Structural Insights into Iron Ions Accumulation in Dps Nanocage
Abstract
1. Introduction
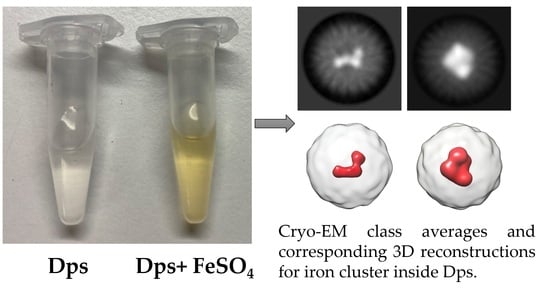
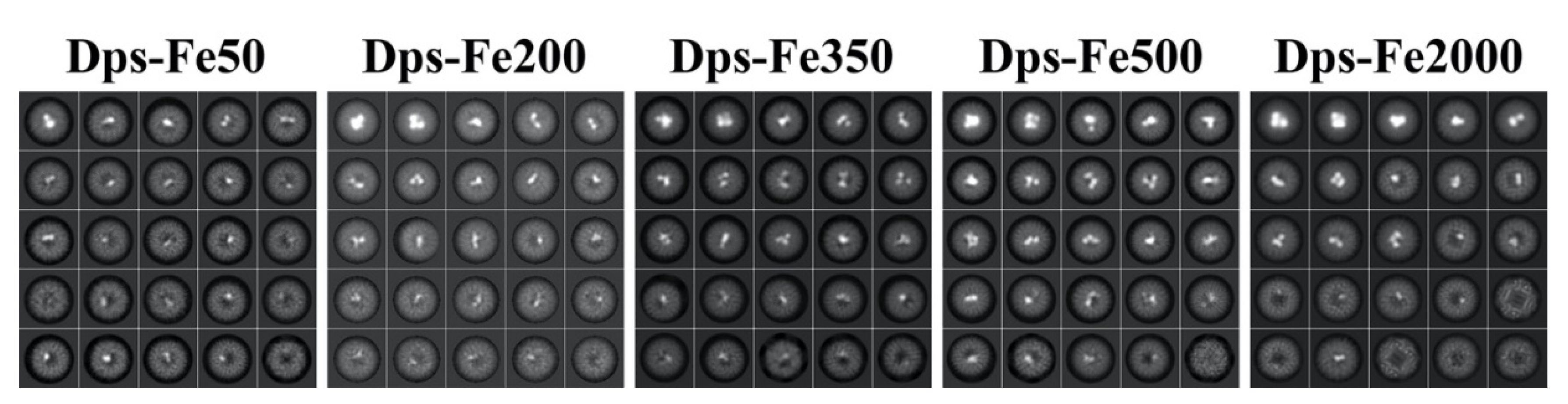
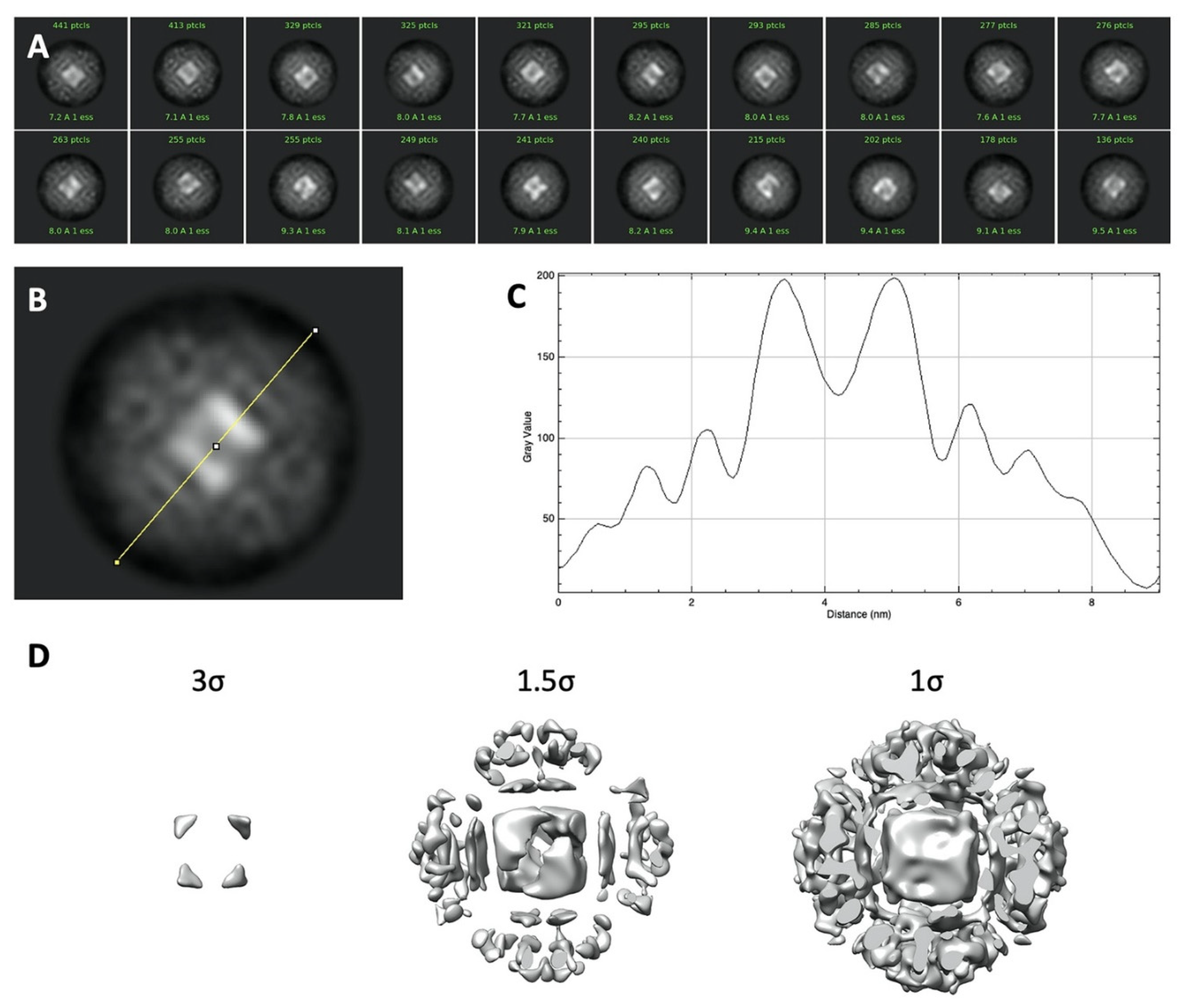
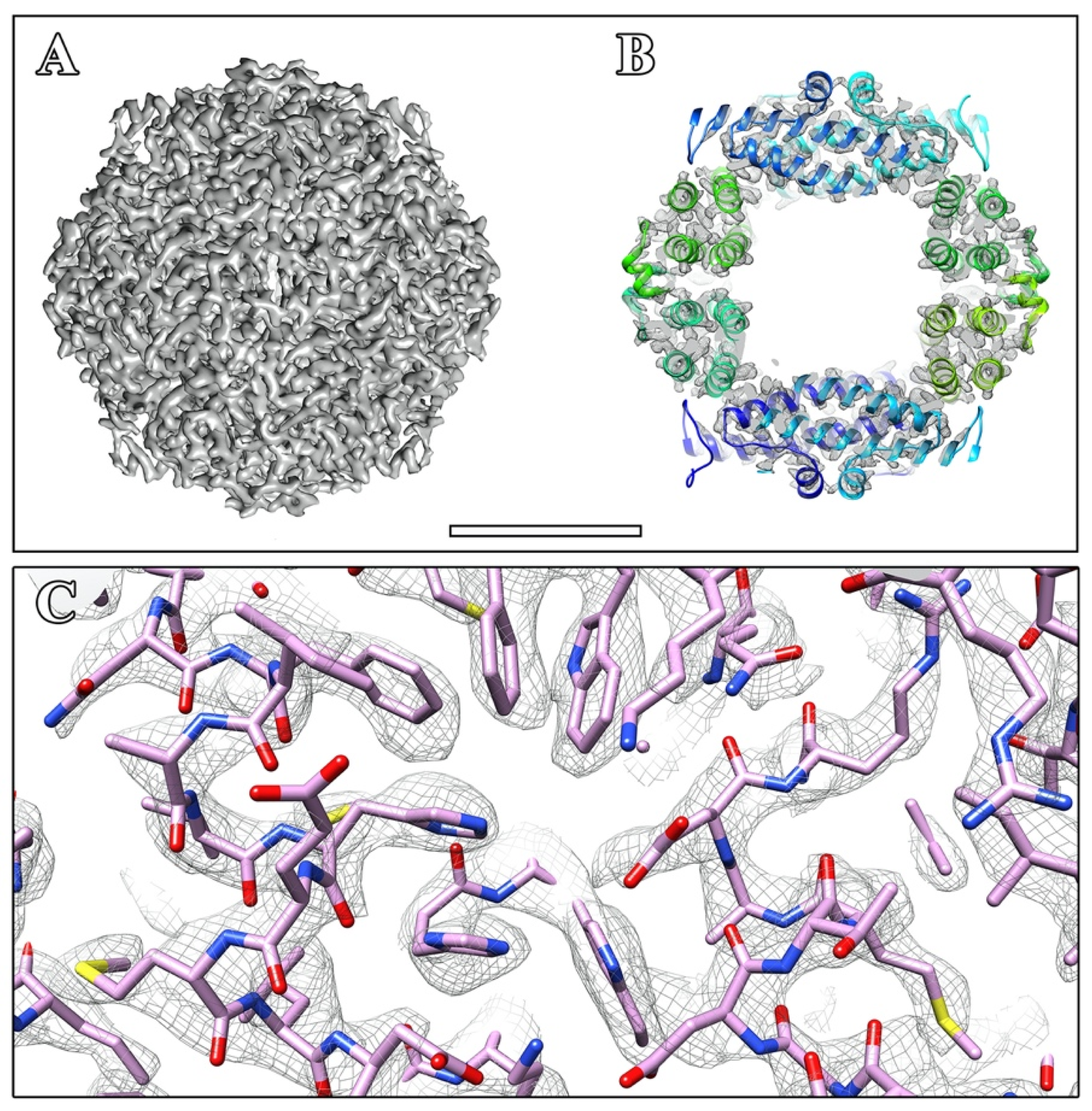
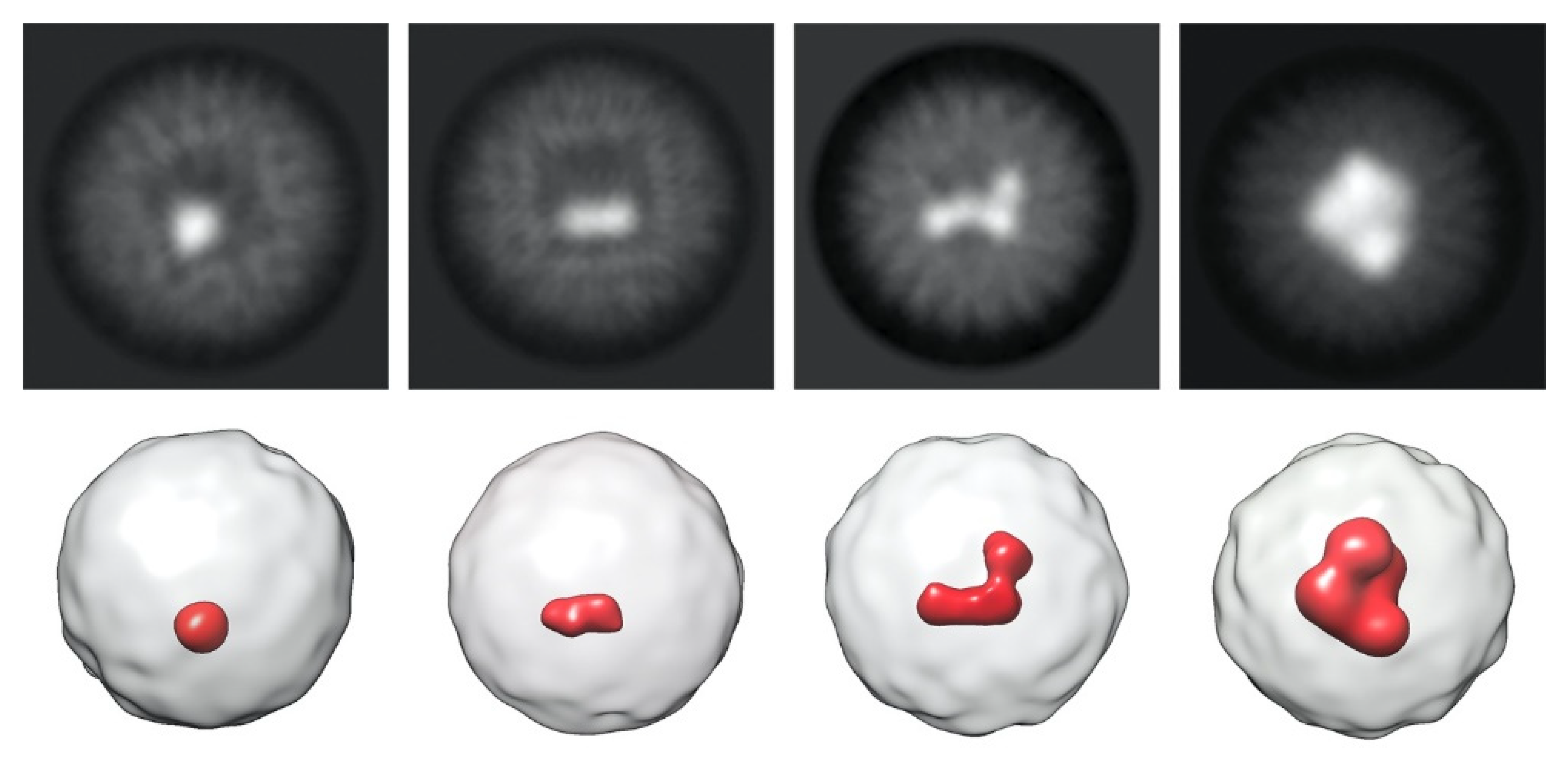
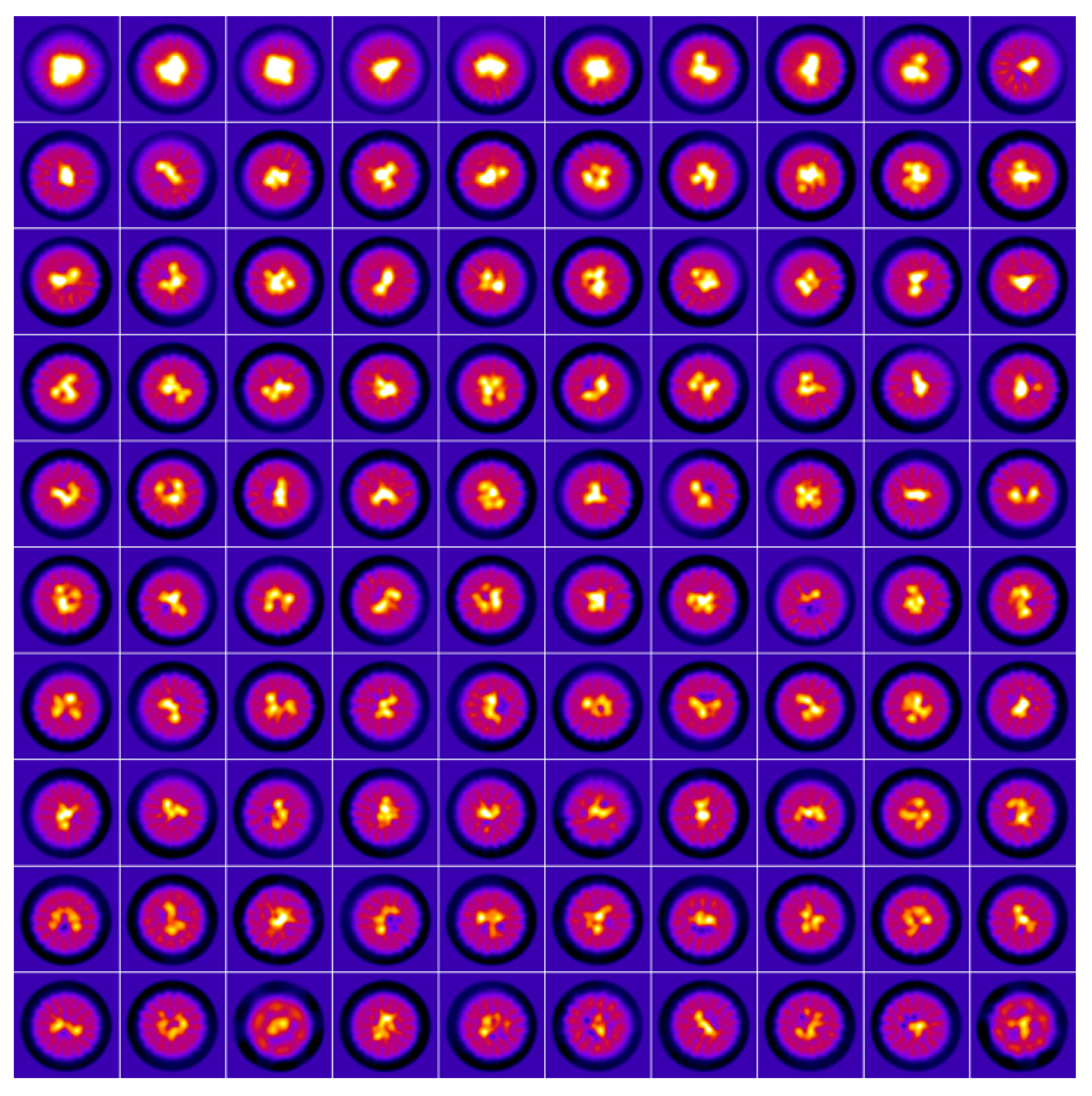
2. Results and Discussion
3. Materials and Methods
3.1. Preparation of Fe-Containing Dps Samples
3.2. Cryo-Electron Microscopy
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
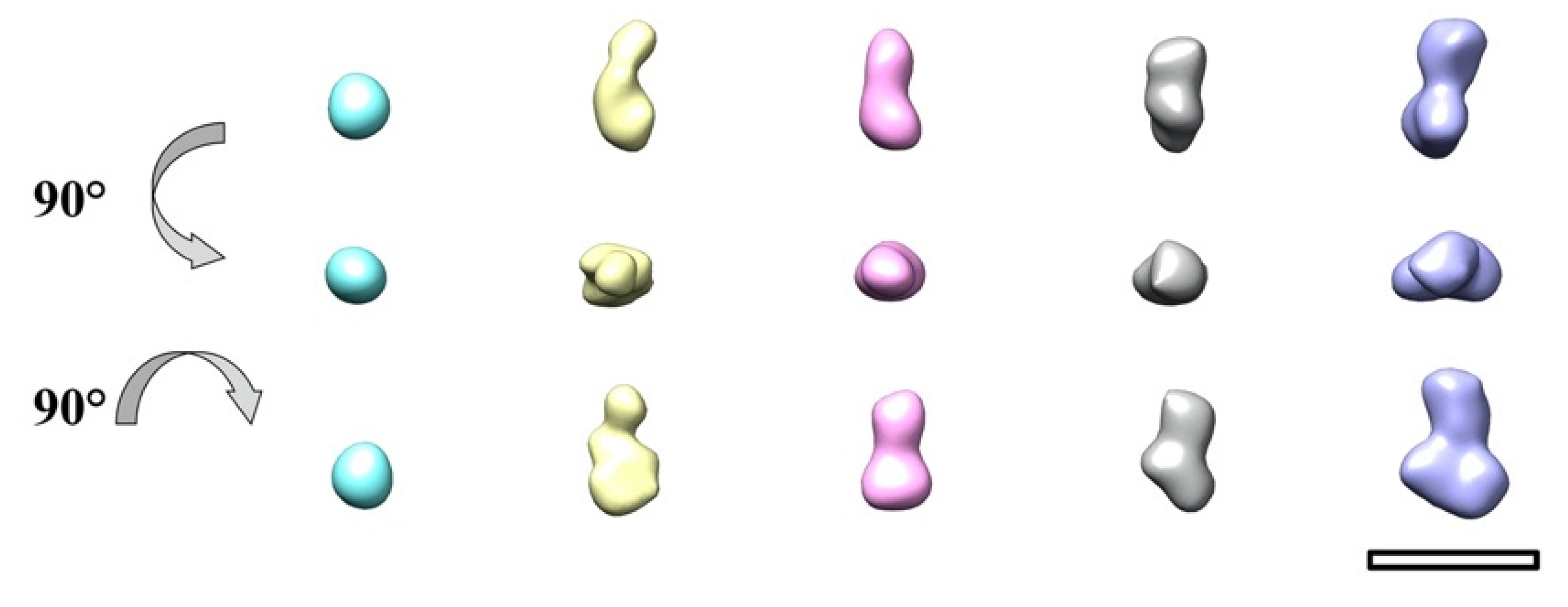
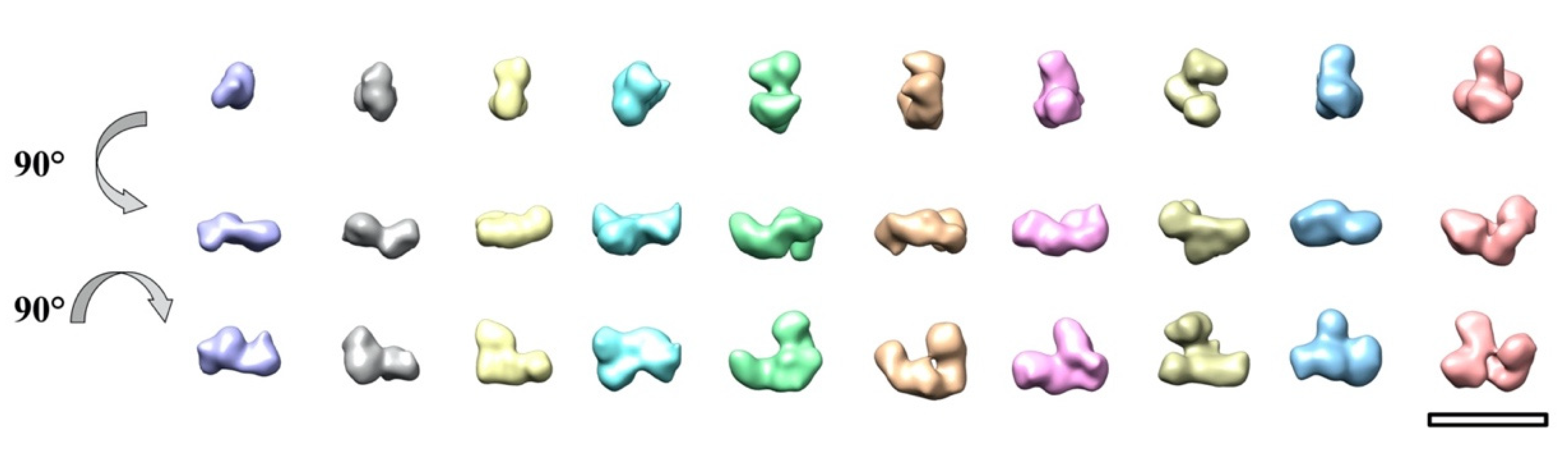
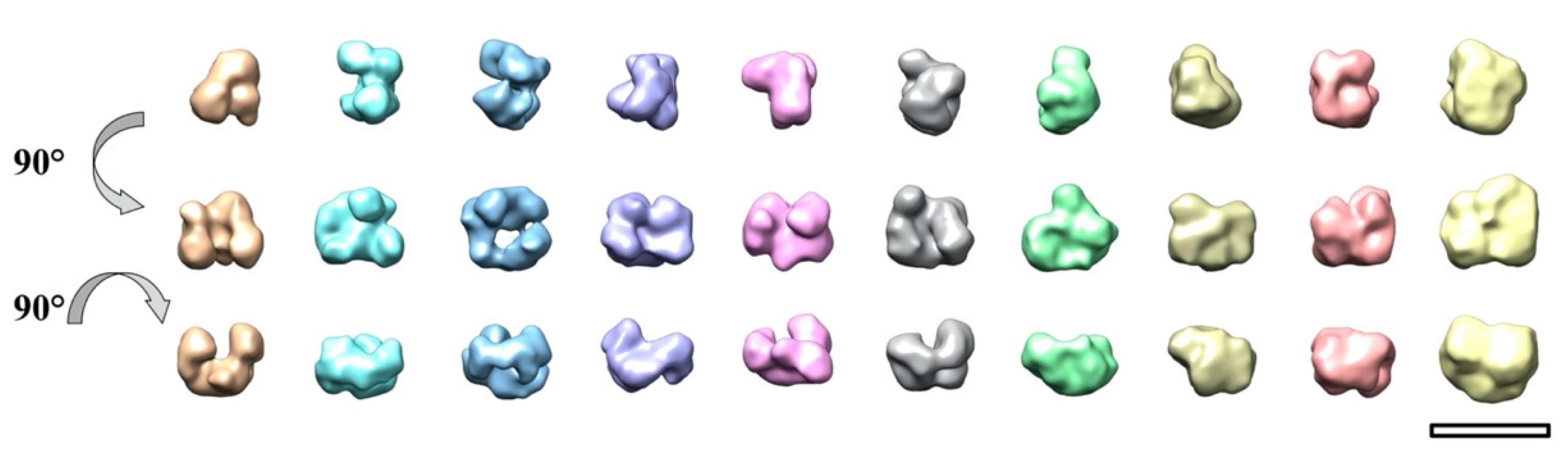
References
- Williams, S.M.; Chatterji, D. An Overview of Dps: Dual Acting Nanovehicles in Prokaryotes with DNA Binding and Ferroxidation Properties. In Subcellular Biochemistry; Subcellular Biochemistry; Harris, J.R., Marles-Wright, J., Eds.; Springer International Publishing: Cham, Switzerland, 2021; Volume 96, pp. 177–216. ISBN 978-3-030-28150-2. [Google Scholar]
- Antipov, S.; Turishchev, S.; Purtov, Y.; Shvyreva, U.; Sinelnikov, A.; Semov, Y.; Preobrazhenskaya, E.; Berezhnoy, A.; Shusharina, N.; Novolokina, N.; et al. The Oligomeric Form of the Escherichia coli Dps Protein Depends on the Availability of Iron Ions. Molecules 2017, 22, 1904. [Google Scholar] [CrossRef] [PubMed]
- Almirón, M.; Link, A.J.; Furlong, D.; Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 1992, 6, 2646–2654. [Google Scholar] [CrossRef]
- Minato, T.; Teramoto, T.; Kakuta, Y.; Ogo, S.; Yoon, K.S. Biochemical and structural characterization of a thermostable Dps protein with His-type ferroxidase centers and outer metal-binding sites. FEBS Open Bio. 2020, 10, 1219–1229. [Google Scholar] [CrossRef] [PubMed]
- Stefanini, S.; Ceci, P.; Ardini, M.; Ilari, A. DNA-Binding Proteins from Starved Cells (Dps Proteins). In Handbook of Metalloproteins; Messerschmidt, A., Ed.; John Wiley & Sons: Hoboken, NJ, USA, 2010; pp. 1–10. [Google Scholar]
- Guerra, J.P.L.; Jacinto, J.P.; Tavares, P. Miniferritins: Small multifunctional protein cages. Coord. Chem. Rev. 2021, 449, 214187. [Google Scholar] [CrossRef]
- Bellapadrona, G.; Ardini, M.; Ceci, P.; Stefanini, S.; Chiancone, E. Dps proteins prevent Fenton-mediated oxidative damage by trapping hydroxyl radicals within the protein shell. Free Radic. Biol. Med. 2010, 48, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Andrews, S.C.; Robinson, A.K.; Rodríguez-Quiñones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef]
- Chiancone, E.; Ceci, P.; Ilari, A.; Ribacchi, F.; Stefanini, S. Iron and proteins for iron storage and detoxification. BioMetals 2004, 17, 197–202. [Google Scholar] [CrossRef]
- Theil, E.C. Ferritin: The protein nanocage and iron biomineral in health and in disease. Inorg. Chem. 2013, 52, 12223–12233. [Google Scholar] [CrossRef]
- Gálvez, N.; Fernández, B.; Sánchez, P.; Cuesta, R.; Ceolín, M.; Clemente-León, M.; Domínguez-Vera, J.M. Comparative Structural and Chemical Studies of Ferritin Cores with Gradual Removal of their Iron Contents. J. Am. Chem. Soc. 2008, 130, 8062–8068. [Google Scholar] [CrossRef]
- Grant, R.; Filman, D.; Finkel, S.; Kolter, R.; Hogle, J.M. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat. Struct. Mol. Biol. 1998, 5, 294–303. [Google Scholar] [CrossRef]
- Lomovskaya, O.L.; Kidwell, J.P.; Matin, A. Characterization of the σ38-dependent expression of a core Escherichia coli starvation gene, pexB. J. Bacteriol. 1994, 176, 3928–3935. [Google Scholar] [CrossRef][Green Version]
- Ilari, A.; Ceci, P.; Ferrari, D.; Rossi, G.L.; Chiancone, E. Iron incorporation into Escherichia coli Dps gives rise to a ferritin-like microcrystalline core. J. Biol. Chem. 2002, 277, 37619–37623. [Google Scholar] [CrossRef]
- Alaleona, F.; Franceschini, S.; Ceci, P.; Ilari, A.; Chiancone, E. Thermosynechoccus elongatus DpsA binds Zn(II) at a unique three histidine-containing ferroxidase center and utilizes O2 as iron oxidant with very high efficiency, unlike the typical Dps proteins. FEBS J. 2010, 277, 903–917. [Google Scholar] [CrossRef]
- Howe, C.; Moparthi, V.K.; Ho, F.M.; Persson, K.; Stensjco, K. The Dps4 from Nostoc punctiforme ATCC 29133 is a member of His-type FOC containing Dps protein class that can be broadly found among cyanobacteria. PLoS ONE 2019, 14, e0218300. [Google Scholar] [CrossRef]
- Zeth, K.; Offermann, S.; Essen, L.O.; Oesterhelt, D. Iron-oxo clusters biomineralizing on protein surfaces: Structural analysis of Halobacterium salinarum DpsA in its low-and high-iron states. Proc. Natl. Acad. Sci. USA 2004, 101, 13780–13785. [Google Scholar] [CrossRef]
- Romao, C.V.; Mitchell, E.P.; McSweeney, S. The crystal structure of Deinococcus radiodurans Dps protein (DR2263) reveals the presence of a novel metal centre in the N terminus. J. Biol. Inorg. Chem. 2006, 11, 891–902. [Google Scholar] [CrossRef]
- Cuypers, M.G.; Mitchell, E.P.; Romao, C.V.; McSweeney, S. The crystal structure of the Dps2 from Deinococcus radiodurans reveals an unusual pore profile with a non-specific metal binding site. J. Mol. Biol. 2007, 371, 787–799. [Google Scholar] [CrossRef]
- Kim, S.G.; Bhattacharyya, G.; Grove, A.; Lee, Y.H. Crystal structure of Dps-1, a functionally distinct Dps protein from Deinococcus radiodurans. J. Mol. Biol. 2006, 361, 105–114. [Google Scholar] [CrossRef]
- Bellapadrona, G.; Chiaraluce, R.; Consalvi, V.; Ilari, A.; Stefanini, S.; Chiancone, E. The mutations Lys 114-Gln and Asp 126-Asn disrupt an intersubunit salt bridge and convert Listeria innocua Dps into its natural mutant Listeria monocytogenes Dps. Effects on protein stability at low pH. Proteins Struct. Funct. Bioinf. 2007, 66, 975–983. [Google Scholar] [CrossRef]
- Franceschini, S.; Ceci, P.; Alaleona, F.; Chiancone, E.; Ilari, A. Antioxidant Dps protein from the thermophilic cyanobacterium Thermosynechococcus elongatus: An intrinsically stable cage-like structure endowed with enhanced stability. FEBS J. 2006, 273, 4913–4928. [Google Scholar] [CrossRef]
- Ceci, P.; Ilari, A.; Falvo, E.; Giangiacomo, L.; Chiancone, E. Reassessment of protein stability, DNA binding, and protection of Mycobacterium smegmatis Dps. J. Biol. Chem. 2005, 280, 34776–34785. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Saraswathi, R.; Chatterji, D.; Vijayan, M. Structural studies on the second Mycobacterium smegmatis Dps: Invariant and variable features of structure, assembly and function. J. Mol. Biol. 2008, 375, 948–959. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.M.; Chatterji, D. Flexible aspartates propel iron to the ferroxidation sites along pathways stabilized by a conserved arginine in Dps proteins from Mycobacterium smegmatis. Metallomics 2007, 9, 685–698. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.; Gupta, S.; Das, S.; Sekar, K.; Chatterji, D.; Vijayan, M. X-ray analysis of Mycobacterium smegmatis Dps and a comparative study involving other Dps and Dps-like molecules. J. Mol. Biol. 2004, 339, 1103–1113. [Google Scholar] [CrossRef]
- Hitchings, M.D.; Townsend, P.; Pohl, E.; Facey, P.D.; Jones, D.H.; Dyson, P.J.; Sol, R.D. A tale of tails: Deciphering the contribution of terminal tails to the biochemical properties of two Dps proteins from Streptomycescoelicolor. Cell Mol. Life Sci. 2014, 71, 4911–4926. [Google Scholar] [CrossRef]
- Zeth, K.; Sancho-Vaello, E.; Okuda, M. Metal positions and translocation pathways of the dodecameric ferritin-like protein Dps. Inorg. Chem. 2019, 58, 11351–11363. [Google Scholar] [CrossRef]
- Stillman, T.J.; Upadhyay, M.; Norte, V.A.; Sedelnikova, S.E.; Carradus, M.; Tzokov, S.; Bullough, P.A.; Shearman, C.A.; Gasson, M.J.; Williams, C.H. The crystal structures of Lactococcus lactis MG1363 Dps proteins reveal the presence of an N-terminal helix that is required for DNA binding. Mol. Microbiol. 2005, 57, 1101–1112. [Google Scholar] [CrossRef]
- Pesek, J.; Buchler, R.; Albrecht, R.; Boland, W.; Zeth, K. Structure and mechanism of iron translocation by a Dps protein from Microbacterium arborescens. J. Biol. Chem. 2011, 286, 34872–34882. [Google Scholar] [CrossRef]
- Ardini, M.; Fiorillo, A.; Fittipaldi, M.; Stefanini, S.; Gatteschi, D.; Ilari, A.; Chiancone, E. Kineococcus radiotolerans Dps forms a heteronuclear Mn–Fe ferroxidase center that may explain the Mn dependent protection against oxidative stress. Bichem. Biophys. Acta. 2013, 1830, 3745–3755. [Google Scholar] [CrossRef]
- Ceci, P.; Ilari, A.; Falvo, E.; Chiancone, E. The Dps protein of Agrobacterium tumefaciens does not bind to DNA but protects it toward oxidative cleavage X-ray crystal structure, iron binding, and hydroxylradical scavenging properties. J. Biol. Chem. 2003, 278, 20319–20326. [Google Scholar] [CrossRef]
- Ren, B.; Tibbelin, G.; Kajino, T.; Asami, O.; Ladenstein, R. The multi-layered structure of Dps with a novel di-nuclear ferroxidase center. J. Mol. Biol. 2003, 329, 467–477. [Google Scholar] [CrossRef]
- Dadinova, L.; Kamyshinsky, R.; Chesnokov, Y.; Mozhaev, A.; Matveev, V.; Gruzinov, A.; Vasiliev, A.; Shtykova, E. Structural Rearrangement of Dps-DNA Complex Caused by Divalent Mg and Fe Cations. Int. J. Mol. Sci. 2021, 22, 6056. [Google Scholar] [CrossRef]
- Bhaskar, S.; Lim, S. Engineering Protein Nanocages as Carriersfor Biomedical Applications. NPG Asia Mater. 2017, 9, e371. [Google Scholar] [CrossRef]
- Kanekiyo, M.; Wei, C.J.; Yassine, H.M.; McTamney, P.M.; Boyington, J.C.; Whittle, J.R.R.; Rao, S.S.; Kong, W.P.; Wang, L.; Nabel, G.J. Self-Assembling Influenza Nanoparticle Vaccines Elicit Broadly Neutralizing H1N1 Antibodies. Nature 2013, 499, 102–106. [Google Scholar] [CrossRef]
- Ren, H.; Zhu, S.; Zheng, G. Nanoreactor Design Based on Self-Assembling Protein Nanocages. Int. J. Mol. Sci. 2019, 20, 592. [Google Scholar] [CrossRef]
- Douglas, T.; Stark, V.T. Nanophase Cobalt Oxyhydroxide Mineral Synthesized within the Protein Cage of Ferritin. Inorg. Chem. 2000, 39, 1828–1830. [Google Scholar] [CrossRef]
- Yamashita, I.; Hayashi, J.; Hara, M. Bio-Template Synthesis of Uniform CdSe Nanoparticles Using Cage-Shaped Protein, Apoferritin. Chem. Lett. 2004, 33, 1158–1159. [Google Scholar] [CrossRef]
- Ueno, T.; Suzuki, M.; Goto, T.; Matsumoto, T.; Nagayama, K.; Watanabe, Y. Size-Selective Olefin Hydrogenation by a Pd Nano-cluster Provided in an Apo-Ferritin Cage. Angew Chem. Int. Ed. Engl. 2004, 43, 2527–2530. [Google Scholar] [CrossRef]
- Douglas, T.; Strable, E.; Willits, D.; Aitouchen, A.; Libera, M.; Young, M. Protein Engineering of a Viral Cage for Constrained Nanomaterials Synthesis. Adv. Mater. 2002, 14, 415–418. [Google Scholar] [CrossRef]
- Narayanan, S.; Shahbazian-Yassar, R.; Tolou Shokuhfar, T. Transmission electron microscopy of the iron oxide core in ferritin proteins: Current status and future directions. J. Phys. D Appl. Phys. 2019, 52, 453001. [Google Scholar] [CrossRef]
- Pan, Y.H.; Sader, K.; Powell, J.J.; Bleloch, A.; Gass, M.; Trinick, J.; Warley, A.; Li, A.; Brydson, R.; Brown, A. 3D morphology of the human hepatic ferritin mineral core: New evidence for a subunit structure revealed by single particle analysis of HAADF-STEM images. J. Struct. Biol. 2009, 166, 22–31. [Google Scholar] [CrossRef]
- Narayanan, S.; Firlar, E.; Rasul, M.G.; Foroozan, T.; Farajpour, N.; Covnot, L.; Shahbazian-Yassar, R.; Shokuhfar, T. On the structure and chemistry of iron oxide cores in human heart and human spleen ferritins using graphene liquid cell electron microscopy. Nanoscale 2019, 36, 16868–16878. [Google Scholar] [CrossRef] [PubMed]
- Jian, N.; Dowle, M.; Horniblow, R.D.; Tselepis, C.; Palmer, R.E. Morphology of the ferritin iron core by aberration corrected scanning transmission electron microscopy. Nanotechnology 2016, 27, 46LT02. [Google Scholar] [CrossRef]
- Yang, X.; Chiancone, E.; Stefanini, S.; Ilari, A.; Chasteen, N.D. Iron oxidation and hydrolysis reactions of a novel ferritin from Listeria innocua. Biochem. J. 2000, 349, 783–786. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.; Ceci, P.; Ilari, A.; Giangiacomo, L.; Laue, T.M.; Chiancone, E.; Chasteen, N.D. Iron and Hydrogen Peroxide Detoxification Properties of DNA- binding Protein from Starved Cells. J. Biol. Chem. 2002, 277, 27689–27696. [Google Scholar] [CrossRef] [PubMed]
- Su, M.; Cavallo, S.; Stefanini, S.; Chiancone, E.; Chasteen, N.D. The So-Called Listeria innocua Ferritin Is a Dps Protein. Iron Incorporation, Detoxification, and DNA Protection Properties. Biochemistry 2005, 44, 5572–5578. [Google Scholar] [CrossRef] [PubMed]
- Dadinova, L.A.; Chesnokov, Y.M.; Kamyshinsky, R.A.; Orlov, I.A.; Petoukhov, M.V.; Mozhaev, A.A.; Soshinskaya, E.Y.; Lazarev, V.N.; Manuvera, V.A.; Orekhov, A.S.; et al. Protective Dps–DNA co-crystallization in stressed cells: An in vitro structural study by small-angle X-ray scattering and cryo-electron tomography. FEBS Lett. 2019, 593, 1360–1371. [Google Scholar] [CrossRef]
- Kamyshinsky, R.; Chesnokov, Y.; Dadinova, L.; Mozhaev, A.; Orlov, I.; Petoukhov, M.; Orekhov, A.; Shtykova, E.; Vasiliev, A. Polymorphic Protective Dps–DNA Co-Crystals by Cryo Electron Tomography and Small Angle X-Ray Scattering. Biomolecules 2020, 10, 39. [Google Scholar] [CrossRef]
- Tegunov, D.; Cramer, P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat. Methods 2019, 16, 1146–1152. [Google Scholar] [CrossRef]
- Punjani, A.; Rubinstein, J.L.; Fleet, D.J.; Brubaker, M.A. cryoSPARC: Algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 2017, 14, 290–296. [Google Scholar] [CrossRef]


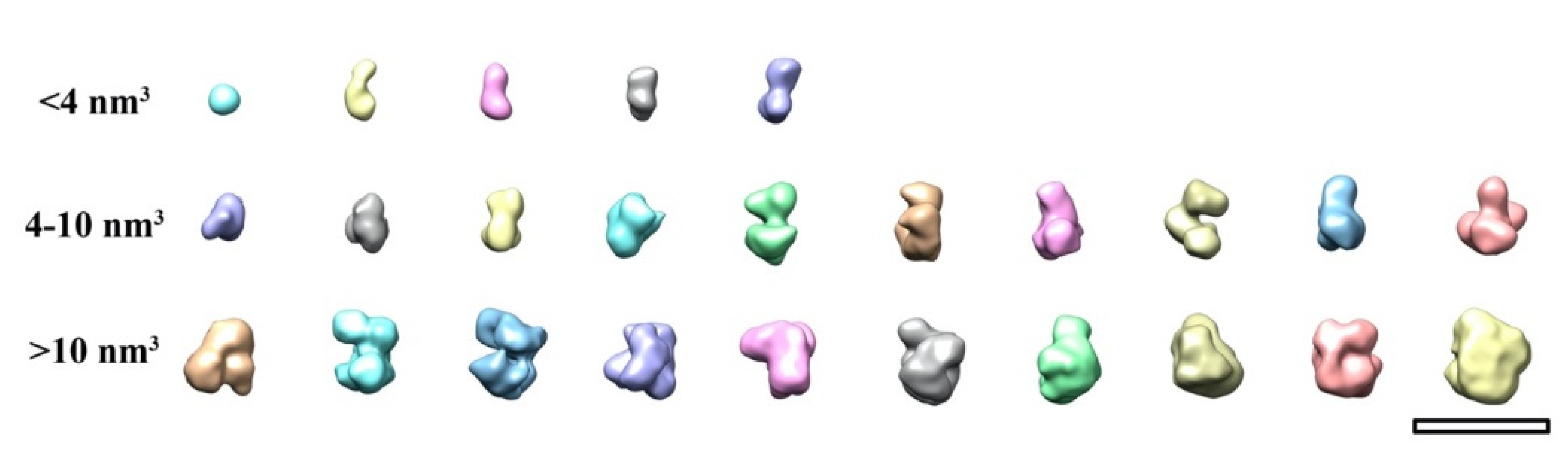


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chesnokov, Y.; Mozhaev, A.; Kamyshinsky, R.; Gordienko, A.; Dadinova, L. Structural Insights into Iron Ions Accumulation in Dps Nanocage. Int. J. Mol. Sci. 2022, 23, 5313. https://doi.org/10.3390/ijms23105313
Chesnokov Y, Mozhaev A, Kamyshinsky R, Gordienko A, Dadinova L. Structural Insights into Iron Ions Accumulation in Dps Nanocage. International Journal of Molecular Sciences. 2022; 23(10):5313. https://doi.org/10.3390/ijms23105313
Chicago/Turabian StyleChesnokov, Yury, Andrey Mozhaev, Roman Kamyshinsky, Alexander Gordienko, and Liubov Dadinova. 2022. "Structural Insights into Iron Ions Accumulation in Dps Nanocage" International Journal of Molecular Sciences 23, no. 10: 5313. https://doi.org/10.3390/ijms23105313
APA StyleChesnokov, Y., Mozhaev, A., Kamyshinsky, R., Gordienko, A., & Dadinova, L. (2022). Structural Insights into Iron Ions Accumulation in Dps Nanocage. International Journal of Molecular Sciences, 23(10), 5313. https://doi.org/10.3390/ijms23105313