Bootstrapping and Pinning down the Root Meristem; the Auxin–PLT–ARR Network Unites Robustness and Sensitivity in Meristem Growth Control
Abstract
1. Introduction
2. Results
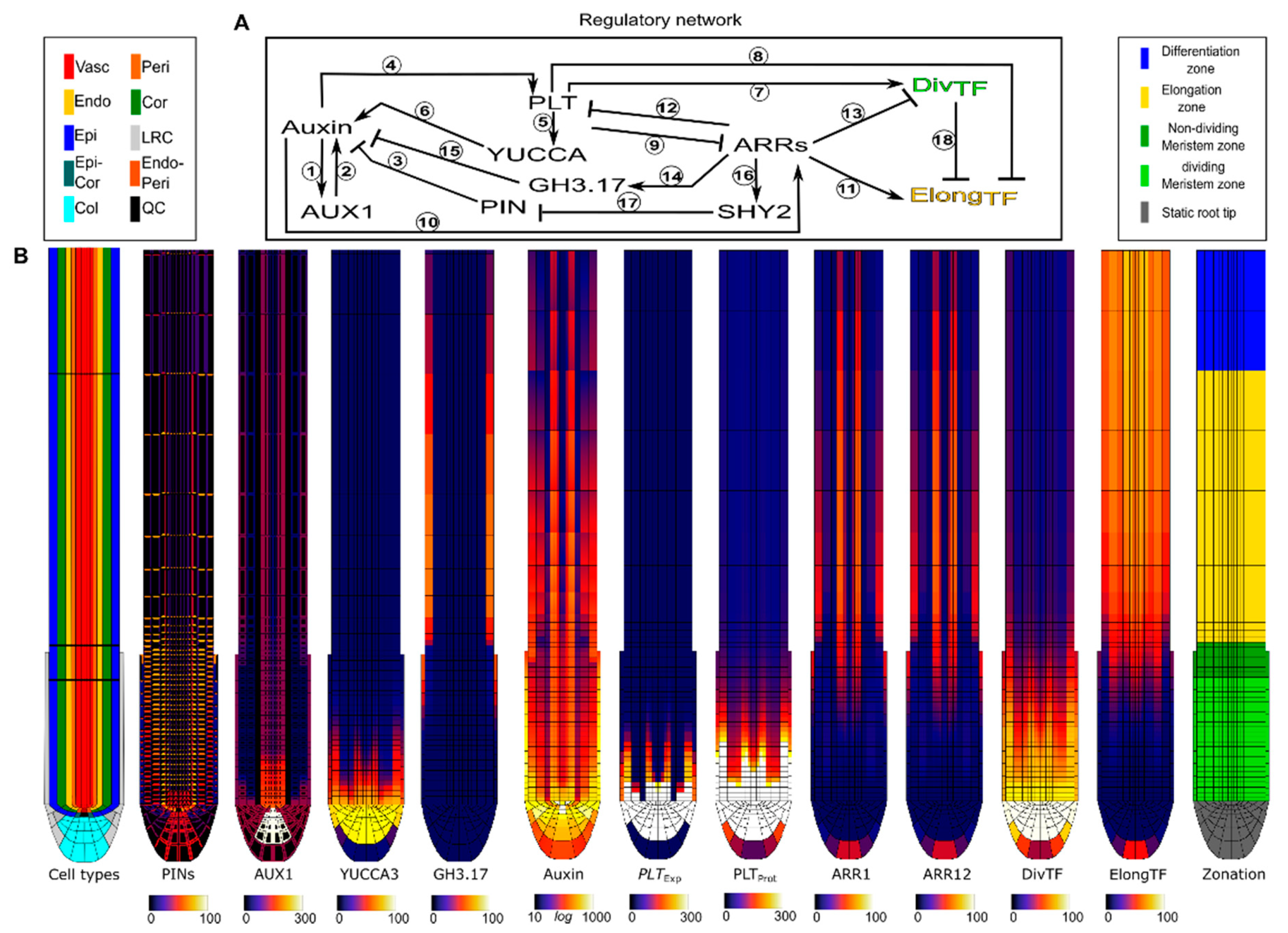
2.1. Introducing the Root Tip Model
| Interaction | Refs | Interaction | Refs | Interaction | Refs |
|---|---|---|---|---|---|
| 1 | [15] | 7 | [1,2,3] | 13 | [14] |
| 2 | [16] | 8 | [1,2,3] | 14 | [11,27] |
| 3 | [17,18] | 9 | [14] | 15 | [11,27] |
| 4 | [3] | 10 | [14] | 16 | [5,6,7] |
| 5 | [24] | 11 | [28] | 17 | [5,6,7] |
| 6 | [21,22,23] | 12 | [14] | 18 | [29] |
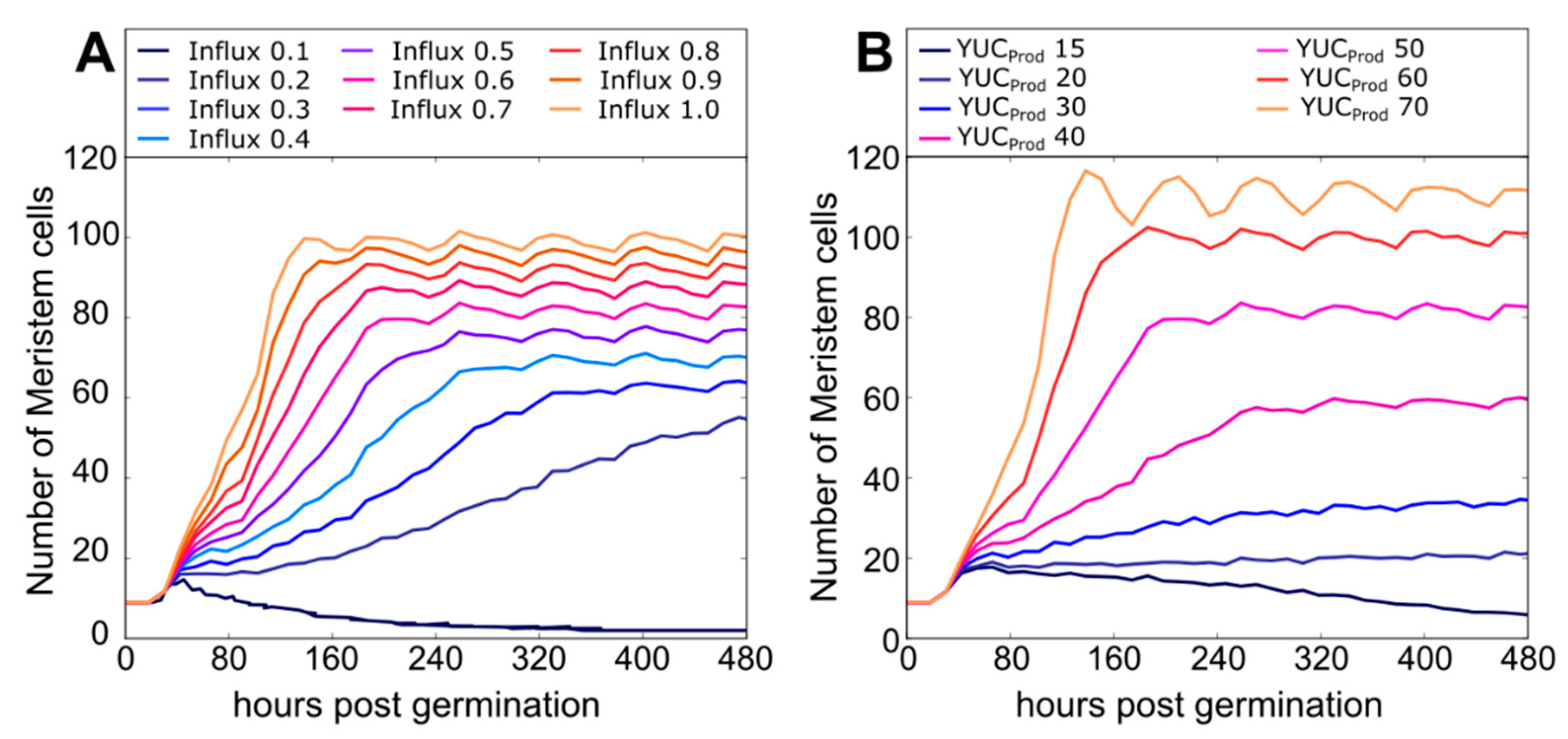
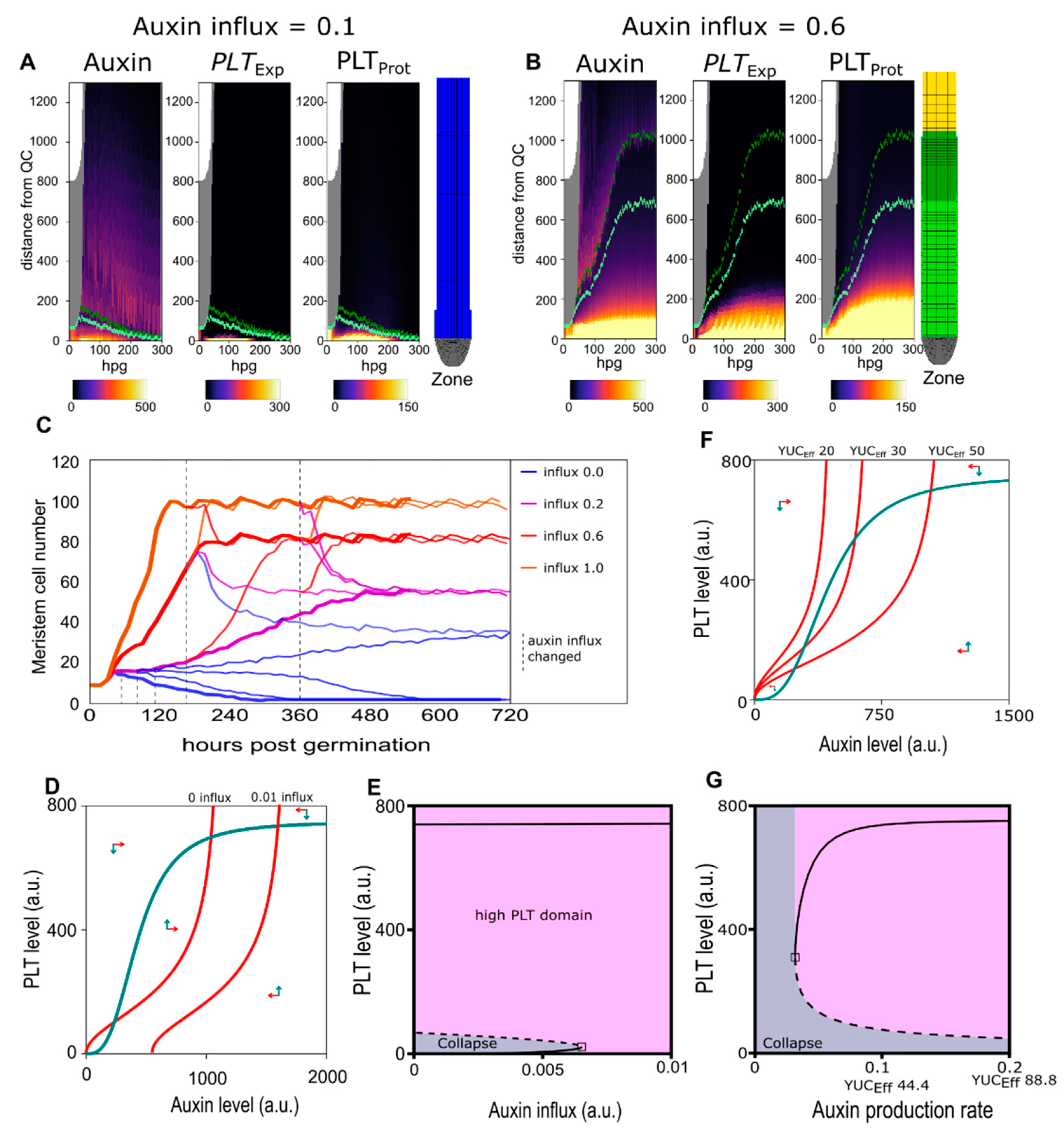
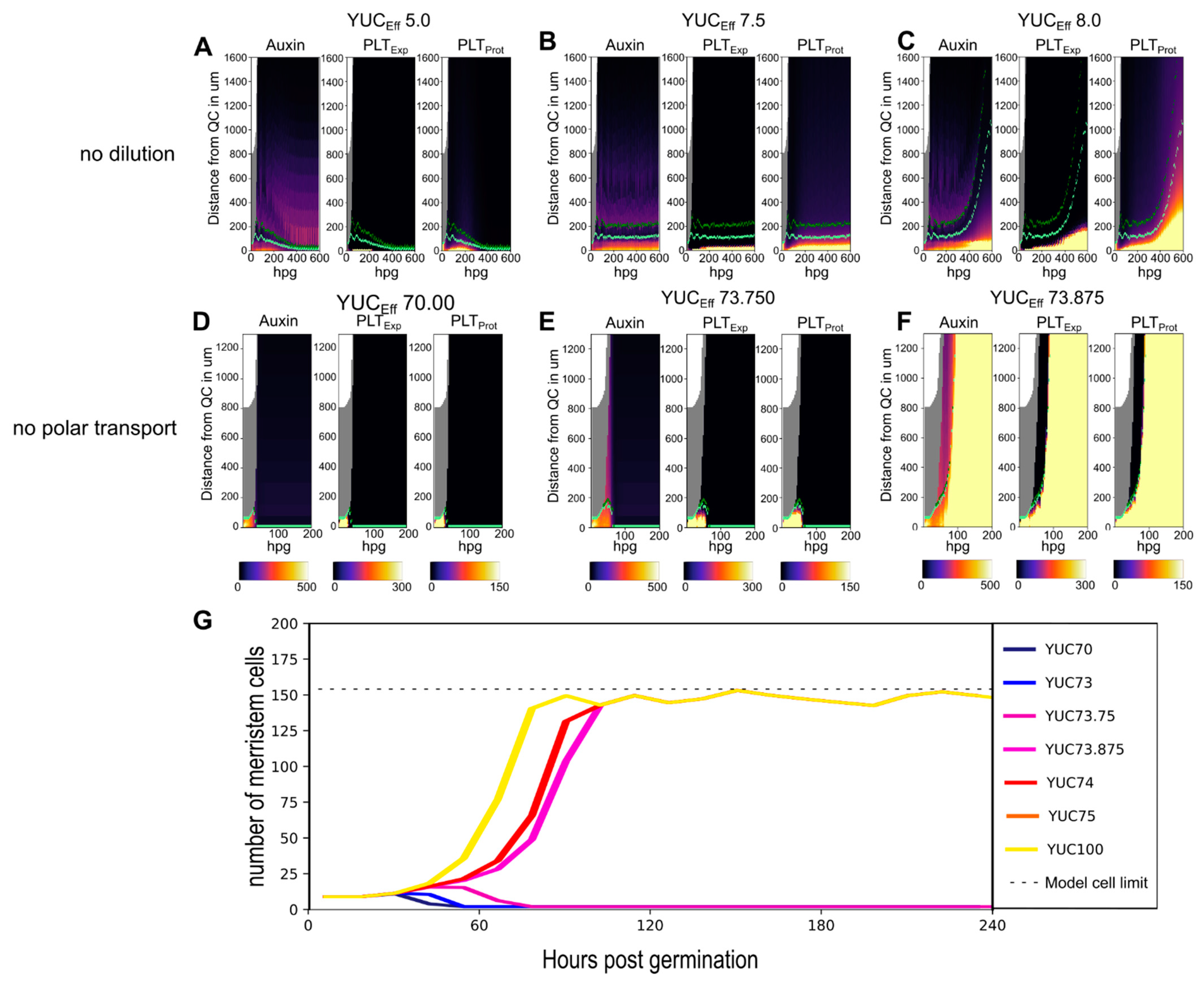
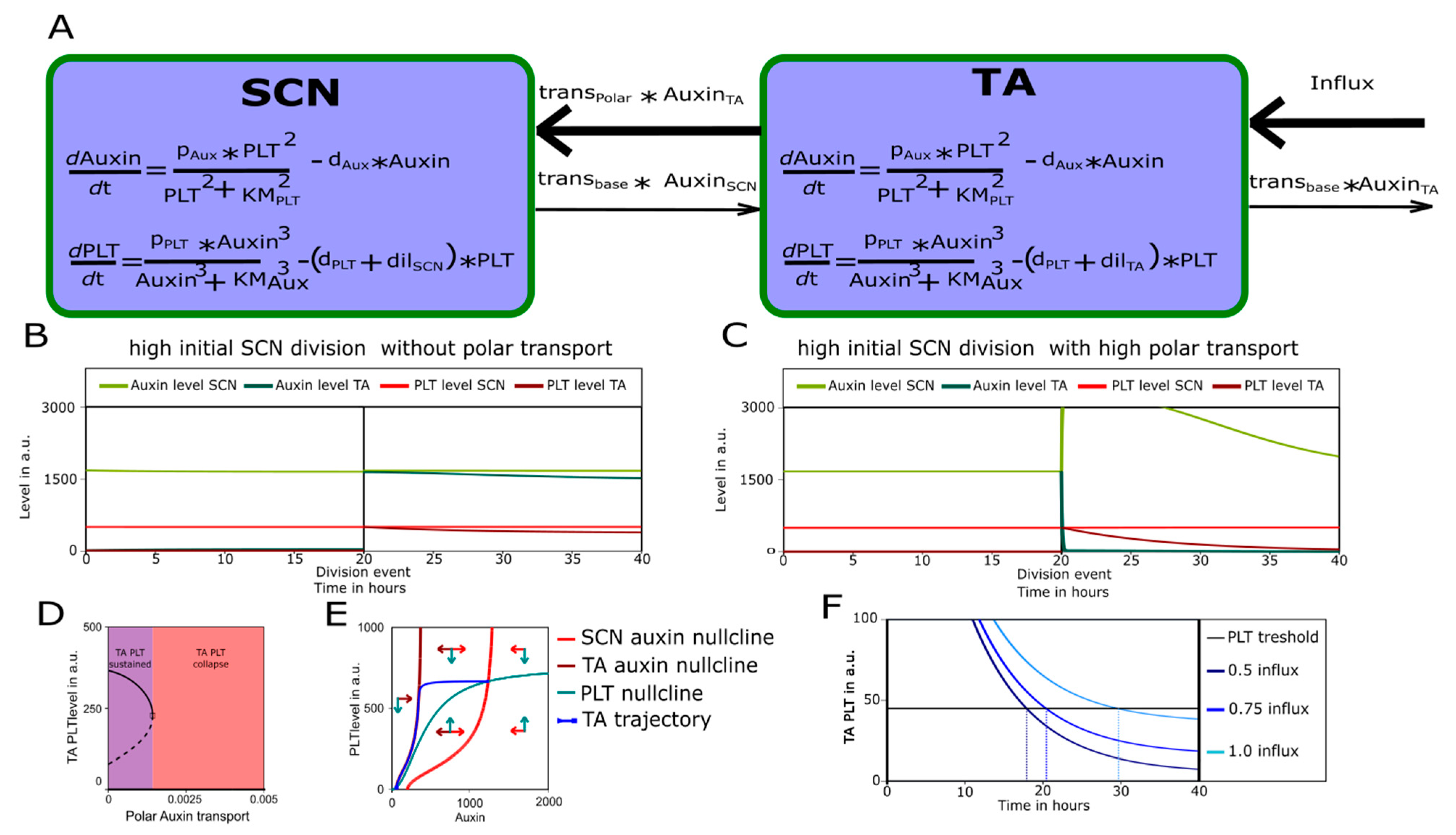
2.2. A Bootstrapping Capable yet Finite Positive Feedback Loop
2.3. Pinning down the High Auxin–High PLT Domain
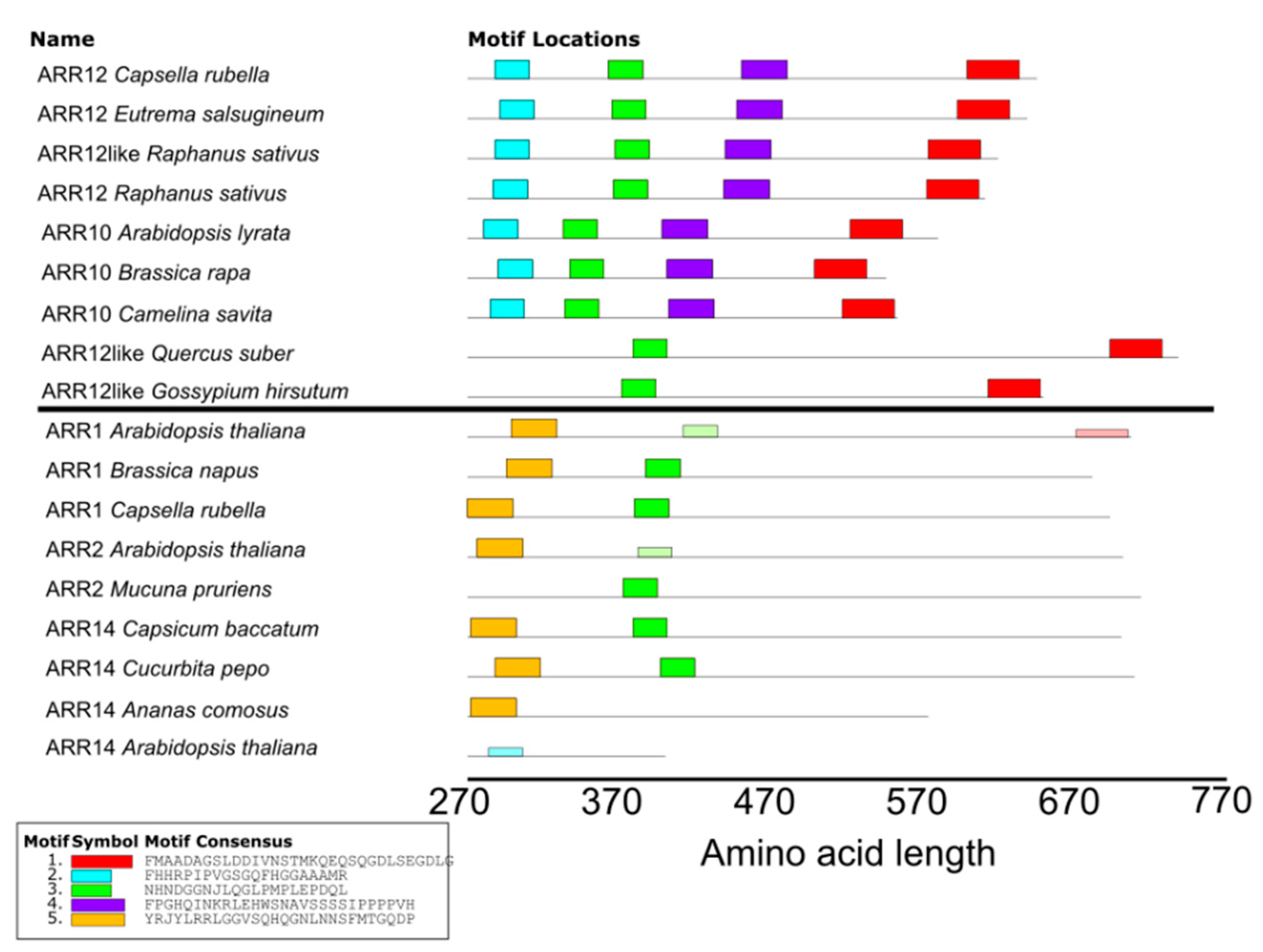
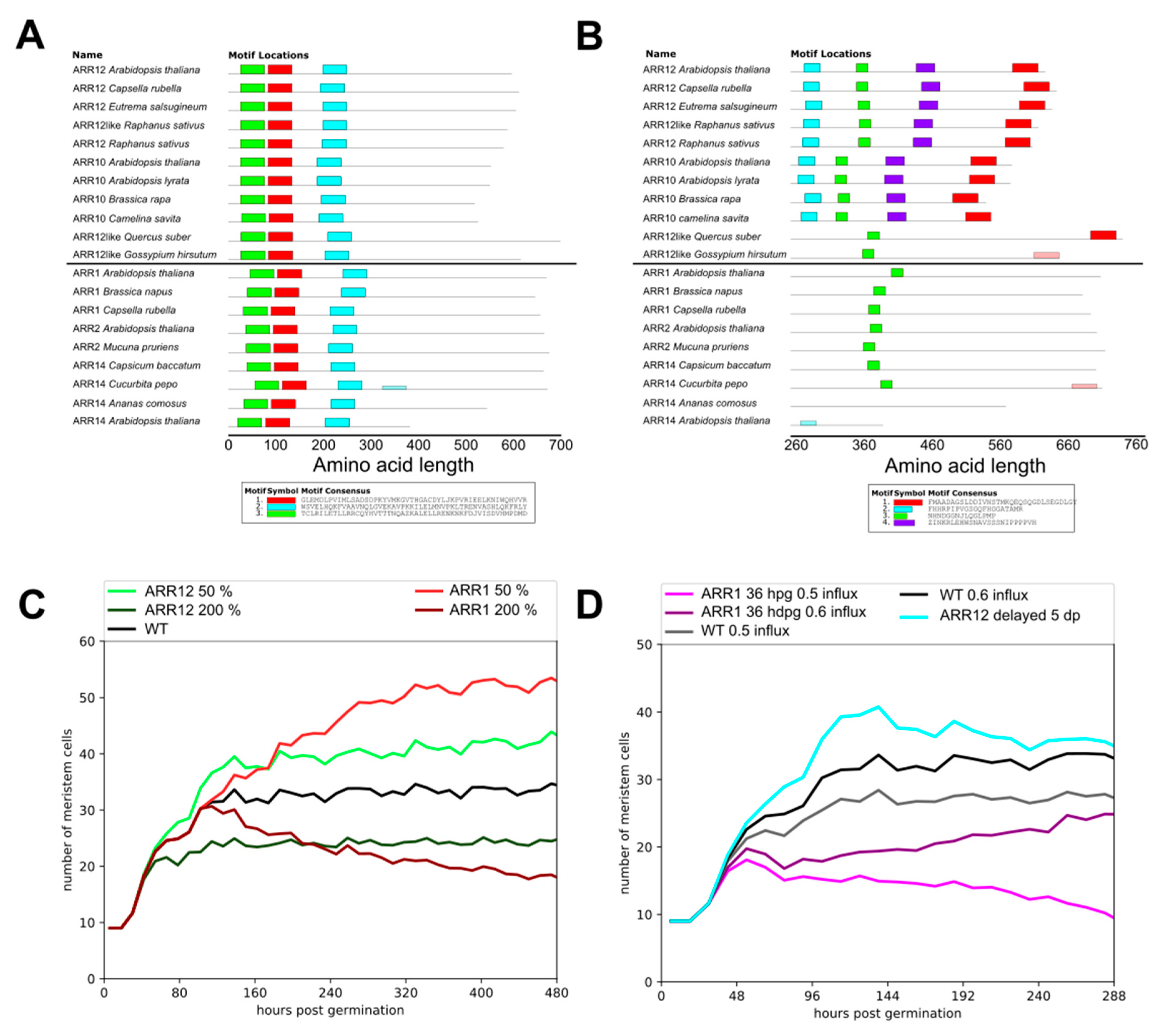
2.4. ARR1–ARR12 Division of Labor
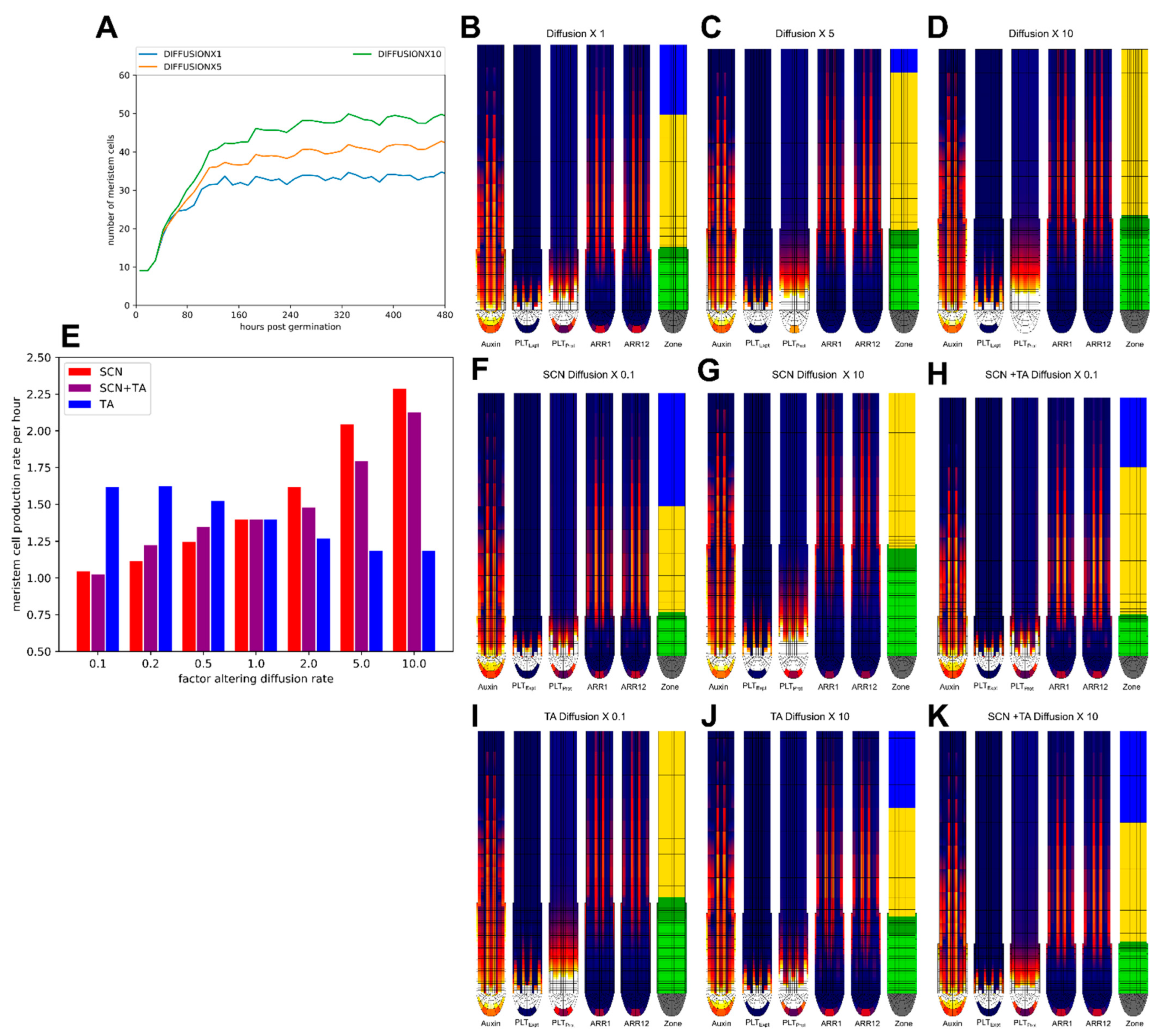
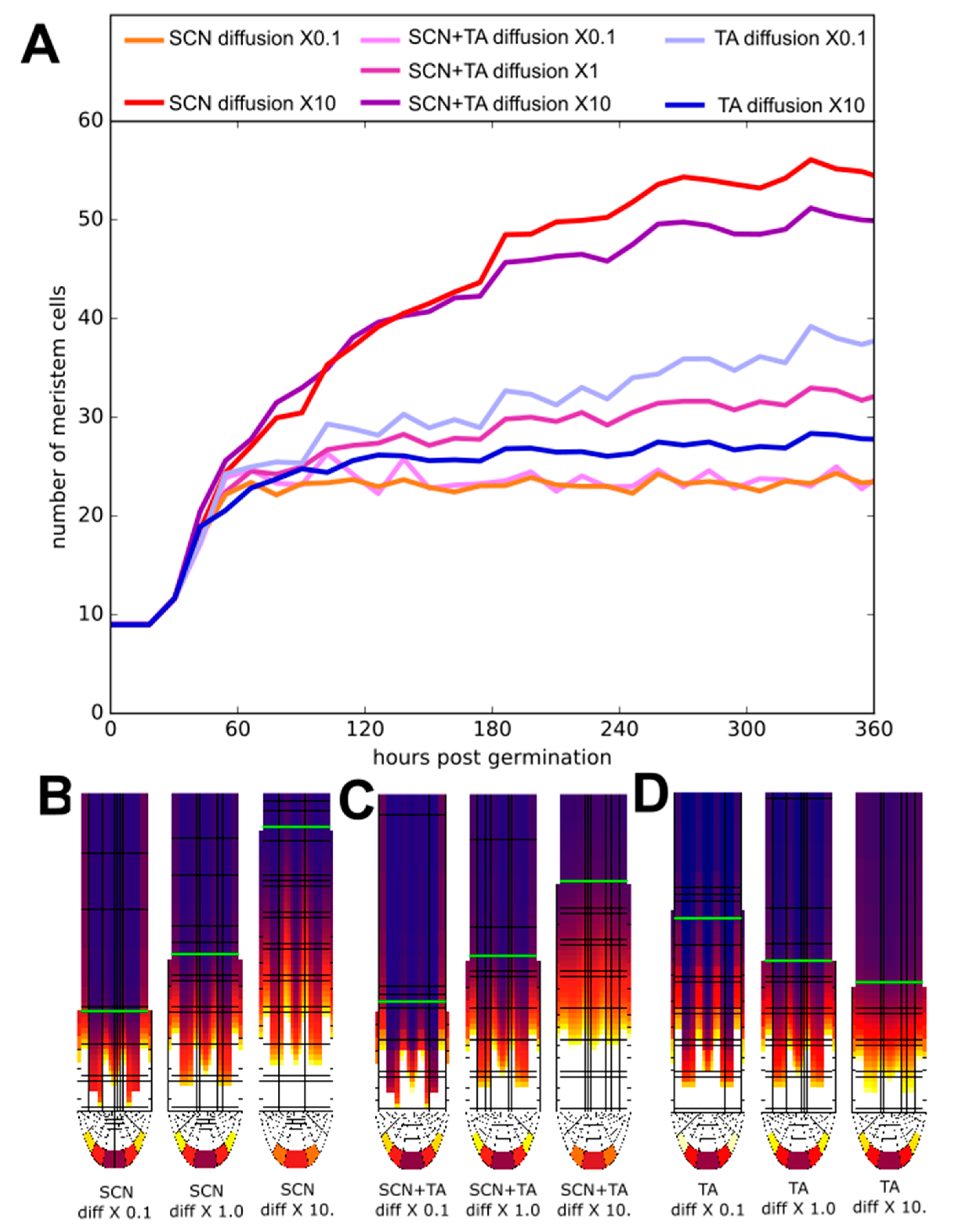
2.5. Impact of PLT Mobility on Meristem Size Depends on Location
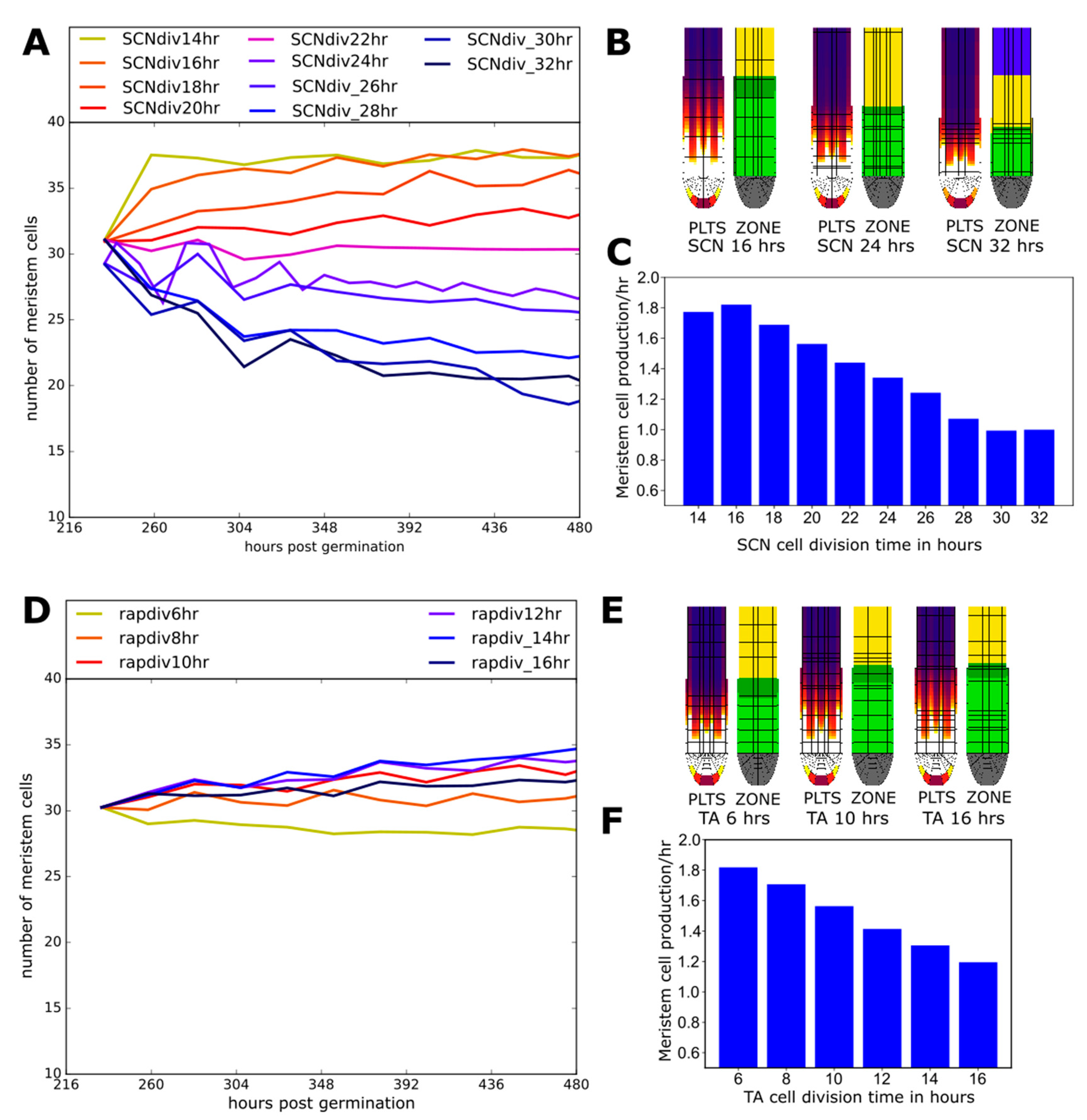
2.6. Effect of Division Rate on Meristem Cell Production Depends on Location
3. Discussion
3.1. Background
3.2. A Pinned down Positive Feedback Loop
3.3. A Bi-Partite Meristem Growth Brake
3.4. The SCN as A PLT Control Center
4. Materials and Methods
4.1. Root Tip Model
4.2. Single-Compartment Model
4.3. Two-Compartment Model
4.4. MEME Analysis
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.-S.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [PubMed]
- Mähönen, A.P.; ten Tusscher, K.; Siligato, R.; Smetana, O.; Díaz-Triviño, S.; Salojärvi, J.; Wachsman, G.; Prasad, K.; Heidstra, R.; Scheres, B. PLETHORA gradient formation mechanism separates auxin responses. Nature 2014, 515, 125–129. [Google Scholar] [CrossRef]
- Ioio, R.D.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins Determine Arabidopsis Root-Meristem Size by Controlling Cell Differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef]
- Ioio, R.D.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A Genetic Framework for the Control of Cell Division and Differentiation in the Root Meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Moubayidin, L.; Perilli, S.; Dello Ioio, R.; Di Mambro, R.; Costantino, P.; Sabatini, S. The Rate of Cell Differentiation Controls the Arabidopsis Root Meristem Growth Phase. Curr. Biol. 2010, 20, 1138–1143. [Google Scholar] [CrossRef]
- Moubayidin, L.; Di Mambro, R.; Sozzani, R.; Pacifici, E.; Salvi, E.; Terpstra, I.; Bao, D.; van Dijken, A.; Dello Ioio, R.; Perilli, S.; et al. Spatial coordination between stem cell activity and cell differentiation in the root meristem. Dev. Cell 2013, 26, 405–415. [Google Scholar] [CrossRef]
- Takatsuka, H.; Umeda, M. ABA inhibits root cell elongation through repressing the cytokinin signaling. Plant Signal. Behav. 2019, 14, e1578632. [Google Scholar] [CrossRef]
- Barbez, E.; Dünser, K.; Gaidora, A.; Lendl, T.; Busch, W. Auxin steers root cell expansion via apoplastic pH regulation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2017, 114, E4884–E4893. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, T.; Korver, R.A.; Testerink, C.; ten Tusscher, K.H.W.J. Modeling halotropism: A key role for root tip architecture and reflux loop remodeling in redistributing auxin. Dev. Camb. Engl. 2016, 143, 3350–3362. [Google Scholar] [CrossRef] [PubMed]
- Di Mambro, R.; De Ruvo, M.; Pacifici, E.; Salvi, E.; Sozzani, R.; Benfey, P.N.; Busch, W.; Novak, O.; Ljung, K.; Di Paola, L.; et al. Auxin minimum triggers the developmental switch from cell division to cell differentiation in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2017, 114, E7641–E7649. [Google Scholar] [CrossRef] [PubMed]
- Blob, B.; Heo, J.; Helariutta, Y. Phloem differentiation: An integrative model for cell specification. J. Plant Res. 2018, 131, 31–36. [Google Scholar] [CrossRef] [PubMed]
- Durgaprasad, K.; Roy, M.V.; Venugopal, A.; Kareem, A.; Raj, K.; Willemsen, V.; Mähönen, A.P.; Scheres, B.; Prasad, K. Gradient Expression of Transcription Factor Imposes a Boundary on Organ Regeneration Potential in Plants. Cell Rep. 2019, 29, 453–463.e3. [Google Scholar] [CrossRef] [PubMed]
- Salvi, E.; Rutten, J.P.; Di Mambro, R.; Polverari, L.; Licursi, V.; Negri, R.; Dello Ioio, R.; Sabatini, S.; Ten Tusscher, K. A Self-Organized PLT/Auxin/ARR-B Network Controls the Dynamics of Root Zonation Development in Arabidopsis thaliana. Dev. Cell 2020, 53, 431–443.e23. [Google Scholar] [CrossRef] [PubMed]
- Laskowski, M.; Biller, S.; Stanley, K.; Kajstura, T.; Prusty, R. Expression Profiling of Auxin-treated Arabidopsis Roots: Toward a Molecular Analysis of Lateral Root Emergence. Plant Cell Physiol. 2006, 47, 788–792. [Google Scholar] [CrossRef] [PubMed]
- Carrier, D.J.; Bakar, N.T.A.; Swarup, R.; Callaghan, R.; Napier, R.M.; Bennett, M.J.; Kerr, I.D. The Binding of Auxin to the Arabidopsis Auxin Influx Transporter AUX1. Plant Physiol. 2008, 148, 529–535. [Google Scholar] [CrossRef] [PubMed]
- Petrášek, J.; Mravec, J.; Bouchard, R.; Blakeslee, J.J.; Abas, M.; Seifertová, D.; Wiśniewska, J.; Tadele, Z.; Kubeš, M.; Čovanová, M.; et al. PIN Proteins Perform a Rate-Limiting Function in Cellular Auxin Efflux. Science 2006, 312, 914–918. [Google Scholar] [CrossRef] [PubMed]
- Wiśniewska, J.; Xu, J.; Seifertová, D.; Brewer, P.B.; Růžička, K.; Blilou, I.; Rouquié, D.; Benková, E.; Scheres, B.; Friml, J. Polar PIN Localization Directs Auxin Flow in Plants. Science 2006, 312, 883. [Google Scholar] [CrossRef]
- Band, L.R.; Wells, D.M.; Fozard, J.A.; Ghetiu, T.; French, A.P.; Pound, M.P.; Wilson, M.H.; Yu, L.; Li, W.; Hijazi, H.I.; et al. Systems Analysis of Auxin Transport in the Arabidopsis Root Apex. Plant Cell 2014, 26, 862–875. [Google Scholar] [CrossRef]
- Grieneisen, V.A.; Xu, J.; Marée, A.F.M.; Hogeweg, P.; Scheres, B. Auxin transport is sufficient to generate a maximum and gradient guiding root growth. Nature 2007, 449, 1008–1013. [Google Scholar] [CrossRef]
- Zhao, Y.; Christensen, S.K.; Fankhauser, C.; Cashman, J.R.; Cohen, J.D.; Weigel, D.; Chory, J. A Role for Flavin Monooxygenase-Like Enzymes in Auxin Biosynthesis. Science 2001, 291, 306–309. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.; Gao, S.; Tian, H.; Wu, W.; Robert, H.S.; Ding, Z. Local Transcriptional Control of YUCCA Regulates Auxin Promoted Root-Growth Inhibition in Response to Aluminium Stress in Arabidopsis. PLOS Genet. 2016, 12, e1006360. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Dai, X.; De-Paoli, H.; Cheng, Y.; Takebayashi, Y.; Kasahara, H.; Kamiya, Y.; Zhao, Y. Auxin Overproduction in Shoots Cannot Rescue Auxin Deficiencies in Arabidopsis Roots. Plant Cell Physiol. 2014, 55, 1072–1079. [Google Scholar] [CrossRef] [PubMed]
- Santuari, L.; Sanchez-Perez, G.F.; Luijten, M.; Rutjens, B.; Terpstra, I.; Berke, L.; Gorte, M.; Prasad, K.; Bao, D.; Timmermans-Hereijgers, J.L.P.M.; et al. The PLETHORA Gene Regulatory Network Guides Growth and Cell Differentiation in Arabidopsis Roots. Plant Cell Online 2016, 28, 2937–2951. [Google Scholar] [CrossRef]
- Moore, S.; Zhang, X.; Mudge, A.; Rowe, J.H.; Topping, J.F.; Liu, J.; Lindsey, K. Spatiotemporal modelling of hormonal crosstalk explains the level and patterning of hormones and gene expression in Arabidopsis thaliana wild-type and mutant roots. New Phytol. 2015, 207, 1110–1122. [Google Scholar] [CrossRef]
- Muraro, D.; Larrieu, A.; Lucas, M.; Chopard, J.; Byrne, H.; Godin, C.; King, J. A multi-scale model of the interplay between cell signalling and hormone transport in specifying the root meristem of Arabidopsis thaliana. J. Theor. Biol. 2016, 404, 182–205. [Google Scholar] [CrossRef]
- Pierdonati, E.; Unterholzner, S.J.; Salvi, E.; Svolacchia, N.; Bertolotti, G.; Dello Ioio, R.; Sabatini, S.; Di Mambro, R. Cytokinin-Dependent Control of GH3 Group II Family Genes in the Arabidopsis Root. Plants 2019, 8, 94. [Google Scholar] [CrossRef]
- Pacifici, E.; Mambro, R.D.; Ioio, R.D.; Costantino, P.; Sabatini, S. Acidic cell elongation drives cell differentiation in the Arabidopsis root. EMBO J. 2018, 37, e99134. [Google Scholar] [CrossRef]
- Ishida, T.; Fujiwara, S.; Miura, K.; Stacey, N.; Yoshimura, M.; Schneider, K.; Adachi, S.; Minamisawa, K.; Umeda, M.; Sugimoto, K. SUMO E3 Ligase HIGH PLOIDY2 Regulates Endocycle Onset and Meristem Maintenance in Arabidopsis. Plant Cell 2009, 21, 2284–2297. [Google Scholar] [CrossRef]
- Ljung, K.; Hull, A.K.; Kowalczyk, M.; Marchant, A.; Celenza, J.; Cohen, J.D.; Sandberg, G. Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol. Biol. 2002, 49, 249–272. [Google Scholar] [CrossRef]
- Blilou, I.; Xu, J.; Wildwater, M.; Willemsen, V.; Paponov, I.; Friml, J.; Heidstra, R.; Aida, M.; Palme, K.; Scheres, B. The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 2005, 433, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Ljung, K.; Hull, A.K.; Celenza, J.; Yamada, M.; Estelle, M.; Normanly, J.; Sandberg, G. Sites and Regulation of Auxin Biosynthesis in Arabidopsis Roots. Plant Cell 2005, 17, 1090–1104. [Google Scholar] [CrossRef] [PubMed]
- Rosa, N.M.; Pfeiffer, A.; Hill, K.; Locascio, A.; Bhalerao, R.P.; Miskolczi, P.; Grønlund, A.L.; Wanchoo-Kohli, A.; Thomas, S.G.; Bennett, M.J.; et al. Genome Wide Binding Site Analysis Reveals Transcriptional Coactivation of Cytokinin-Responsive Genes by DELLA Proteins. PLOS Genet. 2015, 11, e1005337. [Google Scholar]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, S.; Beis, D.; Wolkenfelt, H.; Murfett, J.; Guilfoyle, T.; Malamy, J.; Benfey, P.; Leyser, O.; Bechtold, N.; Weisbeek, P.; et al. An Auxin-Dependent Distal Organizer of Pattern and Polarity in the Arabidopsis Root. Cell 1999, 99, 463–472. [Google Scholar] [CrossRef]
- Bondarenko, V.E.; Bett, G.C.L.; Rasmusson, R.L. A model of graded calcium release and L-type Ca2+ channel inactivation in cardiac muscle. Am. J. Physiol. Heart Circ. Physiol. 2004, 286, H1154–H1169. [Google Scholar] [CrossRef][Green Version]
- Bondarenko, V.E.; Szigeti, G.P.; Bett, G.C.L.; Kim, S.-J.; Rasmusson, R.L. Computer model of action potential of mouse ventricular myocytes. Am. J. Physiol. Heart Circ. Physiol. 2004, 287, H1378–H1403. [Google Scholar] [CrossRef]
- Calderwood, A.; Lloyd, A.; Hepworth, J.; Tudor, E.H.; Jones, D.M.; Woodhouse, S.; Bilham, L.; Chinoy, C.; Williams, K.; Corke, F.; et al. Total FLC transcript dynamics from divergent paralogue expression explains flowering diversity in Brassica napus. New Phytol. 2021, 229, 3534–3548. [Google Scholar] [CrossRef]
- Jaeger, J.; Surkova, S.; Blagov, M.; Janssens, H.; Kosman, D.; Kozlov, K.N.; Myasnikova, E.; Vanario-Alonso, C.E.; Samsonova, M.; Sharp, D.H.; et al. Dynamic control of positional information in the early Drosophila embryo. Nature 2004, 430, 368–371. [Google Scholar] [CrossRef]
- Perrimon, N.; Pitsouli, C.; Shilo, B.-Z. Signaling Mechanisms Controlling Cell Fate and Embryonic Patterning. Cold Spring Harb. Perspect. Biol. 2012, 4, a005975. [Google Scholar] [CrossRef] [PubMed]
- Sokolowski, T.R.; Erdmann, T.; Wolde, P.R. ten Mutual Repression Enhances the Steepness and Precision of Gene Expression Boundaries. PLOS Comput. Biol. 2012, 8, e1002654. [Google Scholar] [CrossRef] [PubMed]
- Mir, M.; Stadler, M.R.; Ortiz, S.A.; Hannon, C.E.; Harrison, M.M.; Darzacq, X.; Eisen, M.B. Dynamic multifactor hubs interact transiently with sites of active transcription in Drosophila embryos. eLife 2018, 7, e40497. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.E.; Toettcher, J.E. Signaling Dynamics Control Cell Fate in the Early Drosophila Embryo. Dev. Cell 2019, 48, 361–370.e3. [Google Scholar] [CrossRef] [PubMed]
- Mellor, N.L.; Voß, U.; Janes, G.; Bennett, M.J.; Wells, D.M.; Band, L.R. Auxin fluxes through plasmodesmata modify root-tip auxin distribution. Development 2020, 147, dev181669. [Google Scholar] [CrossRef]
- Wu, G.; Cameron, J.N.; Ljung, K.; Spalding, E.P. A role for ABCB19-mediated polar auxin transport in seedling photomorphogenesis mediated by cryptochrome 1 and phytochrome B. Plant J. 2010, 62, 179–191. [Google Scholar] [CrossRef]
- Cho, M.; Cho, H.-T. The function of ABCB transporters in auxin transport. Plant Signal. Behav. 2013, 8, e22990. [Google Scholar] [CrossRef]
- Ding, Z.; Friml, J. Auxin regulates distal stem cell differentiation in Arabidopsis roots. Proc. Natl. Acad. Sci. USA 2010, 107, 12046–12051. [Google Scholar] [CrossRef]
- Shimotohno, A.; Heidstra, R.; Blilou, I.; Scheres, B. Root stem cell niche organizer specification by molecular convergence of PLETHORA and SCARECROW transcription factor modules. Genes Dev. 2018, 32, 1085–1100. [Google Scholar] [CrossRef]
- Müller, J.; Toev, T.; Heisters, M.; Teller, J.; Moore, K.L.; Hause, G.; Dinesh, D.C.; Bürstenbinder, K.; Abel, S. Iron-Dependent Callose Deposition Adjusts Root Meristem Maintenance to Phosphate Availability. Dev. Cell 2015, 33, 216–230. [Google Scholar] [CrossRef]
- Benitez-Alfonso, Y.; Faulkner, C.; Pendle, A.; Miyashima, S.; Helariutta, Y.; Maule, A. Symplastic Intercellular Connectivity Regulates Lateral Root Patterning. Dev. Cell 2013, 26, 136–147. [Google Scholar] [CrossRef]
- Hofhuis, H.; Laskowski, M.; Du, Y.; Prasad, K.; Grigg, S.; Pinon, V.; Scheres, B. Phyllotaxis and rhizotaxis in Arabidopsis are modified by three PLETHORA transcription factors. Curr. Biol. 2013, 23, 956–962. [Google Scholar] [CrossRef]
- De Boer, R.J.; Pagie, L. Great Integrator Differential Equations. Available online: http://tbb.bio.uu.nl/rdb/grindC/grind.pdf (accessed on 12 March 2021).
| Parameter | Value | Dimension |
|---|---|---|
| PAux | 0.1125 | []s−1 |
| dAux | 0.0001 | s−1 |
| KMPlts,YUC | 200 | [] |
| Influx | 0 | []s−1 |
| pPlts | 0.01312 | []s−1 |
| KMAuxin,Plts | 435 | [] |
| dPlts | 0.0000175 | s−1 |
| Parameter | Value | Dimension |
|---|---|---|
| TransApolar | 0.00001 | s−1 |
| TransPolar | 0.01 | s−1 |
| Influx | 0 | []s−1 |
| dilSCN | 0.00000875 | s−1 |
| dilTA | 0.0000175 | s−1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rutten, J.P.; Ten Tusscher, K.H. Bootstrapping and Pinning down the Root Meristem; the Auxin–PLT–ARR Network Unites Robustness and Sensitivity in Meristem Growth Control. Int. J. Mol. Sci. 2021, 22, 4731. https://doi.org/10.3390/ijms22094731
Rutten JP, Ten Tusscher KH. Bootstrapping and Pinning down the Root Meristem; the Auxin–PLT–ARR Network Unites Robustness and Sensitivity in Meristem Growth Control. International Journal of Molecular Sciences. 2021; 22(9):4731. https://doi.org/10.3390/ijms22094731
Chicago/Turabian StyleRutten, Jacob P., and Kirsten H. Ten Tusscher. 2021. "Bootstrapping and Pinning down the Root Meristem; the Auxin–PLT–ARR Network Unites Robustness and Sensitivity in Meristem Growth Control" International Journal of Molecular Sciences 22, no. 9: 4731. https://doi.org/10.3390/ijms22094731
APA StyleRutten, J. P., & Ten Tusscher, K. H. (2021). Bootstrapping and Pinning down the Root Meristem; the Auxin–PLT–ARR Network Unites Robustness and Sensitivity in Meristem Growth Control. International Journal of Molecular Sciences, 22(9), 4731. https://doi.org/10.3390/ijms22094731





