OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development
Abstract
1. Introduction
2. Results
2.1. Genotype of OsARF11 Mutant Plants
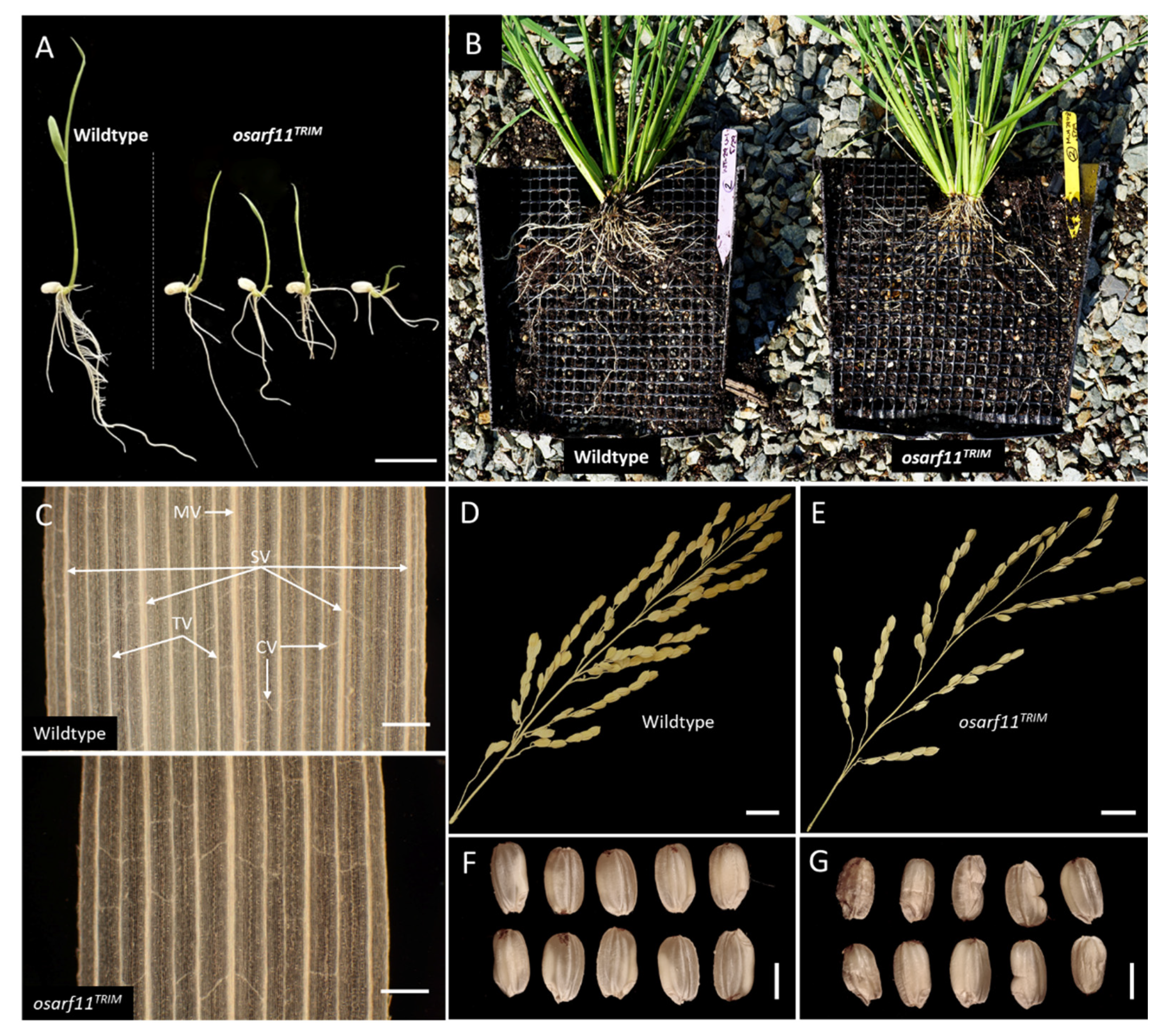
2.2. Morphology of OsARF11TRIM Mutant Plants
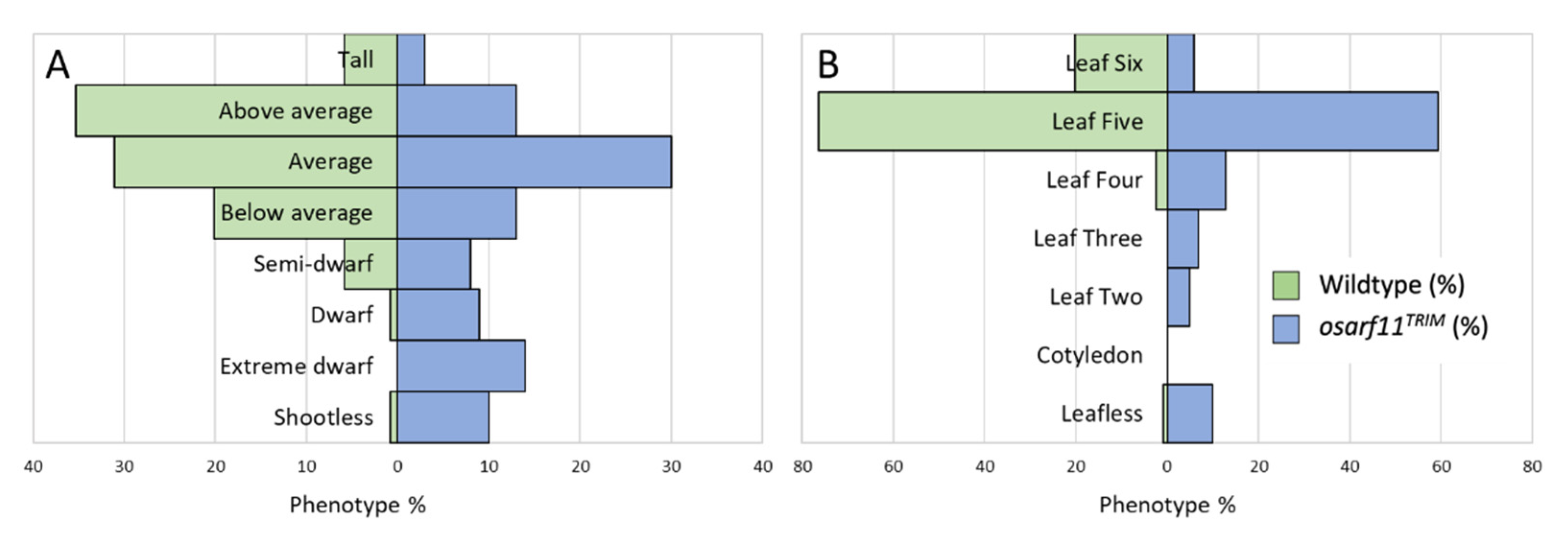
2.2.1. Seedlings
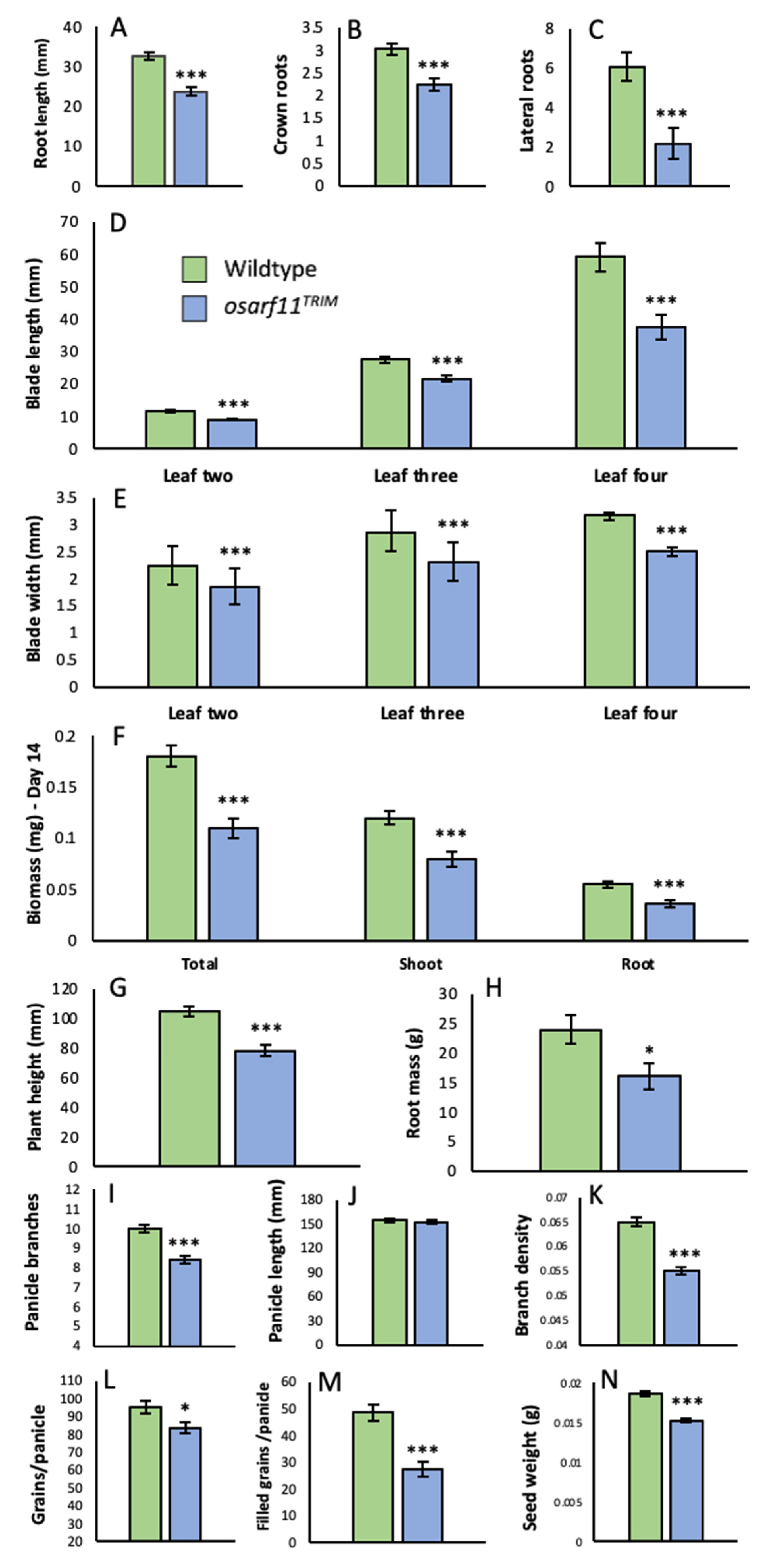
2.2.2. Roots
2.2.3. Panicle and Seed Development
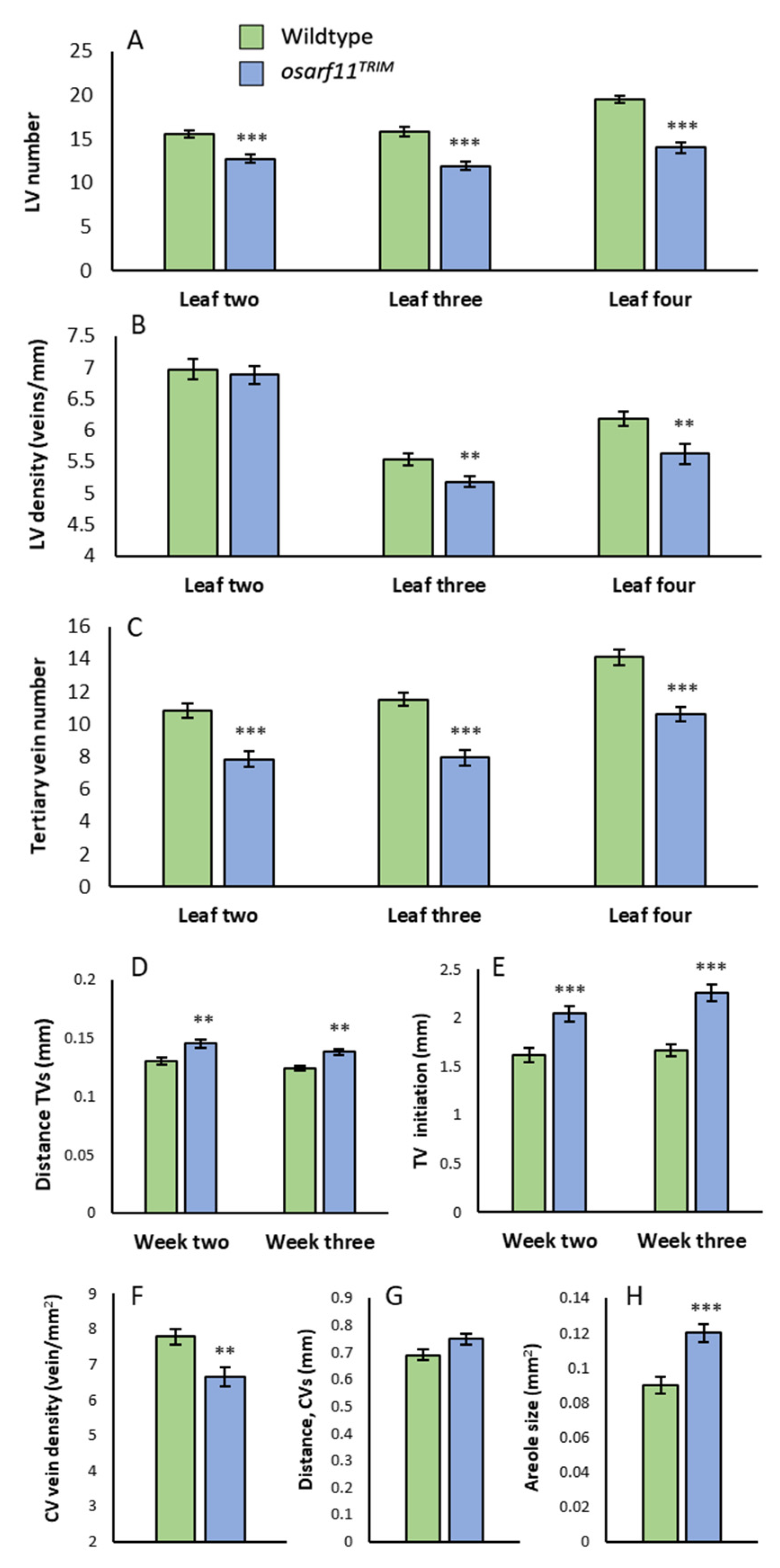
2.3. Leaf Vascular Pattern Formation Is Altered in osarf11 Mutants
2.3.1. Vein Number and Density
2.3.2. Tertiary Veins
2.3.3. Commissural Veins and Areole Size
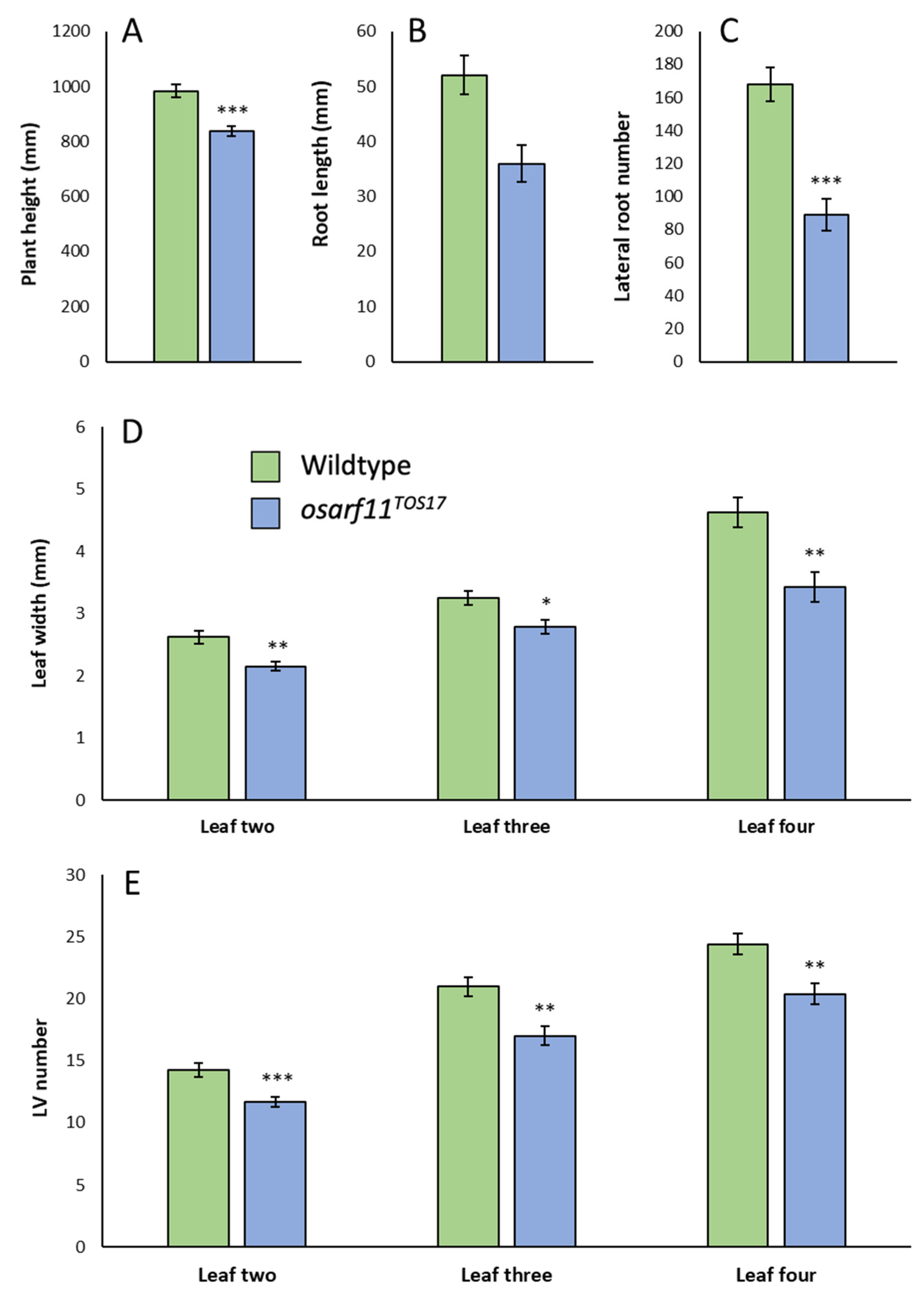
2.4. Morphometric Analyses of OsARF11TOS-17 Mutants
2.5. Loss of OsARF11 Results in Reduced Auxin Perception
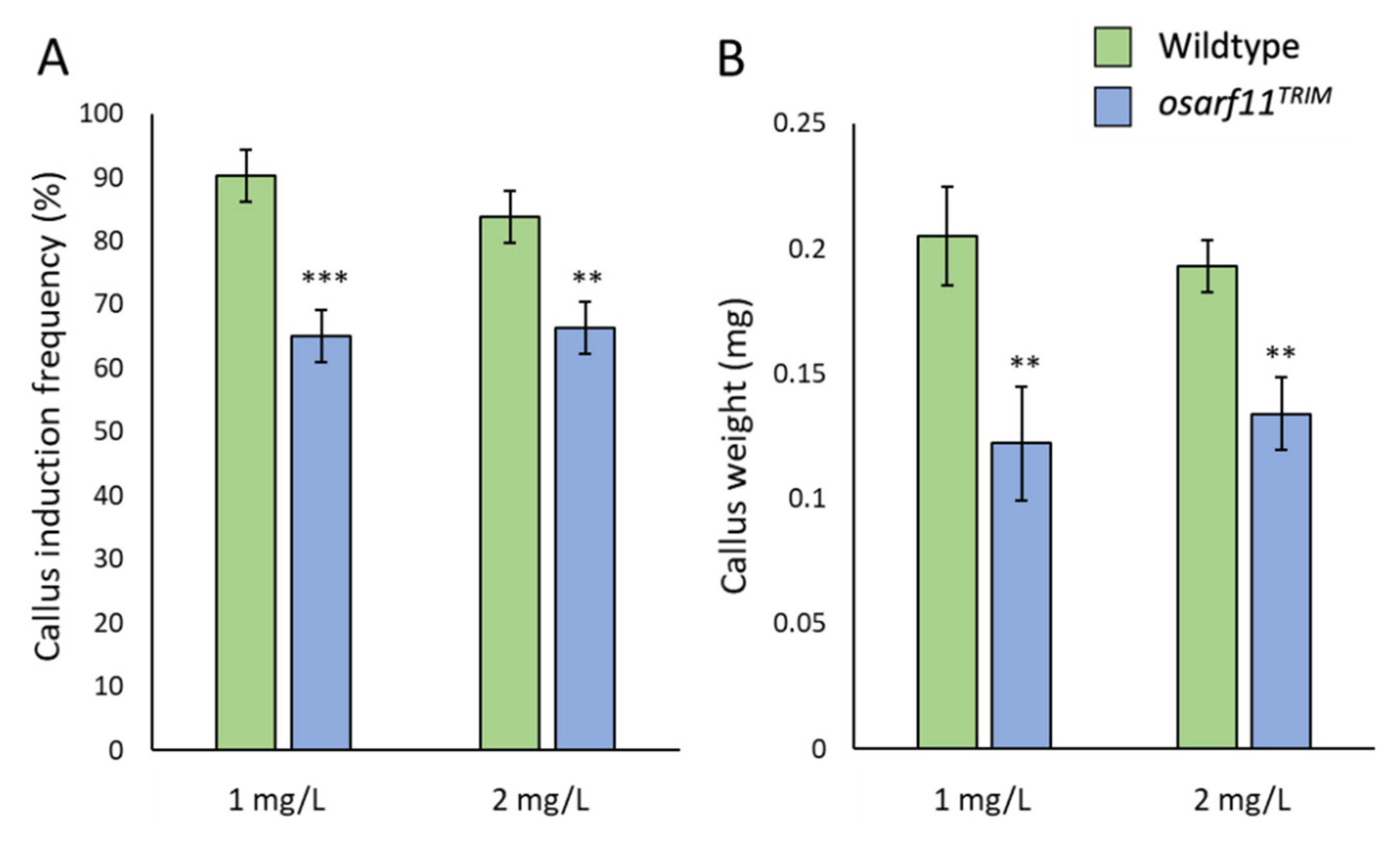
2.5.1. Callus Induction
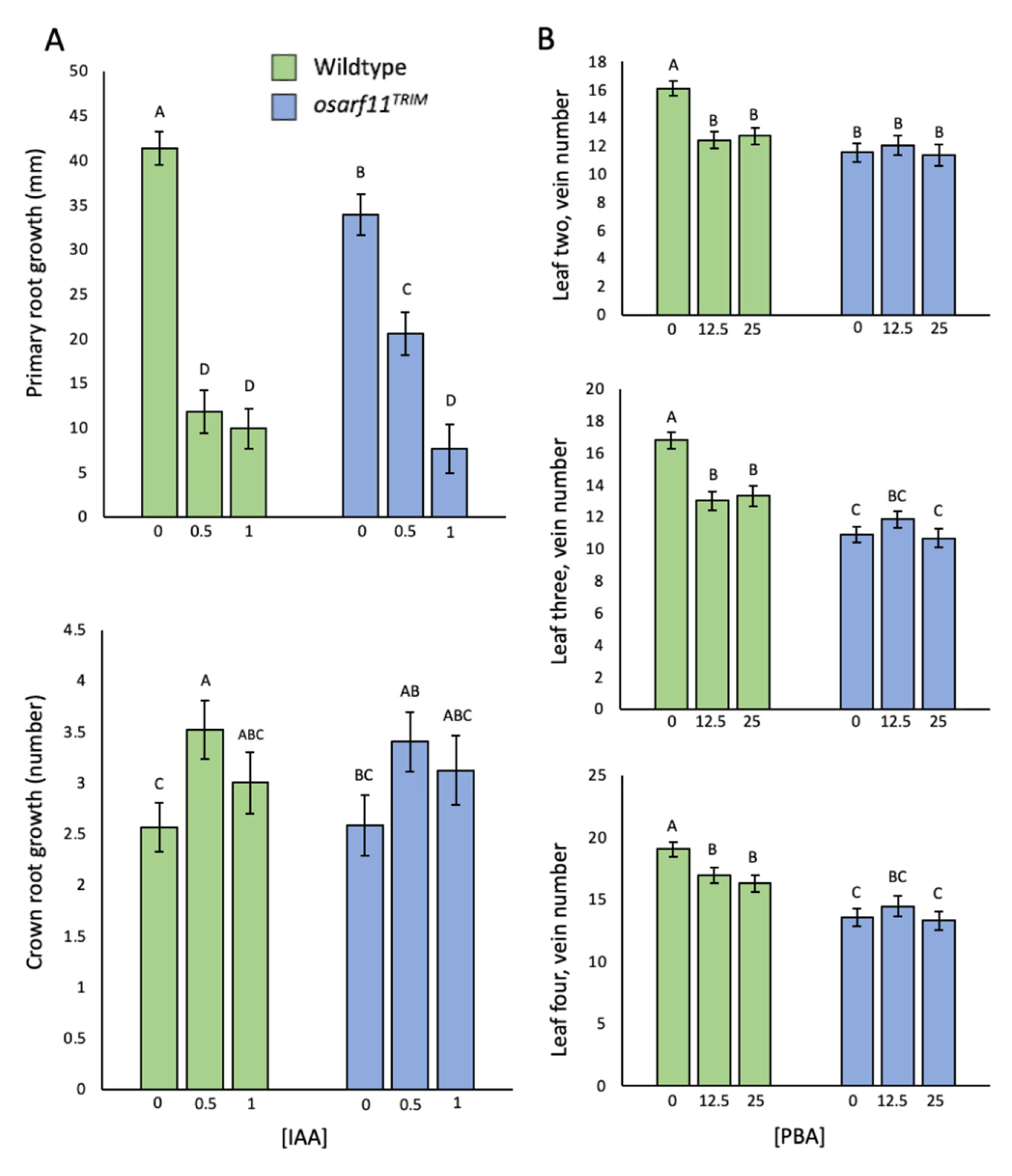
2.5.2. IAA Applications
2.5.3. Phenylboronic Acid Exposure
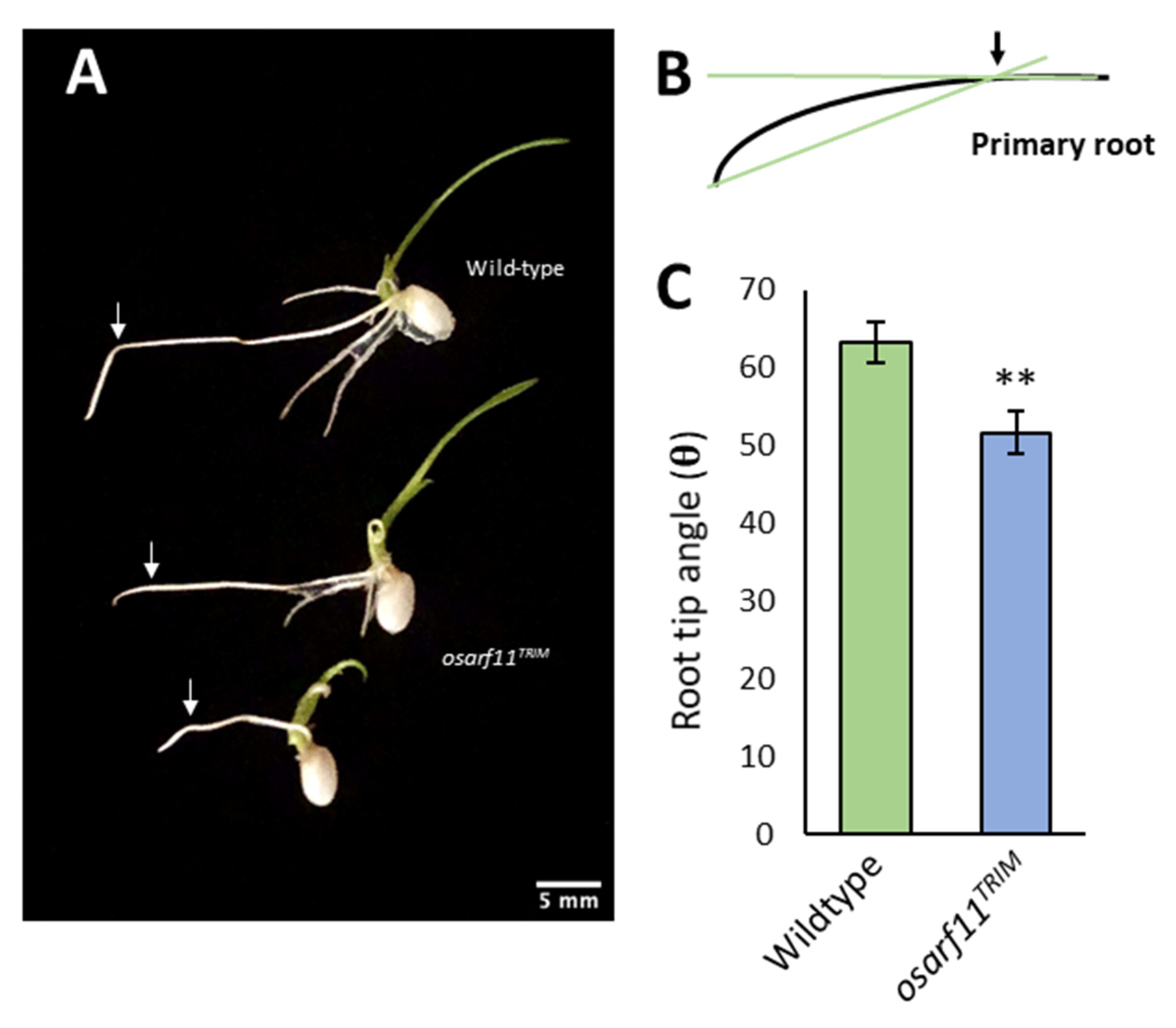
2.6. Gravitropism
3. Discussion
4. Materials and Methods
4.1. Gene Terminology, Mutant Lines, Genotyping, and Growth Conditions
4.2. Tissue Culture Assays
4.3. Microscopy
4.4. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kajala, K.; Covshoff, S.; Karki, S.; Woodfield, H.; Tolley, B.J.; Dionora, M.J.A.; Mogul, R.T.; Mabilangan, A.E.; Danila, F.R.; Hibberd, J.M.; et al. Strategies for engineering a two-celled C 4 photosynthetic pathway into rice. J. Exp. Bot. 2011, 62, 3001–3010. [Google Scholar] [CrossRef]
- Wang, P.; Vlad, D.; Langdale, J.A. Finding the genes to build C4 rice. Curr. Opin. Plant Biol. 2016, 31, 44–50. [Google Scholar] [CrossRef]
- Perrot-Rechenmann, C. Cellular responses to auxin: Division versus expansion. Cold Spring Harb. Perspect. Biol. 2010, 2, a001446. [Google Scholar] [CrossRef] [PubMed]
- Scarpella, E.; Helariutta, Y. Vascular Pattern Formation in Plants. Curr. Top. Dev. Biol. 2010, 91, 221–265. [Google Scholar] [PubMed]
- Aloni, R. The Induction of Vascular Tissues by Auxin. In Plant Hormones; Springer: Dordrecht, The Netherlands, 2010; pp. 485–518. [Google Scholar]
- Smit, M.E.; Weijers, D. The role of auxin signaling in early embryo pattern formation. Curr. Opin. Plant Biol. 2015, 28, 99–105. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, D.; Pesce, E.R.; Stieger, P.; Mandel, T.; Baltensperger, K.; Bennett, M.; Traas, J.; Friml, J.; Kuhlemeier, C. Regulation of phyllotaxis by polar auxin transport. Nature 2003, 426, 255–260. [Google Scholar] [CrossRef] [PubMed]
- Scarpella, E.; Marcos, D.; Friml, J.; Berleth, T. Control of leaf vascular patterning by polar auxin transport. Genes Dev. 2006, 20, 1015–1027. [Google Scholar] [CrossRef]
- Weijers, D.; Wagner, D. Transcriptional Responses to the Auxin Hormone. Annu. Rev. Plant Biol. 2016, 67, 539–574. [Google Scholar] [CrossRef]
- Chandler, J.W. Auxin response factors. Plant Cell Environ. 2016, 39, 1014–1028. [Google Scholar] [CrossRef] [PubMed]
- Vernoux, T.; Brunoud, G.; Farcot, E.; Morin, V.; Van Den Daele, H.; Legrand, J.; Oliva, M.; Das, P.; Larrieu, A.; Wells, D.; et al. The auxin signalling network translates dynamic input into robust patterning at the shoot apex. Mol. Syst. Biol. 2011, 7, 508. [Google Scholar] [CrossRef]
- Calderón Villalobos, L.I.A.; Lee, S.; De Oliveira, C.; Ivetac, A.; Brandt, W.; Armitage, L.; Sheard, L.B.; Tan, X.; Parry, G.; Mao, H.; et al. A combinatorial TIR1/AFB–Aux/IAA co-receptor system for differential sensing of auxin. Nat. Chem. Biol. 2012, 8, 477–485. [Google Scholar] [CrossRef]
- Powers, S.K.; Strader, L.C. Regulation of auxin transcriptional responses. Dev. Dyn. 2020, 249, 483–495. [Google Scholar] [CrossRef]
- Kepinski, S.; Leyser, O. The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 2005, 435, 446–451. [Google Scholar] [CrossRef]
- Sharon, M.; Zheng, C.; Zheng, N.; Calderon-Villalobos, L.I.A.; Estelle, M.; Tan, X.; Robinson, C.V. Mechanism of auxin perception by the TIR1 ubiquitin ligase. Nature 2007, 446, 640–645. [Google Scholar]
- Ulmasov, T. ARF1, a Transcription Factor That Binds to Auxin Response Elements. Science 1997, 276, 1865–1868. [Google Scholar] [CrossRef]
- Freire-Rios, A.; Tanaka, K.; Crespo, I.; Van der Wijk, E.; Sizentsova, Y.; Levitsky, V.; Lindhoud, S.; Fontana, M.; Hohlbein, J.; Roeland Boer, D.; et al. Architecture of DNA elements mediating ARF transcription factor binding and auxin-responsive gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2020, 117, 24557–24566. [Google Scholar] [CrossRef]
- Pierre-Jerome, E.; Moss, B.L.; Lanctot, A.; Hageman, A.; Nemhauser, J.L. Functional analysis of molecular interactions in synthetic auxin response circuits. Proc. Natl. Acad. Sci. USA 2016, 113, 11354–11359. [Google Scholar] [CrossRef]
- Stigliani, A.; Martin-Arevalillo, R.; Lucas, J.; Bessy, A.; Vinos-Poyo, T.; Mironova, V.; Vernoux, T.; Dumas, R.; Parcy, F. Capturing auxin response factors syntax using DNA binding models. Mol. Plant 2019, 12, 822–832. [Google Scholar] [CrossRef]
- Wu, M.F.; Yamaguchi, N.; Xiao, J.; Bargmann, B.; Estelle, M.; Sang, Y.; Wagner, D. Auxin-regulated chromatin switch directs acquisition of flower primordium founder fate. Elife 2015, 4, e09269. [Google Scholar] [CrossRef] [PubMed]
- Roosjen, M.; Paque, S.; Weijers, D. Auxin Response Factors: Output control in auxin biology. J. Exp. Bot. 2018, 69, 179–188. [Google Scholar] [CrossRef]
- Przemeck, G.K.H.; Mattsson, J.; Hardtke, C.S.C.S.; Sung, Z.R.R.; Berleth, T. Studies on the role of the Arabidopsis gene MONOPTEROS in vascular development and plant cell axialization. Planta 1996, 200, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Berleth, T.; Jurgens, G. The role of the monopteros gene in organising the basal body region of the Arabidopsis embryos. Trends Genet. 2008, 9, 299. [Google Scholar]
- Hardtke, C.S.; Ckurshumova, W.; Vidaurre, D.P.; Singh, S.A.; Stamatiou, G.; Tiwari, S.B.; Hagen, G.; Guilfoyle, T.J.; Berleth, T. Overlapping and non-redundant functions of the Arabidopsis auxin response factors MONOPTEROS and NONPHOTOTROPIC HYPOCOTYL 4. Development 2004, 131, 1089–1100. [Google Scholar] [CrossRef]
- Hardtke, C.S.; Berleth, T. The Arabidopsis gene MONOPTEROS encodes a transription factor mediating embryo axis formation and vascular development. EMBO J. 1998, 17, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Wenzel, C.L.; Schuetz, M.; Yu, Q.; Mattsson, J. Dynamics of MONOPTEROS and PIN-FORMED1 expression during leaf vein pattern formation in Arabidopsis thaliana. Plant J. 2007, 49, 387–398. [Google Scholar] [CrossRef] [PubMed]
- Schuetz, M.; Berleth, T.; Mattsson, J. Multiple MONOPTEROS-dependent pathways are involved in leaf initiation. Plant Physiol. 2008, 148, 870–880. [Google Scholar] [CrossRef] [PubMed]
- Schlereth, A.; Möller, B.; Liu, W.; Kientz, M.; Flipse, J.; Rademacher, E.H.; Schmid, M.; Jürgens, G.; Weijers, D. MONOPTEROS controls embryonic root initiation by regulating a mobile transcription factor. Nature 2010, 464, 913–916. [Google Scholar] [CrossRef]
- Bhatia, N.; Bozorg, B.; Larsson, A.; Ohno, C.; Jönsson, H.; Heisler, M.G. Auxin Acts through MONOPTEROS to Regulate Plant Cell Polarity and Pattern Phyllotaxis. Curr. Biol. 2016, 26, 3202–3208. [Google Scholar] [CrossRef]
- Nemhauser, J.L.; Feldman, L.J.; Zambryski, P.C. Auxin and ETTIN in Arabidopsis gynoecium morphogenesis. Development 2000, 127, 3877–3888. [Google Scholar]
- Simonini, S.; Deb, J.; Moubayidin, L.; Stephenson, P.; Valluru, M.; Freire-Rios, A.; Sorefan, K.; Weijers, D.; Friml, J.; Østergaard, L. A noncanonical auxin-sensing mechanism is required for organ morphogenesis in arabidopsis. Genes Dev. 2016, 30, 2286–2296. [Google Scholar] [CrossRef]
- Ellis, C.M.; Nagpal, P.; Young, J.C.; Hagen, G.; Guilfoyle, T.J.; Reed, J.W. AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsisthaliana. Development 2005, 132, 4563–4574. [Google Scholar] [CrossRef]
- Shin, R.; Burch, A.Y.; Huppert, K.A.; Tiwari, S.B.; Murphy, A.S.; Guilfoyle, T.J.; Schachtman, D.P. The Arabidopsis Transcription Factor MYB77 Modulates Auxin Signal Transduction. Plant Cell Online 2007, 19, 2440–2453. [Google Scholar] [CrossRef]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 1–9. [Google Scholar] [CrossRef]
- Perez-Torres, C.-A.; Lopez-Bucio, J.; Cruz-Ramirez, A.; Ibarra-Laclette, E.; Dharmasiri, S.; Estelle, M.; Herrera-Estrella, L. Phosphate Availability Alters Lateral Root Development in Arabidopsis by Modulating Auxin Sensitivity via a Mechanism Involving the TIR1 Auxin Receptor. Plant Cell 2008, 20, 3258–3272. [Google Scholar] [CrossRef]
- Sato, Y.; Nishimura, A.; Ito, M.; Ashikari, M.; Hirano, H.-Y.; Matsuoka, M. Auxin response factor family in rice. Genes Genet. Syst. 2001, 76, 373–380. [Google Scholar] [CrossRef][Green Version]
- Wang, D.; Pei, K.; Fu, Y.; Sun, Z.; Li, S.; Liu, H.; Tang, K.; Han, B.; Tao, Y. Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 2007, 394, 13–24. [Google Scholar] [CrossRef]
- Attia, K.A.; Abdelkhalik, A.F.; Ammar, M.H.; Wei, C.; Yang, J.; Lightfoot, D.A.; El-Sayed, W.M.; El-Shemy, H.A. Antisense phenotypes reveal a functional expression of OsARF1, an auxin response factor, in transgenic rice. Curr. Issues Mol. Biol. 2009, 11, 11. [Google Scholar]
- Shen, C.; Yue, R.; Sun, T.; Zhang, L.; Yang, Y.; Wang, H. OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 2015, 231, 148–158. [Google Scholar] [CrossRef]
- Shen, C.; Yue, R.; Yang, Y.; Zhang, L.; Sun, T.; Tie, S.; Wang, H. OsARF16 is involved in cytokinin-mediated inhibition of phosphate transport and phosphate signaling in rice (Oryza sativa L.). PLoS ONE 2014, 9, e112906. [Google Scholar] [CrossRef]
- Wang, S.; Zhang, S.; Sun, C.; Xu, Y.; Chen, Y.; Yu, C.; Qian, Q.; Jiang, D.A.; Qi, Y. Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 2014, 201, 91–103. [Google Scholar] [CrossRef]
- Qi, Y.; Wang, S.; Shen, C.; Zhang, S.; Chen, Y.; Xu, Y.; Liu, Y.; Wu, Y.; Jiang, D. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef]
- Huang, J.; Li, Z.; Zhao, D. Deregulation of the OsmiR160 target gene OsARF18 causes growth and developmental defects with an alteration of auxin signaling in rice. Sci. Rep. 2016, 6, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Inukai, Y. Characterization of a Tos17 Insertion Mutant of Rice Auxin Signal Transcription Factor Gene, OsARF24. Am. J. Plant Sci. 2013, 4, 84–91. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, T.; Liu, S.; Liu, X.; Jiang, L.; Wan, J. Disruption of OsARF19 is Critical for Floral Organ Development and Plant Architecture in Rice (Oryza sativa L.). Plant Mol. Biol. Report. 2016, 34, 748–760. [Google Scholar] [CrossRef]
- Zhang, S.; Wang, S.; Xu, Y.; Yu, C.; Shen, C.; Qian, Q.; Geisler, M.; Jiang, D.A.; Qi, Y. The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3-5 and OsBRI1. Plant Cell Environ. 2015, 38, 638–654. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.H.; Zhou, L.J.; Xu, P.; Xue, H.W. SPOC domain-containing protein Leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PLoS Genet. 2018, 14, e1007829. [Google Scholar] [CrossRef]
- Li, Y.; Li, J.; Chen, Z.; Wei, Y.; Qi, Y.; Wu, C. OsmiR167a-targeted auxin response factors modulate tiller angle via fine-tuning auxin distribution in rice. Plant Biotechnol. J. 2020, 18, 2015–2026. [Google Scholar] [CrossRef]
- Hsing, Y.I.; Chern, C.G.; Fan, M.J.; Lu, P.C.; Chen, K.T.; Lo, S.F.; Sun, P.K.; Ho, S.L.; Lee, K.W.; Wang, Y.C.; et al. A rice gene activation/knockout mutant resource for high throughput functional genomics. Plant Mol. Biol. 2007, 63, 351–364. [Google Scholar] [CrossRef]
- Hirochika, H. Contribution of the Tos17 retrotransposon to rice functional genomics. Curr. Opin. Plant Biol. 2001, 4, 118–122. [Google Scholar] [CrossRef]
- Song, Y.; Wang, L.; Xiong, L. Comprehensive expression profiling analysis of OsIAA gene family in developmental processes and in response to phytohormone and stress treatments. Planta 2009, 229, 577–591. [Google Scholar] [CrossRef]
- Waese, J.; Fan, J.; Pasha, A.; Yu, H.; Fucile, G.; Shi, R.; Cumming, M.; Kelley, L.A.; Sternberg, M.J.; Krishnakumar, V.; et al. ePlant: Visualizing and exploring multiple levels of data for hypothesis generation in plant biology. Plant Cell 2017, 29, 1806–1821. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Yang, C.Y.; Miao, R.; Zhou, C.L.; Cao, P.H.; Lan, J.; Zhu, X.J.; Mou, C.L.; Huang, Y.S.; Liu, S.J.; et al. DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 2018, 11, 46. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, T.; Morinaka, Y.; Inukai, Y.; Kitano, H.; Fujioka, S. Auxin signal transcription factor regulates expression of the brassinosteroid receptor gene in rice. Plant J. 2013, 73, 676–688. [Google Scholar] [CrossRef] [PubMed]
- Qi, J.; Qian, Q.; Bu, Q.; Li, S.; Chen, Q.; Sun, J.; Liang, W.; Zhou, Y.; Chu, C.; Li, X.; et al. Mutation of the rice narrow leaf1 gene, which encodes a novel protein, affects vein patterning and polar auxin transport. Plant Physiol. 2008, 147, 1947–1959. [Google Scholar] [CrossRef] [PubMed]
- Matthes, M.; Torres-Ruiz, R.A. Boronic acid treatment phenocopies monopteros by affecting PIN1 membrane stability and polar auxin transport in Arabidopsis thaliana embryos. Development 2016, 143, 4053–4062. [Google Scholar] [CrossRef] [PubMed]
- Band, L.R.; Wells, D.M.; Larrieu, A.; Sun, J.; Middleton, A.M.; French, A.P.; Brunoud, G.; Sato, E.M.; Wilson, M.H.; Peŕet, B.; et al. Root gravitropism is regulated by a transient lateral auxin gradient controlled by a tipping-point mechanism. Proc. Natl. Acad. Sci. USA 2012, 109, 4668–4673. [Google Scholar] [CrossRef]
- Ottenschläger, I.; Wolff, P.; Wolverton, C.; Bhalerao, R.P.; Sandberg, G.; Ishikawa, H.; Evans, M.; Palme, K. Gravity-regulated differential auxin transport from columella to lateral root cap cells. Proc. Natl. Acad. Sci. USA 2003, 100, 2987–2991. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Matsuda, Y.; Ozawa, K.; Nishimura, T.; Koshiba, T.; Fraaije, M.W.; Sekiguchi, H. NARROW LEAF 7 controls leaf shape mediated by auxin in rice. Mol. Genet. Genom. 2008, 279, 499–507. [Google Scholar] [CrossRef]
- Xu, Y.; Hu, D.; Hou, X.; Shen, J.; Liu, J.; Cen, X.; Fu, J.; Li, X.; Hu, H.; Xiong, L. OsTMF attenuates cold tolerance by affecting cell wall properties in rice. New Phytol. 2020, 227, 498–512. [Google Scholar] [CrossRef] [PubMed]
- Du, M.; Spalding, E.P.; Gray, W.M. Rapid Auxin-Mediated Cell Expansion. Annu. Rev. Plant Biol. 2020, 71, 379–402. [Google Scholar] [CrossRef]
- Wang, R.; Zhang, Y.; Kieffer, M.; Yu, H.; Kepinski, S.; Estelle, M. HSP90 regulates temperature-dependent seedling growth in Arabidopsis by stabilizing the auxin co-receptor F-box protein TIR1. Nat. Commun. 2016, 7, 10269. [Google Scholar] [CrossRef]
- Inukai, Y.; Sakamoto, T.; Ueguchi-Tanaka, M.; Shibata, Y.; Gomi, K.; Umemura, I.; Hasegawa, Y.; Ashikari, M.; Kitano, H.; Matsuoka, M. Crown rootless1, which is essential for crown root formation in rice, is a target of an Auxin Response Factor in auxin signaling. Plant Cell 2005, 17, 1387–1396. [Google Scholar] [CrossRef]
- Mariyamma, N.P.; Hou, H.; Carland, F.M.; Nelson, T.; Schultz, E.A. Localization of Arabidopsis FORKED1 to a RABA-positive compartment suggests a role in secretion. J. Exp. Bot. 2017, 68, 3375–3390. [Google Scholar] [CrossRef]
- Prabhakaran Mariyamma, N.; Clarke, K.J.; Yu, H.; Wilton, E.E.; Van Dyk, J.; Hou, H.; Schultz, E.A. Members of the Arabidopsis FORKED1-LIKE gene family act to localize PIN1 in developing veins. J. Exp. Bot. 2018, 69, 4773–4790. [Google Scholar] [CrossRef]
- Sieburth, L.E.; Muday, G.K.; King, E.J.; Benton, G.; Kim, S.; Metcalf, K.E.; Meyers, L.; Seamen, E.; Van Norman, J.M. SCARFACE encodes an ARF-GAP that is required for normal auxin efflux and vein patterning in Arabidopsis. Plant Cell 2006, 18, 1396–1411. [Google Scholar] [CrossRef]
- Baylis, T.; Cierlik, I.; Sundberg, E.; Mattsson, J. SHORT INTERNODES/STYLISH genes, regulators of auxin biosynthesis, are involved in leaf vein development in Arabidopsis thaliana. New Phytol. 2013, 197, 737–750. [Google Scholar] [CrossRef]
- Nelson, T.; Dengler, N. Leaf vascular pattern formation. Plant Cell 1997, 9, 1121–1135. [Google Scholar] [CrossRef]
- Pantin, F.; Simonneau, T.; Muller, B. Coming of leaf age: Control of growth by hydraulics and metabolics during leaf ontogeny. New Phytol. 2012, 196, 349–366. [Google Scholar] [CrossRef]
- Feldman, A.B.; Leung, H.; Baraoidan, M.; Elmido-Mabilangan, A.; Canicosa, I.; Quick, W.P.; Sheehy, J.; Murchie, E.H. Increasing leaf vein density via mutagenesis in rice results in an enhanced rate of photosynthesis, smaller cell sizes and can reduce interveinal mesophyll cell number. Front. Plant Sci. 2017, 8, 1883. [Google Scholar] [CrossRef]
- Nardini, A.; Õunapuu-Pikas, E.; Savi, T. When smaller is better: Leaf hydraulic conductance and drought vulnerability correlate to leaf size and venation density across four Coffea arabica genotypes. Funct. Plant Biol. 2014, 41, 972–982. [Google Scholar] [CrossRef]
- Scoffoni, C.; Chatelet, D.S.; Pasquet-Kok, J.; Rawls, M.; Donoghue, M.J.; Edwards, E.J.; Sack, L. Hydraulic basis for the evolution of photosynthetic productivity. Nat. Plants 2016, 2, 1–8. [Google Scholar] [CrossRef]
- Mattsson, J.; Ckurshumova, W.; Berleth, T. Auxin Signaling in Arabidopsis Leaf Vascular Development. Plant Physiol. 2003, 131, 1327–1339. [Google Scholar] [CrossRef]
- Wenzel, C.L.; Hester, Q.; Mattsson, J. Identification of genes expressed in vascular tissues using NPA-induced vascular overgrowth in Arabidopsis. Plant Cell Physiol. 2008, 49, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Geldner, N.; Anders, N.; Wolters, H.; Keicher, J.; Kornberger, W.; Muller, P.; Delbarre, A.; Ueda, T.; Nakano, A.; Jürgens, G. The Arabidopsis GNOM ARF-GEF mediates endosomal recycling, auxin transport, and auxin-dependent plant growth. Cell 2003, 112, 219–230. [Google Scholar] [CrossRef]
- Mayer, U.; Ruiz, R.A.T.; Berleth, T.; Miseéra, S.; Jürgens, G. Mutations affecting body organization in the Arabidopsis embryo. Nature 1991, 353, 402–407. [Google Scholar] [CrossRef]
- Sakai, H.; Lee, S.S.; Tanaka, T.; Numa, H.; Kim, J.; Kawahara, Y.; Wakimoto, H.; Yang, C.C.; Iwamoto, M.; Abe, T.; et al. Rice annotation project database (RAP-DB): An integrative and interactive database for rice genomics. Plant Cell Physiol. 2013, 54, e6. [Google Scholar] [CrossRef]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sims, K.; Abedi-Samakush, F.; Szulc, N.; Macias Honti, M.G.; Mattsson, J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. Int. J. Mol. Sci. 2021, 22, 4089. https://doi.org/10.3390/ijms22084089
Sims K, Abedi-Samakush F, Szulc N, Macias Honti MG, Mattsson J. OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. International Journal of Molecular Sciences. 2021; 22(8):4089. https://doi.org/10.3390/ijms22084089
Chicago/Turabian StyleSims, Katherine, Fatemeh Abedi-Samakush, Nicole Szulc, Monika Gyongyi Macias Honti, and Jim Mattsson. 2021. "OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development" International Journal of Molecular Sciences 22, no. 8: 4089. https://doi.org/10.3390/ijms22084089
APA StyleSims, K., Abedi-Samakush, F., Szulc, N., Macias Honti, M. G., & Mattsson, J. (2021). OsARF11 Promotes Growth, Meristem, Seed, and Vein Formation during Rice Plant Development. International Journal of Molecular Sciences, 22(8), 4089. https://doi.org/10.3390/ijms22084089






