrossRef]
- Kim, S.; Gwon, D.; Kim, J.A.; Choi, H.; Jang, C.Y. Bisphenol A disrupts mitotic progression via disturbing spindle attachment to kinetochore and centriole duplication in cancer cell lines. Toxicol. Vitr. 2019, 59, 115–125. [Google Scholar] [CrossRef]
- McGrogan, B.; Phelan, S.; Fitzpatrick, P.; Maguire, A.; Prencipe, M.; Brennan, D.; Doyle, E.; O’Grady, A.; Kay, E.; Furlong, F.; et al. Spindle assembly checkpoint protein expression correlates with cellular proliferation and shorter time to recurrence in ovarian cancer. Hum. Pathol. 2014, 45, 1509–1519. [Google Scholar] [CrossRef]
- Shah, K.; Al-Haidari, A.; Sun, J.; Kazi, J.U. T cell receptor (TCR) signaling in health and disease. Signal. Transduct. Target. Ther. 2021, 6, 412. [Google Scholar] [CrossRef]
- Yoshino, S.; Yamaki, K.; Li, X.; Sai, T.; Yanagisawa, R.; Takano, H.; Taneda, S.; Hayashi, H.; Mori, Y. Prenatal exposure to bisphenol A up-regulates immune responses, including T helper 1 and T helper 2 responses, in mice. Immunology 2004, 112, 489–495. [Google Scholar] [CrossRef]
- Alnouti, Y.; Klaassen, C.D. Tissue Distribution and Ontogeny of Sulfotransferase Enzymes in Mice. Toxicol. Sci. 2006, 93, 242–255. [Google Scholar] [CrossRef] [PubMed]
- Wolin, S.L.; Maquat, L.E. Cellular RNA Surveillance in Health and Disease. Science 2019, 366, 822. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Ren, C.; Yang, L. Effect of eukaryotic translation initiation factor 4A3 in malignant tumors. Oncol. Lett. 2021, 21, 358. [Google Scholar] [CrossRef] [PubMed]
- Wu, A.H.; Huang, Y.L.; Zhang, L.Z.; Tian, G.; Liao, Q.Z.; Chen, S.L. MiR-572 prompted cell proliferation of human ovarian cancer cells by suppressing PPP2R2C expression. Biomed. Pharmacother. 2016, 77, 92–97. [Google Scholar] [CrossRef]
- Adhikari, D.; Busayavalasa, K.; Zhang, J.; Hu, M.; Risal, S.; Bayazit, M.B.; Singh, M.; Diril, M.K.; Kaldis, P.; Liu, K. Inhibitory phosphorylation of Cdk1 mediates prolonged prophase I arrest in female germ cells and is essential for female reproductive lifespan. Cell Res. 2016, 26, 1212–1225. [Google Scholar] [CrossRef]
- Leland, S.; Nagarajan, P.; Polyzos, A.; Thomas, S.; Samaan, G.; Donnell, R.; Marchetti, F.; Venkatachalam, S. Heterozygosity for a Bub1 mutation causes female-specific germ cell aneuploidy in mice. Proc. Natl. Acad. Sci. USA 2009, 106, 12776. [Google Scholar] [CrossRef] [PubMed]
- Salehnia, M.; Zavareh, S. The Effects of Progesterone on Oocyte Maturationand Embryo Development. Int. J. Fertil. Steril. 2013, 7, 7. [Google Scholar]
- Zhang, Q.H.; Yuen, W.S.; Adhikari, D.; Flegg, J.A.; FitzHarris, G.; Conti, M.; Sicinski, P.; Nabti, I.; Marangos, P.; Carroll, J. Cyclin A2 modulates kinetochore-microtubule attachment in meiosis II. J. Cell Biol. 2017, 216, 3133–3143. [Google Scholar] [CrossRef]
- Li, J.; Qian, W.P.; Sun, Q.Y. Cyclins regulating oocyte meiotic cell cycle progression. Biol. Reprod. 2019, 101, 878–881. [Google Scholar] [CrossRef]
- Karasu, M.E.; Bouftas, N.; Keeney, S.; Wassmann, K. Cyclin B3 promotes anaphase I onset in oocyte meiosis. J. Cell Biol. 2019, 218, 1265. [Google Scholar] [CrossRef]
- Li, Y.; Wang, L.; Zhang, L.; He, Z.; Feng, G.; Sun, H.; Wang, J.; Li, Z.; Liu, C.; Han, J.; et al. Cyclin B3 is required for metaphase to anaphase transition in oocyte meiosis I. J. Cell Biol. 2019, 218, 1553. [Google Scholar] [CrossRef] [PubMed]
- Hunt, P.A.; Lawson, C.; Gieske, M.; Murdoch, B.; Smith, H.; Marre, A.; Hassold, T.; VandeVoort, C.A. Bisphenol A alters early oogenesis and follicle formation in the fetal ovary of the rhesus monkey. Proc. Natl. Acad. Sci. USA 2012, 109, 17525–17530. [Google Scholar] [CrossRef] [PubMed]
- Ptak, A.; Hoffmann, M.; Rak, A. The Ovary as a Target Organ for Bisphenol A Toxicity. In Bisphenol A Exposure and Health Risks; IntechOpen: London, UK, 2017. [Google Scholar] [CrossRef]
- Shelby, M.D. NTP-CERHR monograph on the potential human reproductive and developmental effects of bisphenol A. NTP CERHR MON 2008, 22, v–vii. [Google Scholar]
- Schecter, A.; Malik, N.; Haffner, D.; Smith, S.; Harris, T.R.; Paepke, O.; Birnbaum, L. Bisphenol A (BPA) in U.S. food. Environ. Sci. Technol. 2010, 44, 9425–9430. [Google Scholar] [CrossRef]
- Vandenberg, L.N.; Hauser, R.; Marcus, M.; Olea, N.; Welshons, W.V. Human exposure to bisphenol A (BPA). Reprod. Toxicol. 2007, 24, 139–177. [Google Scholar] [CrossRef]
- Kawa, I.A.; Fatima, Q.; Mir, S.A.; Jeelani, H.; Manzoor, S.; Rashid, F. Endocrine disrupting chemical Bisphenol A and its potential effects on female health. Diabetes Metab. Syndr. 2021, 15, 803–811. [Google Scholar] [CrossRef]
- Costa, J.; Mackay, R.; de Aguiar Greca, S.C.; Corti, A.; Silva, E.; Karteris, E.; Ahluwalia, A. The Role of the 3Rs for Understanding and Modeling the Human Placenta. J. Clin. Med. 2021, 10, 3444. [Google Scholar] [CrossRef]
- Ricciardelli, C.; Lokman, N.A.; Sabit, I.; Gunasegaran, K.; Bonner, W.M.; Pyragius, C.E.; Macpherson, A.M.; Oehler, M.K. Novel ex vivo ovarian cancer tissue explant assay for prediction of chemosensitivity and response to novel therapeutics. Cancer Lett. 2018, 421, 51–58. [Google Scholar] [CrossRef]
- Patisaul, H.B.; Fenton, S.E.; Aylor, D. Animal Models of Endocrine Disruption. Best Pract. Res. Clin. Endocrinol. Metab. 2018, 32, 283. [Google Scholar] [CrossRef]
- Rajapakse, N.; Silva, E.; Kortenkamp, A. Combining xenoestrogens at levels below individual no-observed-effect concentrations dramatically enhances steroid hormone action. Environ. Health Perspect. 2002, 110, 917. [Google Scholar] [CrossRef]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Pathan, M.; Keerthikumar, S.; Ang, C.S.; Gangoda, L.; Quek, C.Y.; Williamson, N.A.; Mouradov, D.; Sieber, O.M.; Simpson, R.J.; Salim, A.; et al. FunRich: An open access standalone functional enrichment and interaction network analysis tool. Proteomics 2015, 15, 2597–2601. [Google Scholar] [CrossRef] [PubMed]

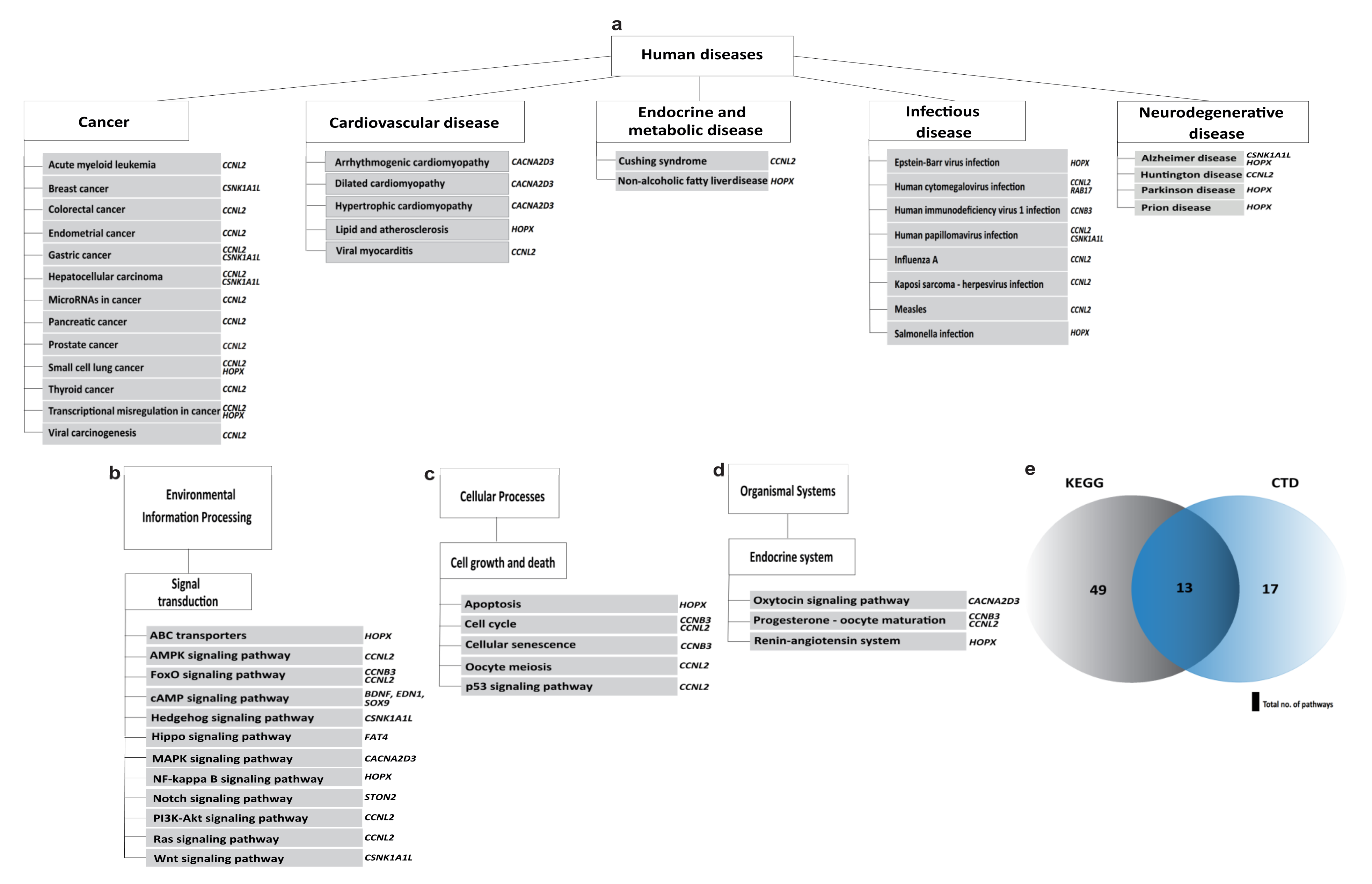
| Pathways | Associated Genes |
|---|---|
| Arrhythmogenic right ventricular cardiomyopathy | CACNA2D3 |
| Breast cancer | CSNK1A1L |
| Cell cycle | CCNB3, CCNL2 |
| Dilated cardiomyopathy | CACNA2D3 |
| FoxO signalling pathway | CCNB3, CCNL2 |
| Hedgehog signalling pathway | CSNK1A1L |
| Hippo signalling pathway | FAT4 |
| Hypertrophic cardiomyopathy (HCM) | CACNA2D3 |
| MAPK signalling pathway | CACNA2D3 |
| Oxytocin signalling pathway | CACNA2D3 |
| Progesterone-mediated oocyte maturation | CCNB3, CCNL2 |
| p53 signalling pathway | CCNB3, CCNL2 |
| Wnt signalling pathway | CSNK1A1L |
| Samples | Total Reads |
|---|---|
| Control | 75,835,336 |
| BPA 10 nM | 82,440,001 |
| BPA 100 nM | 65,361,410 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zahra, A.; Kerslake, R.; Kyrou, I.; Randeva, H.S.; Sisu, C.; Karteris, E. Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary. Int. J. Mol. Sci. 2022, 23, 5334. https://doi.org/10.3390/ijms23105334
Zahra A, Kerslake R, Kyrou I, Randeva HS, Sisu C, Karteris E. Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary. International Journal of Molecular Sciences. 2022; 23(10):5334. https://doi.org/10.3390/ijms23105334
Chicago/Turabian StyleZahra, Aeman, Rachel Kerslake, Ioannis Kyrou, Harpal S. Randeva, Cristina Sisu, and Emmanouil Karteris. 2022. "Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary" International Journal of Molecular Sciences 23, no. 10: 5334. https://doi.org/10.3390/ijms23105334
APA StyleZahra, A., Kerslake, R., Kyrou, I., Randeva, H. S., Sisu, C., & Karteris, E. (2022). Impact of Environmentally Relevant Concentrations of Bisphenol A (BPA) on the Gene Expression Profile in an In Vitro Model of the Normal Human Ovary. International Journal of Molecular Sciences, 23(10), 5334. https://doi.org/10.3390/ijms23105334








