Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex
Abstract
:1. Introduction
2. Results
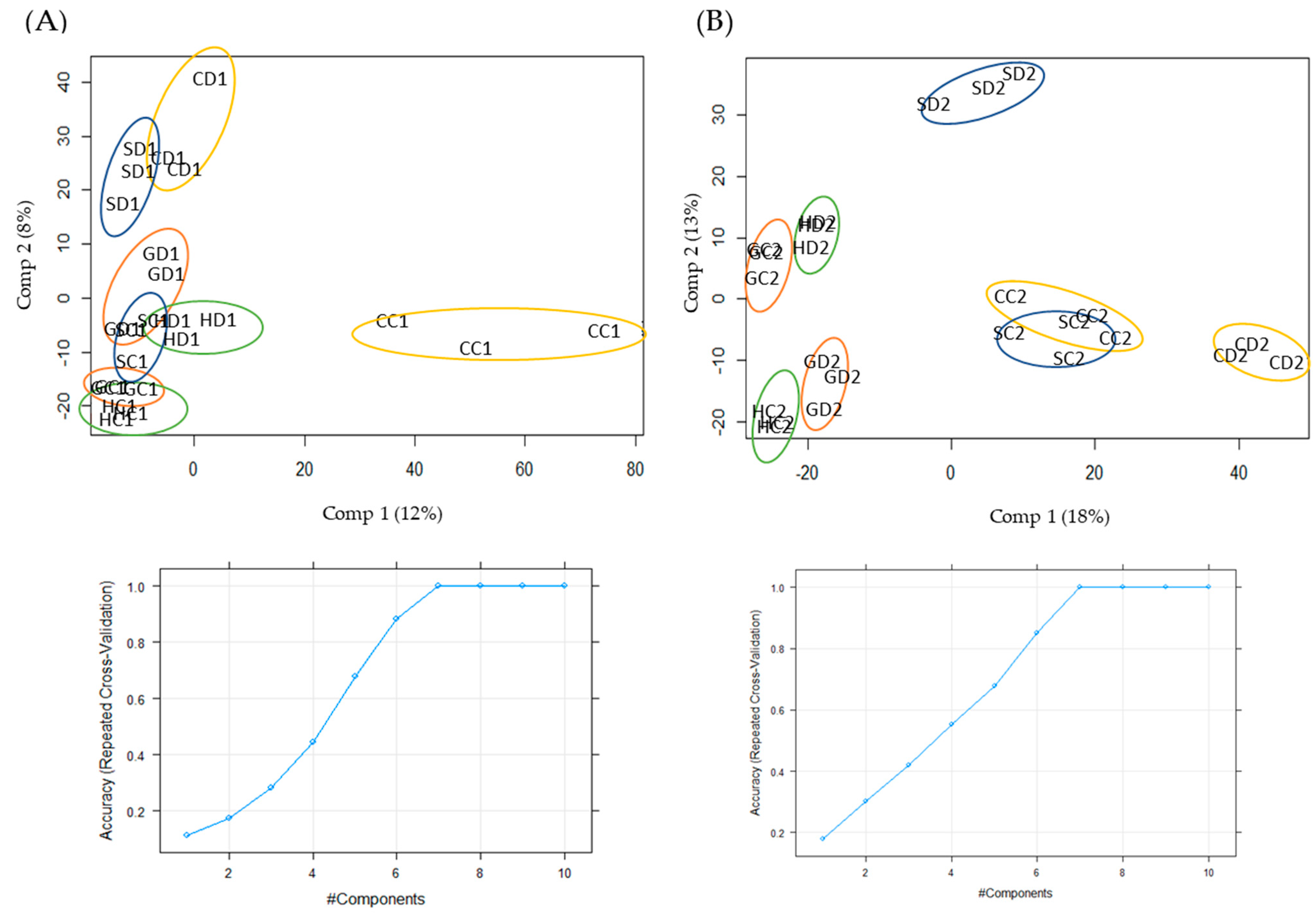
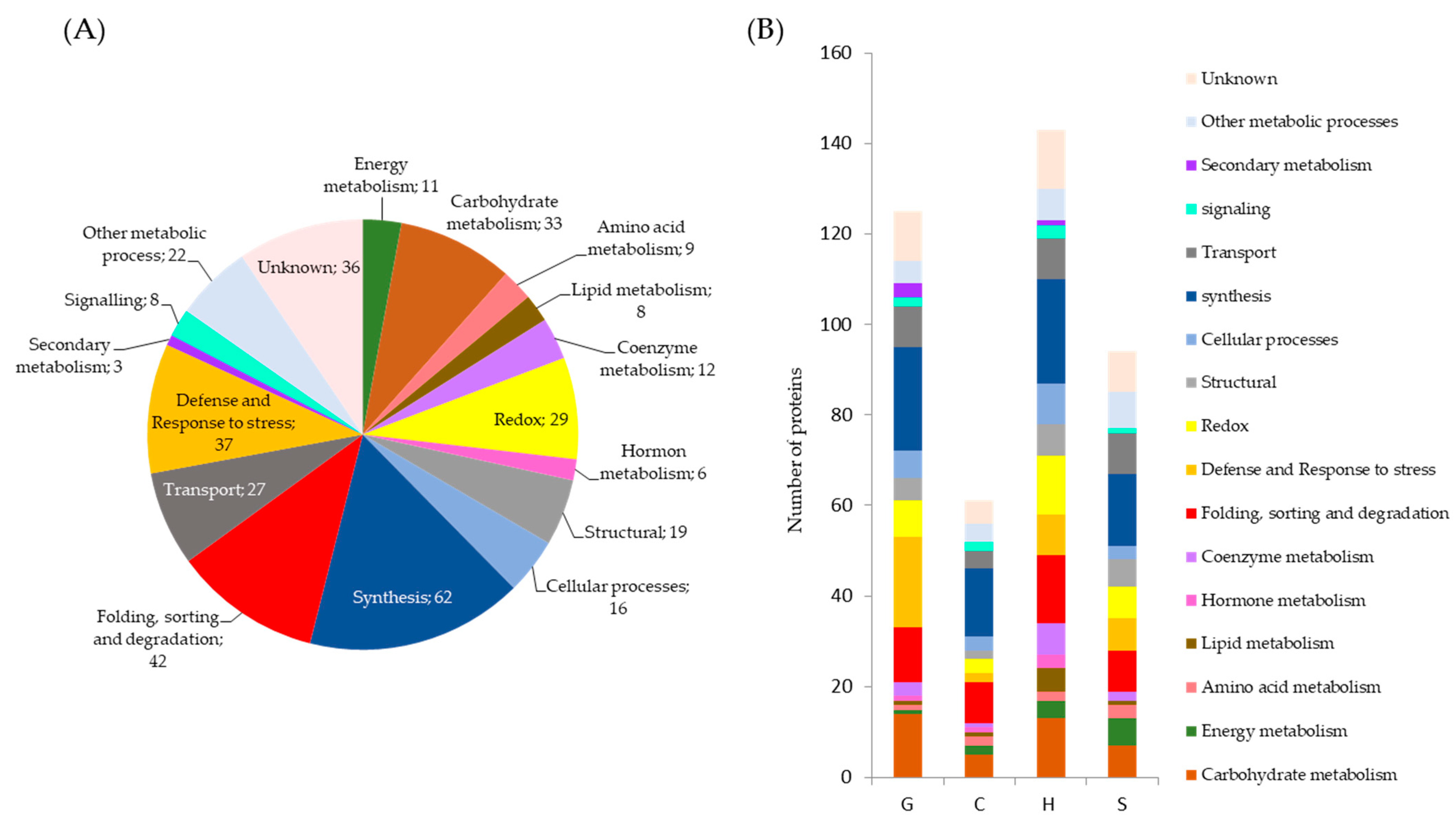
2.1. Qualitative and Quantitative Analysis of Drought Stress Responsive Proteins
2.2. Targeted Data Analysis for Selection of Peptides as Putative Markers of Drought Tolerance
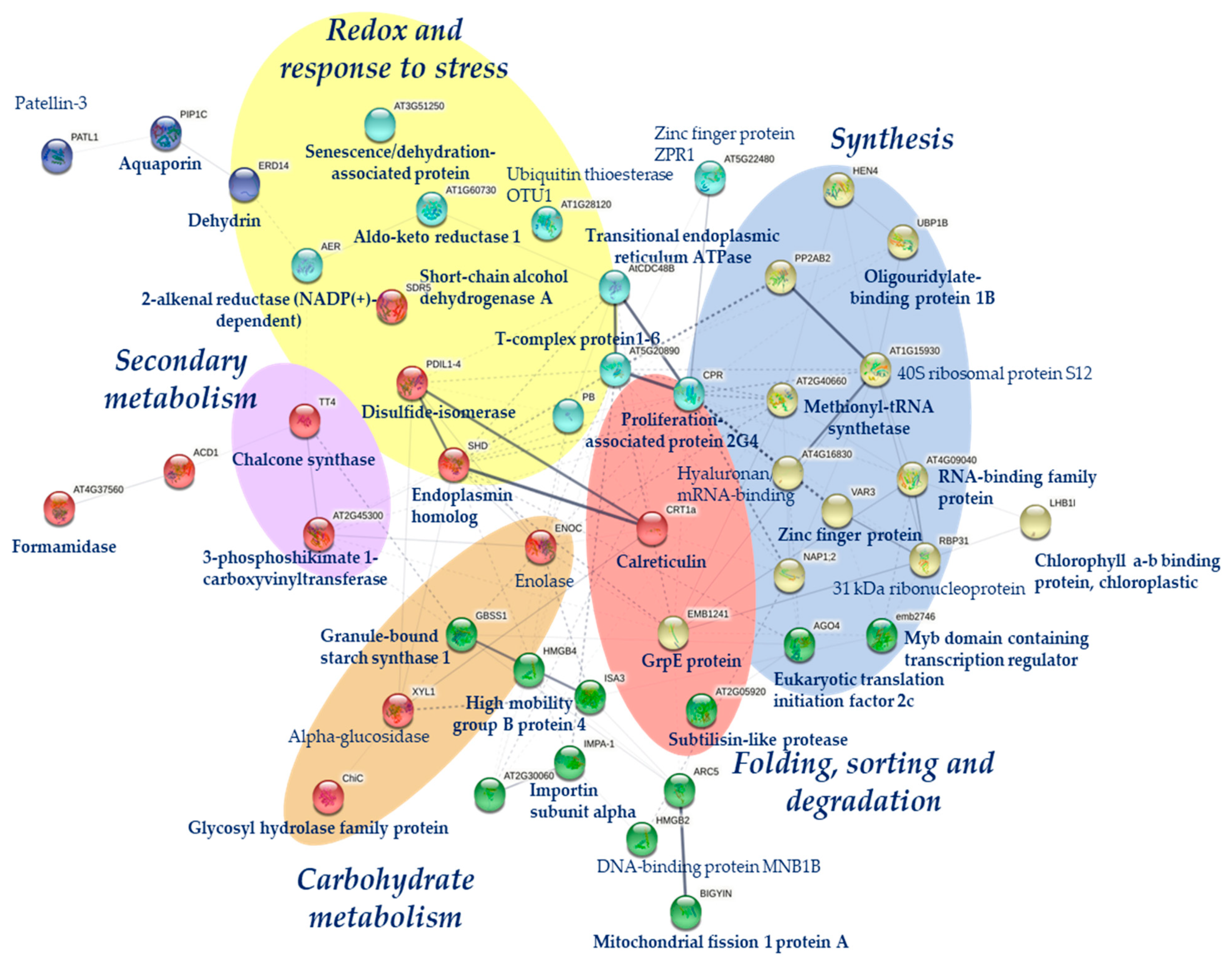
3. Discussion
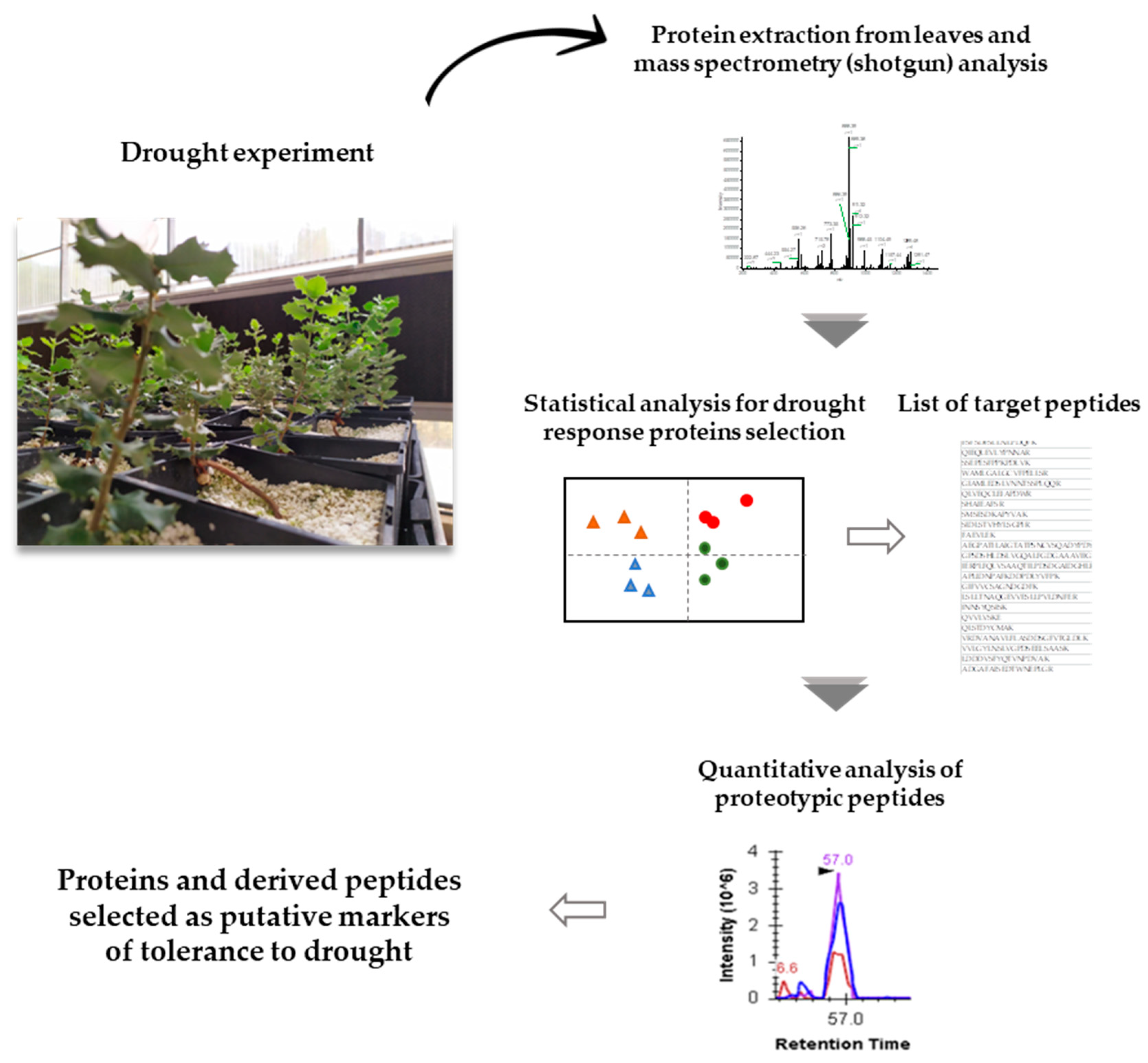
4. Materials and Methods
4.1. Proteomic Dataset
4.2. Protein Extraction and Mass Spectrometry Analysis
4.3. Protein Identification and Quantification
4.4. Statistical Analysis of Data and Selection of Target Proteins
4.5. Targeted Post-Acquisition Data Analysis for Selection of Putative Peptides Markers
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Olea, L.; San Miguel-Ayanz, A. The Spanish dehesa. A traditional Mediterranean silvopastoral system linking production and nature conservation. In Proceedings of the 21st General Meeting of the European Grassland Federation, Badajoz, Spain, 3–6 April 2006; pp. 1–15. [Google Scholar]
- Abril, N.; Gion, J.M.; Kerner, R.; Muller-Starck, G.; Cerrillo, R.M.; Plomion, C.; Renaut, J.; Valledor, L.; Jorrin-Novo, J.V. Proteomics research on forest trees, the most recalcitrant and orphan plant species. Phytochemistry 2011, 72, 1219–1242. [Google Scholar] [CrossRef]
- Crescente, M.F.; Gratani, L.; Larcher, W. Shoot growth efficiency and production of Quercus ilex L. in different climates. Flora Morphol. Distrib. Funct. Ecol. Plants 2002, 197, 2–9. [Google Scholar] [CrossRef]
- Echevarria-Zomeno, S.; Ariza, D.; Jorge, I.; Lenz, C.; Del Campo, A.; Jorrin, J.V.; Navarro, R.M. Changes in the protein profile of Quercus ilex leaves in response to drought stress and recovery. J. Plant. Physiol. 2009, 166, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Keenan, T.; Maria Serra, J.; Lloret, F.; Ninyerola, M.; Sabate, S. Predicting the future of forests in the Mediterranean under climate change, with niche- and process-based models: CO2 matters! Glob. Chang. Biol. 2011, 17, 565–579. [Google Scholar] [CrossRef]
- Castro-Diez, P.; Villar-Salvador, P.; Pérez-Rontomé, C.; Maestro-Martínez, M.; Montserrat-Martí, G. Leaf morphology and leaf chemical composition in three Quercus (Fagaceae) species along a rainfall gradient in NE Spain. Trees 1997, 11, 127–134. [Google Scholar] [CrossRef]
- Lumaret, R.; López de Heredia, U.; Soto, A. Origin and genetic variability. In Cork Oak Woodlands on the Edge: Ecology, Adaptive Management and Restoration; Island Press: Washington, DC, USA, 2009; pp. 25–32. [Google Scholar]
- Valero-Galván, J.; Navarro Cerrillo, R.M.; Romero-Rodríguez, M.C.; Ariza-Mateos, D.; Jorrín-Novo, J.V. Estudio de la respuesta al estrés hídrico en dos poblaciones de encina (Quercus ilex subsp. ballota (Desf.) Samp.) mediante una aproximación de proteómica comparativa basada en electroforesis bidimensional. In Proteómica; Jorrín, J.V., Vázquez, J., Eds.; SEPROT; Argos Press: Córdoba, Spain, 2010; Volume 5, pp. 156–157. Available online: http://www.seprot.es/wp-content/uploads/2016/05/Proteomica_Vol5.pdf (accessed on 1 March 2021).
- Valero Galvan, J.; Valledor, L.; Navarro Cerrillo, R.M.; Gil Pelegrin, E.; Jorrin-Novo, J.V. Studies of variability in Holm oak (Quercus ilex subsp. ballota [Desf.] Samp.) through acorn protein profile analysis. J. Proteom. 2011, 74, 1244–1255. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Valiente, J.A.; Lorenzo, Z.; Soto, A.; Valladares, F.; Gil, L.; Aranda, I. Elucidating the role of genetic drift and natural selection in cork oak differentiation regarding drought tolerance. Mol. Ecol. 2009, 18, 3803–3815. [Google Scholar] [CrossRef] [PubMed]
- Jorge, I.; Navarro, R.M.; Lenz, C.; Ariza, D.; Jorrin, J. Variation in the holm oak leaf proteome at different plant developmental stages, between provenances and in response to drought stress. Proteomics 2006, 6 (Suppl. S1), S207–S214. [Google Scholar] [CrossRef]
- Valero-Galvan, J.; Gonzalez-Fernandez, R.; Navarro-Cerrillo, R.M.; Gil-Pelegrin, E.; Jorrin-Novo, J.V. Physiological and proteomic analyses of drought stress response in Holm oak provenances. J. Proteome Res. 2013, 12, 5110–5123. [Google Scholar] [CrossRef]
- Jorrín-Novo, J.; Navarro-Cerrillo, R.M. Variabilidad y respuesta a distintos estreses en poblaciones de encina (Quercus ilex L.) en Andalucía mediante una aproximación proteómica. Ecosistemas 2014, 23, 99–107. [Google Scholar] [CrossRef]
- Rico, L.; Ogaya, R.; Terradas, J.; Penuelas, J. Community structures of N2 -fixing bacteria associated with the phyllosphere of a Holm oak forest and their response to drought. Plant. Biol. 2014, 16, 586–593. [Google Scholar] [CrossRef] [PubMed]
- Rivas-Ubach, A.; Barbeta, A.; Sardans, J.; Guenther, A.; Ogaya, R.; Oravec, M.; Urban, O.; Peñuelas, J. Topsoil depth substantially influences the responses to drought of the foliar metabolomes of Mediterranean forests. Perspect Plant. Ecol. Evol. Syst. 2016, 21, 41–54. [Google Scholar] [CrossRef] [Green Version]
- Guerrero-Sanchez, V.M.; Maldonado-Alconada, A.M.; Amil-Ruiz, F.; Jorrin-Novo, J.V. Holm Oak (Quercus ilex) Transcriptome. De novo Sequencing and Assembly Analysis. Front. Mol. Biosci. 2017, 4, 70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrero-Sanchez, V.M.; Maldonado-Alconada, A.M.; Amil-Ruiz, F.; Verardi, A.; Jorrin-Novo, J.V.; Rey, M.D. Ion Torrent and lllumina, two complementary RNA-seq platforms for constructing the holm oak (Quercus ilex) transcriptome. PLoS ONE 2019, 14, e0210356. [Google Scholar] [CrossRef] [PubMed]
- Fernández i Marti, A.; Romero-Rodríguez, C.; Navarro-Cerrillo, R.; Abril, N.; Jorrín-Novo, J.; Dodd, R. Population Genetic Diversity of Quercus ilex subsp. ballota (Desf.) Samp. Reveals Divergence in Recent and Evolutionary Migration Rates in the Spanish Dehesas. Forests 2018, 9, 337. [Google Scholar] [CrossRef] [Green Version]
- Lopez-Hidalgo, C.; Guerrero-Sanchez, V.M.; Gomez-Galvez, I.; Sanchez-Lucas, R.; Castillejo-Sanchez, M.A.; Maldonado-Alconada, A.M.; Valledor, L.; Jorrin-Novo, J.V. A Multi-Omics Analysis Pipeline for the Metabolic Pathway Reconstruction in the Orphan Species Quercus ilex. Front. Plant. Sci. 2018, 9, 935. [Google Scholar] [CrossRef] [Green Version]
- Natali, L.; Vangelisti, A.; Guidi, L.; Remorini, D.; Cotrozzi, L.; Lorenzini, G.; Nali, C.; Pellegrini, E.; Trivellini, A.; Vernieri, P.; et al. How Quercus ilex L. saplings face combined salt and ozone stress: A transcriptome analysis. BMC Genom. 2018, 19, 872. [Google Scholar] [CrossRef] [Green Version]
- Romero-Rodriguez, M.C.; Jorrin-Novo, J.V.; Castillejo, M.A. Toward characterizing germination and early growth in the non-orthodox forest tree species Quercus ilex through complementary gel and gel-free proteomic analysis of embryo and seedlings. J. Proteom. 2019, 197, 60–70. [Google Scholar] [CrossRef]
- Lopez-Hidalgo, C.; Trigueros, M.; Menendez, M.; Jorrin-Novo, J.V. Phytochemical composition and variability in Quercus ilex acorn morphotypes as determined by NIRS and MS-based approaches. Food Chem. 2021, 338, 127803. [Google Scholar] [CrossRef]
- Simova-Stoilova, L.P.; Romero-Rodriguez, M.C.; Sanchez-Lucas, R.; Navarro-Cerrillo, R.M.; Medina-Aunon, J.A.; Jorrin-Novo, J.V. 2-DE proteomics analysis of drought treated seedlings of Quercus ilex supports a root active strategy for metabolic adaptation in response to water shortage. Front. Plant. Sci. 2015, 6, 627. [Google Scholar] [CrossRef] [Green Version]
- Gomez-Galvez, I.; Sanchez-Lucas, R.; San-Eufrasio, B.; de Francisco, L.E.R.; Maldonado-Alconada, A.M.; Fuentes-Almagro, C.; Castillejo, M.A. Optimizing Shotgun Proteomics Analysis for a Confident Protein Identification and Quantitation in Orphan Plant Species: The Case of Holm Oak (Quercus ilex). Methods Mol. Biol. 2020, 2139, 157–168. [Google Scholar] [CrossRef]
- Plomion, C.; Aury, J.M.; Amselem, J.; Alaeitabar, T.; Barbe, V.; Belser, C.; Berges, H.; Bodenes, C.; Boudet, N.; Boury, C.; et al. Decoding the oak genome: Public release of sequence data, assembly, annotation and publication strategies. Mol. Ecol. Resour. 2016, 16, 254–265. [Google Scholar] [CrossRef] [PubMed]
- Ramos, A.M.; Usie, A.; Barbosa, P.; Barros, P.M.; Capote, T.; Chaves, I.; Simoes, F.; Abreu, I.; Carrasquinho, I.; Faro, C.; et al. The draft genome sequence of cork oak. Sci. Data 2018, 5, 180069. [Google Scholar] [CrossRef]
- Gillet, L.C.; Navarro, P.; Tate, S.; Rost, H.; Selevsek, N.; Reiter, L.; Bonner, R.; Aebersold, R. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: A new concept for consistent and accurate proteome analysis. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Domon, B.; Aebersold, R. Options and considerations when selecting a quantitative proteomics strategy. Nat. Biotechnol. 2010, 28, 710–721. [Google Scholar] [CrossRef]
- Escandon, M.; Jorrin-Novo, J.V.; Castillejo, M.A. Application and optimization of label-free shotgun approaches in the study of Quercus ilex. J. Proteom. 2021, 233, 104082. [Google Scholar] [CrossRef]
- Navarro-Cerrillo, R.M.; Ruiz Gomez, F.J.; Cabrera-Puerto, R.J.; Sánchez-Cuesta, R.; Palacios Rodriguez, G.; Quero Pérez, J.L. Growth and physiological sapling responses of eleven Quercus ilex ecotypes under identical environmental conditions. Ecol. Manag. 2018, 415–416, 58–69. [Google Scholar] [CrossRef]
- San-Eufrasio, B.; Sánchez-Lucas, R.; López-Hidalgo, C.; Guerrero-Sánchez, V.M.; Castillejo, M.Á.; Maldonado-Alconada, A.M.; Jorrín-Novo, J.V.; Rey, M.-D. Responses and Differences in Tolerance to Water Shortage under Climatic Dryness Conditions in Seedlings from Quercus spp. and Andalusian Q. ilex Populations. Forests 2020, 11, 707. [Google Scholar] [CrossRef]
- Lohse, M.; Nagel, A.; Herter, T.; May, P.; Schroda, M.; Zrenner, R.; Tohge, T.; Fernie, A.R.; Stitt, M.; Usadel, B. Mercator: A fast and simple web server for genome scale functional annotation of plant sequence data. Plant. Cell Environ. 2014, 37, 1250–1258. [Google Scholar] [CrossRef] [Green Version]
- Rodiger, A.; Baginsky, S. Tailored Use of Targeted Proteomics in Plant-Specific Applications. Front. Plant. Sci. 2018, 9, 1204. [Google Scholar] [CrossRef] [Green Version]
- Chawade, A.; Alexandersson, E.; Bengtsson, T.; Andreasson, E.; Levander, F. Targeted Proteomics Approach for Precision Plant Breeding. J. Proteome Res. 2016, 15, 638–646. [Google Scholar] [CrossRef] [Green Version]
- Buts, K.; Michielssens, S.; Hertog, M.L.; Hayakawa, E.; Cordewener, J.; America, A.H.; Nicolai, B.M.; Carpentier, S.C. Improving the identification rate of data independent label-free quantitative proteomics experiments on non-model crops: A case study on apple fruit. J. Proteom. 2014, 105, 31–45. [Google Scholar] [CrossRef] [PubMed]
- Riebel, M.; Fronk, P.; Distler, U.; Tenzer, S.; Decker, H. Proteomic profiling of German Dornfelder grape berries using data-independent acquisition. Plant. Physiol. Biochem. 2017, 118, 64–70. [Google Scholar] [CrossRef]
- Martin, L.B.; Sherwood, R.W.; Nicklay, J.J.; Yang, Y.; Muratore-Schroeder, T.L.; Anderson, E.T.; Thannhauser, T.W.; Rose, J.K.; Zhang, S. Application of wide selected-ion monitoring data-independent acquisition to identify tomato fruit proteins regulated by the CUTIN DEFICIENT2 transcription factor. Proteomics 2016, 16, 2081–2094. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mata, C.I.; Fabre, B.; Parsons, H.T.; Hertog, M.; Van Raemdonck, G.; Baggerman, G.; Van de Poel, B.; Lilley, K.S.; Nicolai, B.M. Ethylene Receptors, CTRs and EIN2 Target Protein Identification and Quantification Through Parallel Reaction Monitoring During Tomato Fruit Ripening. Front. Plant. Sci. 2018, 9, 1626. [Google Scholar] [CrossRef]
- Bose, U.; Byrne, K.; Howitt, C.A.; Colgrave, M.L. Targeted proteomics to monitor the extraction efficiency and levels of barley alpha-amylase trypsin inhibitors that are implicated in non-coeliac gluten sensitivity. J. Chromatogr. A 2019, 1600, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Bromilow, S.N.; Gethings, L.A.; Langridge, J.I.; Shewry, P.R.; Buckley, M.; Bromley, M.J.; Mills, E.N. Comprehensive Proteomic Profiling of Wheat Gluten Using a Combination of Data-Independent and Data-Dependent Acquisition. Front. Plant. Sci. 2016, 7, 2020. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, J.G.; Schilling, B. Clinical applications of quantitative proteomics using targeted and untargeted data-independent acquisition techniques. Expert. Rev. Proteom. 2017, 14, 419–429. [Google Scholar] [CrossRef] [PubMed]
- Castillejo, M.A.; Fondevilla-Aparicio, S.; Fuentes-Almagro, C.; Rubiales, D. Quantitative Analysis of Target Peptides Related to Resistance Against Ascochyta Blight (Peyronellaea pinodes) in Pea. J. Proteome Res. 2020, 19, 1000–1012. [Google Scholar] [CrossRef] [PubMed]
- Vaseva, I.; Sabotič, J.; Šuštar-Vozlič, J.; Meglič, V.; Kidrič, M.; Demirevska, K.; Simova-Stoilova, L. The response of plants to drought stress: The role of dehydrins, chaperones, proteases and protease inhibitors in maintaining cellular protein function. In Droughts: New Research; Neves, D.F., Sanz, J.D., Eds.; Nova Science Publishers, Inc.: New York, NY, USA, 2012; Volume 1, pp. 1–45. [Google Scholar]
- Sergeant, K.; Spiess, N.; Renaut, J.; Wilhelm, E.; Hausman, J.F. One dry summer: A leaf proteome study on the response of oak to drought exposure. J. Proteom. 2011, 74, 1385–1395. [Google Scholar] [CrossRef]
- González-Cruz, J.; Pastenes, C. Water-stress-induced thermotolerance of photosynthesis in bean (Phaseolus vulgaris L.) plants: The possible involvement of lipid composition and xanthophyll cycle pigments. Environ. Exp. Bot. 2012, 77, 127–140. [Google Scholar] [CrossRef]
- Cuellar-Ortiz, S.M.; De La Paz Arrieta-Montiel, M.; Acosta-Gallegos, J.; Covarrubias, A.A. Relationship between carbohydrate partitioning and drought resistance in common bean. Plant. Cell Environ. 2008, 31, 1399–1409. [Google Scholar] [CrossRef] [PubMed]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot. 2012, 63, 1593–1608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thalmann, M.; Santelia, D. Starch as a determinant of plant fitness under abiotic stress. New Phytol. 2017, 214, 943–951. [Google Scholar] [CrossRef] [Green Version]
- Kaplan, F.; Guy, C.L. RNA interference of Arabidopsis beta-amylase8 prevents maltose accumulation upon cold shock and increases sensitivity of PSII photochemical efficiency to freezing stress. Plant. J. 2005, 44, 730–743. [Google Scholar] [CrossRef]
- Yin, Y.G.; Kobayashi, Y.; Sanuki, A.; Kondo, S.; Fukuda, N.; Ezura, H.; Sugaya, S.; Matsukura, C. Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. ‘Micro-Tom’) fruits in an ABA- and osmotic stress-independent manner. J. Exp. Bot. 2010, 61, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Skirycz, A.; De Bodt, S.; Obata, T.; De Clercq, I.; Claeys, H.; De Rycke, R.; Andriankaja, M.; Van Aken, O.; Van Breusegem, F.; Fernie, A.R.; et al. Developmental stage specificity and the role of mitochondrial metabolism in the response of Arabidopsis leaves to prolonged mild osmotic stress. Plant. Physiol. 2010, 152, 226–244. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.J.; Liu, L.F.; Chen, C.K.; Chen, L.W. Regulations of granule-bound starch synthase I gene expression in rice leaves by temperature and drought stress. Biol. Plant. 2006, 50, 537–541. [Google Scholar] [CrossRef]
- Prathap, V.; Tyagi, A. Correlation between expression and activity of ADP glucose pyrophosphorylase and starch synthase and their role in starch accumulation during grain filling under drought stress in rice. Plant. Physiol. Biochem. 2020, 157, 239–243. [Google Scholar] [CrossRef]
- Sghaier-Hammami, B.; Valero-Galvan, J.; Romero-Rodriguez, M.C.; Navarro-Cerrillo, R.M.; Abdelly, C.; Jorrin-Novo, J. Physiological and proteomics analyses of Holm oak (Quercus ilex subsp. ballota [Desf.] Samp.) responses to Phytophthora cinnamomi. Plant. Physiol. Biochem. 2013, 71, 191–202. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of Phenylpropanoid Pathway and the Role of Polyphenols in Plants under Abiotic Stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [Green Version]
- Nogués, I.; Llusià, J.; Ogaya, R.; Munné-Bosch, S.; Sardans, J.; Peñuelas, J.; Loreto, F. Physiological and antioxidant responses of Quercus ilex to drought in two different seasons. Plant. Biosyst. Int. J. Deal. All Asp. Plant Biol. 2013, 148, 268–278. [Google Scholar] [CrossRef]
- Jafarnia, S.; Akbarinia, M.; Hosseinpour, B.; Modarres Sanavi, S.A.M.; Salami, S.A. Effect of drought stress on some growth, morphological, physiological, and biochemical parameters of two different populations of Quercus brantii. Iforest Biogeosciences For. 2018, 11, 212–220. [Google Scholar] [CrossRef]
- Ghanbary, E.; Tabari Kouchaksaraei, M.; Zarafshar, M.; Bader, K.M.; Mirabolfathy, M.; Ziaei, M. Differential physiological and biochemical responses of Quercus infectoria and Q. libani to drought and charcoal disease. Physiol. Plant 2020, 168, 876–892. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Ban, L.; Wen, H.; Wang, Z.; Dzyubenko, N.; Chapurin, V.; Gao, H.; Wang, X. An aquaporin protein is associated with drought stress tolerance. Biochem. Biophys. Res. Commun. 2015, 459, 208–213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillejo, M.A.; Iglesias-Garcia, R.; Wienkoop, S.; Rubiales, D. Label-free quantitative proteomic analysis of tolerance to drought in Pisum sativum. Proteomics 2016, 16, 2776–2787. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, W.; Vignani, R.; Scali, M.; Cresti, M. A universal and rapid protocol for protein extraction from recalcitrant plant tissues for proteomic analysis. Electrophoresis 2006, 27, 2782–2786. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of proteins utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Valledor, L.; Weckwerth, W. An Improved Detergent-Compatible Gel-Fractionation LC-LTQ-Orbitrap-MS Workflow for Plant and Microbial Proteomics. In Plant Proteomics. Methods in Molecular Biology (Methods and Protocols); Jorrin-Novo, J.K.S., Weckwerth, W., Wienkoop, S., Eds.; Humana Press: Totowa, NJ, USA, 2014; Volume 1072. [Google Scholar] [CrossRef]
- Al Shweiki, M.R.; Monchgesang, S.; Majovsky, P.; Thieme, D.; Trutschel, D.; Hoehenwarter, W. Assessment of Label-Free Quantification in Discovery Proteomics and Impact of Technological Factors and Natural Variability of Protein Abundance. J. Proteome Res. 2017, 16, 1410–1424. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: Improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Caret: Classification and Regression Training. R package version 6.0-86. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 1 March 2021).
- Altschu, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Chang, C.-Y.; Picotti, P.; Hüttenhain, R.; Heinzelmann-Schwarz, V.; Jovanovic, M.; Aebersold, R.; Vitek, O. Protein Significance Analysis in Selected Reaction Monitoring (SRM) Measurements. Mol. Cell Proteom. 2012, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suo, J.; Zhao, Q.; Zhang, Z.; Chen, S.; Cao, J.; Liu, G.; Wei, X.; Wang, T.; Yang, C.; Dai, S. Cytological and Proteomic Analyses of Osmunda cinnamomea Germinating Spores Reveal Characteristics of Fern Spore Germination and Rhizoid Tip Growth. Mol. Cell Proteom. 2015, 14, 2510–2534. [Google Scholar] [CrossRef] [PubMed] [Green Version]

| Protein ID | Peptide Sequences | Precursor m/z | Protein Description | Protein Function | Experimental Condition Showing Significant Change a | |||
|---|---|---|---|---|---|---|---|---|
| qilexprot_45247 | DAWDTSVLVEMK | 697,333 | Granule-bound starch synthase 1, chloroplastic/amyloplastic | Carbohydrate metabolism | S | G | ||
| FSFSDFSLLNLPDQFK | 952,971 | S | G | |||||
| QIEQLEVLYPNNAR | 843,940 | S | G | |||||
| qilexprot_71384 | SSEPESFPPKPDLVK | 828,924 | Glycosyl hydrolase family protein with chitinase insertion domain | Carbohydrate metabolism | H | S | ||
| qilexprot_18873 | WAMLGALGCVFPELLSR | 968,491 | Chlorophyll a-b binding protein, chloroplastic | Photosynthesis | H | S | ||
| qilexprot_32784 | GIAMLEDSLVNNTSSPLQQR | 1087,046 | Mitochondrial fission 1 protein A | Cellular processes | C | S | ||
| QLVEQCLEIAPDWR | 878,934 | C | S | |||||
| qilexprot_49492 | SHAIEAFSR | 509,256 | T-complex protein 1 subunit beta | Cellular processes | S | G | ||
| qilexprot_42362 | SMSESDKAPYVAK | 714,834 | High mobility group B protein 4 | Cellular processes | H | S | ||
| qilexprot_14200 | SIDLSTVHYLSGPIR | 552,964 | Formamidase | Other metabolic process | C | H | ||
| qilexprot_39395 | FAEVLEK | 418,228 | 3-phosphoshikimate 1-carboxyvinyltransferase | Other metabolic process | H | G | ||
| qilexprot_29542 | AEGPATILAIGTATPSNCVSQADYPDYYFR | 1083,173 | Chalcone synthase | Secondary metabolism | H | G | ||
| GPSDSHLDSLVGQALFGDGAAAVIIGADPDTK | 1031,844 | H | G | |||||
| IERPLFQLVSAAQTILPDSDGAIDGHLR | 1011,205 | H | G | |||||
| qilexprot_49771 | APLIDNPAFKDDPDLYVFPK | 1138,082 | Calreticulin | Folding, sorting and degradation | H | S | ||
| qilexprot_25223 | GIFVVCSAGNDGDFK | 793,366 | Subtilisin-like protease | Folding, sorting and degradation | C | S | G | |
| qilexprot_13677 | LSLLTNAQGEVVESLLPVLDNFER | 1328,700 | GrpE protein | Folding, sorting and degradation | C | H | S | |
| INNSYQSISK | 577,300 | C | H | S | ||||
| qilexprot_55000 | QVVLVSKE | 451,268 | 2-alkenal reductase (NADP(+)-dependent) | Redox | C | H | ||
| qilexprot_1533 | QLSTDYCMAK | 616,764 | Short-chain alcohol dehydrogenase A | Redox | C | S | ||
| VRDVANAVLFLASDDSGFVTGLDLK | 874,459 | C | S | |||||
| qilexprot_55764 | VVLGYLNSLVGPDSEELSAASK | 1124,585 | Protein disulfide-isomerase | Redox | H | S | ||
| LDDDVSFYQTVNPDVAK | 963,456 | H | S | |||||
| qilexprot_3345 | ADGAFAISEDTWNEPLGR | 974,952 | Endoplasmin homolog | Response to stress | H | S | ||
| EVTEEEYTK | 564,255 | H | S | |||||
| FYHSLAK | 433,228 | H | S | |||||
| YLNFLMGLVDSDTLPLNVSR | 1142,084 | H | S | |||||
| IAEEDPDEANDKDK | 794,849 | H | S | |||||
| qilexprot_72159 | NKDEHETTTTTTPGGNEGAVESK | 801,364 | Dehydrin | Response to stress | H | G | ||
| EEEMASEFEK | 614,752 | H | G | |||||
| qilexprot_48029 | ELAENLFPEDDTVSQTPTLQSSENVLVR | 1044,179 | Senescence/dehydration-associated protein AT3g51250 | Response to stress | C | S | ||
| qilexprot_8871 | KGCTPSQLALAWVHHQGK | 673,013 | Probable aldo-keto reductase 1 | Response to stress | H | S | ||
| qilexprot_19464 | VNWAYASGQR | 576,300 | Oligouridylate-binding protein | mRNA processing | C | G | ||
| SVVELTNGSSEDGK | 711,300 | C | G | |||||
| qilexprot_70616 | LITVTASENPDSR | 701,900 | BnaC03g49780D protein | mRNA processing | H | G | ||
| qilexprot_26698 | DKPESDGADLANK | 680,319 | Zinc finger protein VAR3, chloroplastic | mRNA processing | H | S | ||
| SVASNAIEWTGNASGSSVPDK | 524,745 | H | S | |||||
| qilexprot_2527 | IEDIDAYAPK | 567,784 | Myb domain containing transcription regulator | Synthesis | C | S | ||
| qilexprot_7552 | IVDVCEIGDSFIR | 761,800 | Proliferation-associated protein 2G4 | Synthesis | H | S | ||
| ALQLVVSECKPK | 686,383 | H | S | |||||
| qilexprot_56656 | ISFSGIDGKPEDVLNPK | 605,650 | Probable methionyl-tRNA synthetase | Synthesis | H | G | ||
| qilexprot_70881 | VQDTYDTELAGK | 670,319 | Eukaryotic translation initiation factor 2c | Synthesis | H | G | ||
| qilexprot_71168 | EDENRLDEVGYDDVGGVR | 679,305 | Transitional endoplasmic reticulum ATPase | Synthesis | H | G | ||
| qilexprot_68980 | EFFGSENNSLVSAQVIFHENPR | 840,737 | RNA-binding family protein | Synthesis | H | S | ||
| qilexprot_68567 | SPPINEVVQSGVVPR | 789,400 | Importin subunit alpha | Transport | H | S | ||
| qilexprot_4392 | GLYENSGGGANVVNHGYTK | 968,957 | Aquaporin | Transport | H | G | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
San-Eufrasio, B.; Bigatton, E.D.; Guerrero-Sánchez, V.M.; Chaturvedi, P.; Jorrín-Novo, J.V.; Rey, M.-D.; Castillejo, M.Á. Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex. Int. J. Mol. Sci. 2021, 22, 3191. https://doi.org/10.3390/ijms22063191
San-Eufrasio B, Bigatton ED, Guerrero-Sánchez VM, Chaturvedi P, Jorrín-Novo JV, Rey M-D, Castillejo MÁ. Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex. International Journal of Molecular Sciences. 2021; 22(6):3191. https://doi.org/10.3390/ijms22063191
Chicago/Turabian StyleSan-Eufrasio, Bonoso, Ezequiel Darío Bigatton, Victor M. Guerrero-Sánchez, Palak Chaturvedi, Jesús V. Jorrín-Novo, María-Dolores Rey, and María Ángeles Castillejo. 2021. "Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex" International Journal of Molecular Sciences 22, no. 6: 3191. https://doi.org/10.3390/ijms22063191
APA StyleSan-Eufrasio, B., Bigatton, E. D., Guerrero-Sánchez, V. M., Chaturvedi, P., Jorrín-Novo, J. V., Rey, M.-D., & Castillejo, M. Á. (2021). Proteomics Data Analysis for the Identification of Proteins and Derived Proteotypic Peptides of Potential Use as Putative Drought Tolerance Markers for Quercus ilex. International Journal of Molecular Sciences, 22(6), 3191. https://doi.org/10.3390/ijms22063191







