OsCRP1, a Ribonucleoprotein Gene, Regulates Chloroplast mRNA Stability That Confers Drought and Cold Tolerance
Abstract
1. Introduction
2. Results
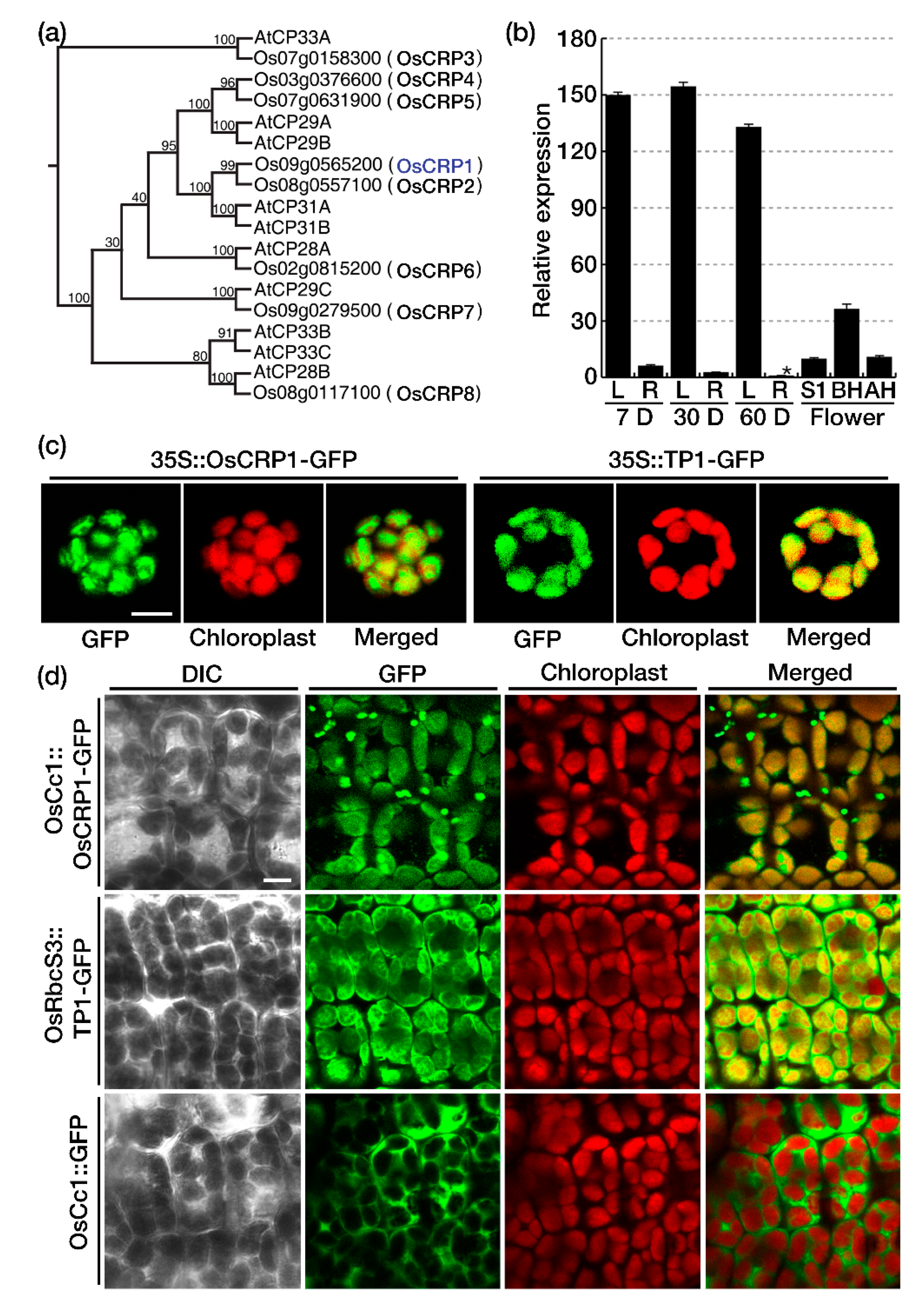
2.1. OsCRP1 Is a Rice Nuclear-Encoded and Chloroplast Targeting Ribonucleoprotein
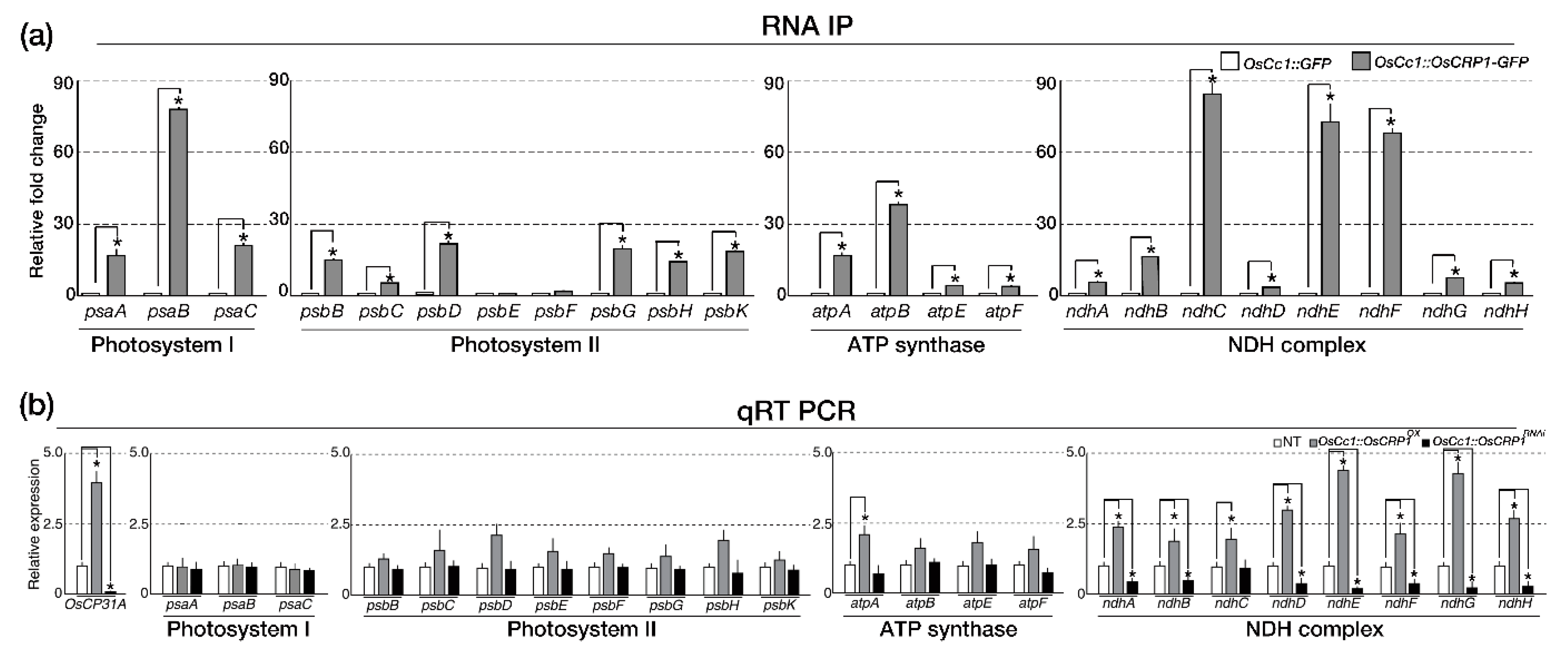
2.2. OsCRP1 Is Required for the Accumulation of Chloroplast mRNAs
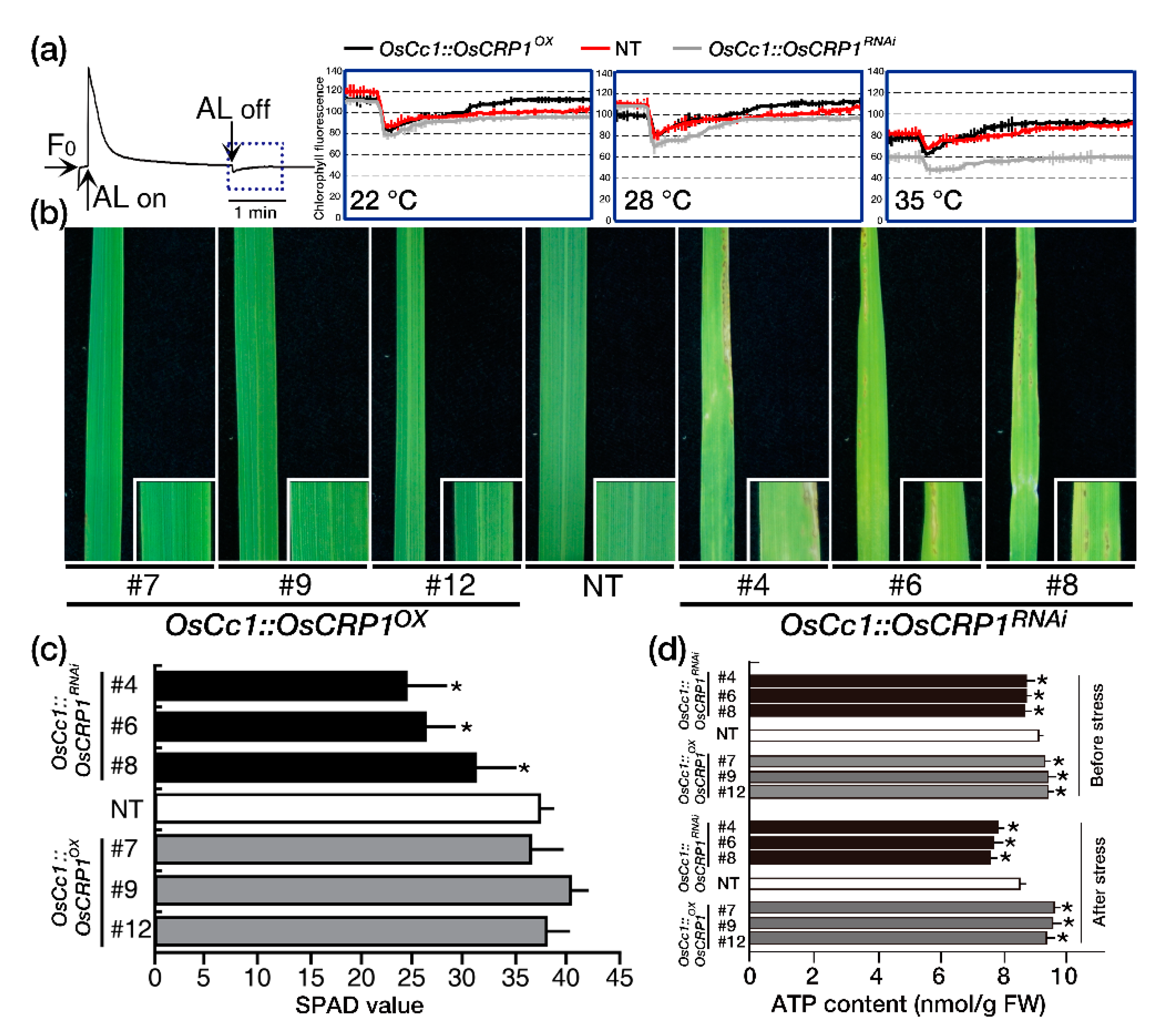
2.3. Down-Regulation of OsCRP1 Results in Chlorosis under Light Stress Conditions
2.4. Overexpression of OsCRP1 Confers Cold Stress Tolerance
2.5. Overexpression of OsCRP1 Confers Drought Stress Tolerance
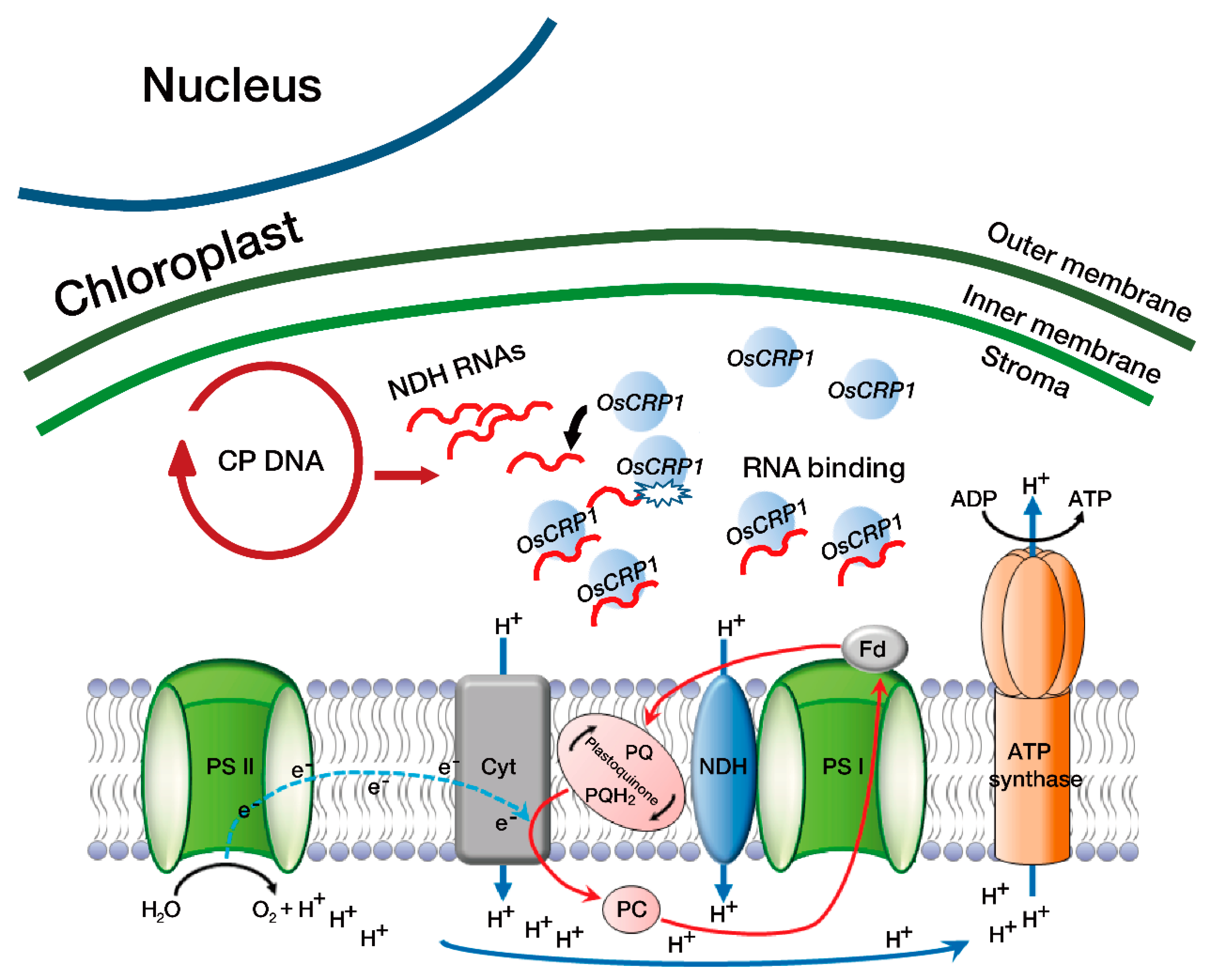
3. Discussion
4. Materials and Methods
4.1. Plasmid Construction and Agrobacterium-Mediated Rice Transformation
4.2. Subcellular Localization of OsCP31A
4.3. qRT-PCR Analysis
4.4. Stress Treatments and Tolerance Evaluation
4.5. RNA-Immunoprecipitation (RIP) Analysis
4.6. RNA-Seq
4.7. Analysis of NDH-Dependent CET
4.8. Determination of ATP Content
4.9. Accession Numbers
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Schmitz-Linneweber, C.; Barkan, A. RNA splicing and RNA editing in chloroplasts. In Cell and Molecular Biology of Plastids; Bock, R., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 213–248. [Google Scholar]
- Kroeger, T.S.; Watkins, K.P.; Friso, G.; van Wijk, K.J.; Barkan, A. A plant-specific RNA-binding domain revealed through analysis of chloroplast group II intron splicing. Proc. Natl. Acad. Sci. USA 2009, 106, 4537–4542. [Google Scholar] [CrossRef] [PubMed]
- Stern, D.B.; Goldschmidt-Clermont, M.; Hanson, M.R. Chloroplast RNA metabolism. Annu. Rev. Plant Biol. 2010, 61, 125–155. [Google Scholar] [CrossRef] [PubMed]
- Barkan, A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant Physiol. 2011, 155, 1520–1532. [Google Scholar] [CrossRef] [PubMed]
- Kotera, E.; Tasaka, M.; Shikanai, T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature 2005, 433, 326–330. [Google Scholar] [CrossRef] [PubMed]
- Okuda, K.; Myouga, F.; Motohashi, R.; Shinozaki, K.; Shikanai, T. Conserved domain structure of pentatricopeptide repeat proteins involved in chloroplast RNA editing. Proc. Natl. Acad. Sci. USA 2007, 104, 8178–8183. [Google Scholar] [CrossRef]
- Jacobs, J.; Kuck, U. Function of chloroplast RNA-binding proteins. Cell Mol. Life Sci. 2011, 68, 735–748. [Google Scholar] [CrossRef]
- Germain, A.; Hotto, A.M.; Barkan, A.; Stern, D.B. RNA processing and decay in plastids. Wiley Interdiscip. Rev. RNA 2013, 4, 295–316. [Google Scholar] [CrossRef]
- Li, Y.; Sugiura, M. Nucleic acid-binding specificities of tobacco chloroplast ribonucleoproteins. Nucleic Acids Res. 1991, 19, 2893–2896. [Google Scholar] [CrossRef]
- Li, Y.; Ye, L.; Sugita, M.; Sugiura, M. Tobacco nuclear gene for the 31 kd chloroplast ribonucleoprotein: Genomic organization, sequence analysis and expression. Nucleic Acids Res. 1991, 19, 2987–2991. [Google Scholar] [CrossRef][Green Version]
- Ye, L.; Sugiura, M. Domains required for nucleic acid binding activities in chloroplast ribonucleoproteins. Nucleic Acids Res. 1992, 20, 6275–6279. [Google Scholar] [CrossRef]
- Schuster, G.; Gruissem, W. Chloroplast mRNA 3 end processing requires a nuclear-encoded RNA-binding protein. EMBO J. 1991, 10, 1493–1502. [Google Scholar] [CrossRef]
- Lorkovic, Z.J.; Barta, A. Genome analysis: RNA recognition motif (RRM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arab. Thaliana. Nucleic Acids Res. 2002, 30, 623–635. [Google Scholar] [CrossRef]
- Ruwe, H.; Kupsch, C.; Teubner, M.; Schmitz-Linneweber, C. The RNA-recognition motif in chloroplasts. J. Plant Physiol. 2011, 168, 1361–1371. [Google Scholar] [CrossRef]
- Tillich, M.; Hardel, S.L.; Kupsch, C.; Armbruster, U.; Delannoy, E.; Gualberto, J.M.; Lehwark, P.; Leister, D.; Small, I.D.; Schmitz-Linneweber, C. Chloroplast ribonucleoprotein CP31A is required for editing and stability of specific chloroplast mRNAs. Proc. Natl. Acad. Sci. USA 2009, 106, 6002–6007. [Google Scholar] [CrossRef]
- Kupsch, C.; Ruwe, H.; Gusewski, S.; Tillich, M.; Small, I.; Schmitz-Linnewebera, C. Arabidopsis chloroplast RNA binding proteins CP31A and CP29A associate with large transcript pools and confer cold stress tolerance by influencing multiple chloroplast RNA processing steps. Plant Cell 2012, 24, 4266–4280. [Google Scholar] [CrossRef] [PubMed]
- Churin, Y.; Hess, W.; Boerner, T. Cloning and characterization of three cDNAs encoding chloroplast RNA binding proteins from barley: Differential regulation of expression by light and plastid development. Curr. Genet. 1999, 36, 173–181. [Google Scholar] [CrossRef] [PubMed]
- Kleffmann, T.; von Zychlinski, A.; Russenberger, D.; Hirsch-Hoffmann, M.; Gehrig, P.; Gruissem, W.; Baginsky, S. Proteome dynamicsduring plastid differentiation in rice. Plant Physiol. 2007, 143, 912–923. [Google Scholar] [CrossRef] [PubMed]
- Yamori, W.; Shikanai, T. Physiological functions of cyclic electron transport around Photosystem I in sustaining photosynthesis and plant growth. Annu. Rev. Plant Biol. 2016, 67, 81–106. [Google Scholar] [CrossRef] [PubMed]
- Seelert, H.; Poetsch, A.; Dencher, N.A.; Engel, A.; Stahlberg, H.; Müller, D.J. Structural biology: Proton-powered turbine of a plant motor. Nature 2000, 405, 418–419. [Google Scholar] [CrossRef]
- Wang, C.; Yamamoto, H.; Shikanai, T. Role of cyclic electron transport around photosystem I in regulating proton motive force. Biochim. Biophys. Acta. 2015, 1847, 931–938. [Google Scholar] [CrossRef]
- Yamori, W.; Makino, A.; Shikanai, T. A physiological role of cyclic electron transport around photosystem I in sustaining photosynthesis under fluctuating light in rice. Sci. Rep. 2016, 6, 20147. [Google Scholar] [CrossRef]
- Munekage, Y.; Hashimoto, M.; Miyake, C.; Tomizawa, K.I.; Endo, T.; Tasaka, M.; Shikanai, T. Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 2004, 429, 579–582. [Google Scholar] [CrossRef] [PubMed]
- Burrows, P.A.; Sazanov, L.A.; Svab, Z.; Maliga, P.; Nixon, P.J. Identification of a functional respiratory complex in chloroplasts through analysis of tobacco mutants containing disrupted plastid ndh genes. EMBO J. 1998, 17, 868–876. [Google Scholar] [CrossRef]
- Shikanai, T.; Endo, T.; Hashimoto, T.; Yamada, Y.; Asada, K.; Yokota, A. Directed disruption of the tobacco ndhB gene impairs cyclic electron flow around photosystem I. Proc. Natl. Acad. Sci. USA 1998, 95, 9705–9709. [Google Scholar] [CrossRef]
- DalCorso, G.; Pesaresi, P.; Masiero, S.; Aseeva, E.; Schünemann, D.; Finazzi, G.; Joliot, P.; Barbato, R.; Leister, D. A complex containing PGRL1 and PGR5 is involved in the switch between linear and cyclic electron flow in Arabidopsis. Cell 2008, 132, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Munekage, Y.; Hojo, M.; Meurer, J.; Endo, T.; Tasaka, M.; Shikanai, T. PGR5 is involved in cyclic electron flow around photosystem I and is essential for photoprotection in Arabidopsis. Cell 2002, 110, 361–371. [Google Scholar] [CrossRef]
- Yamori, W.; Sakata, N.; Suzuki, Y.; Shikanai, T.; Makino, A. Cyclic electron flow around photosystem I via chloroplast NAD (P) H dehydrogenase (NDH) complex performs a significant physiological role during photosynthesis and plant growth at low temperature in rice. Plant J. 2011, 68, 966–976. [Google Scholar] [CrossRef]
- Yamori, W.; Shikanai, T.; Makino, A. Photosystem I cyclic electron flow via chloroplast NADH dehydrogenase-like complex performs a physiological role for photosynthesis at low light. Sci. Rep. 2015, 5, 13908. [Google Scholar] [CrossRef] [PubMed]
- Munne-Bosch, S.; Shikanai, T.; Asada, K. Enhanced ferredoxin dependent cyclic electron flow around photosystem I and α-tocopherol quinone accumulation in water-stressed ndhB inactived tobacco mutants. Planta 2005, 222, 502–511. [Google Scholar] [CrossRef]
- He, Y.; Fu, J.; Yu, C.; Wang, X.; Jiang, Q.; Hong, J.; Lu, K.; Xue, G.; Yan, C.; James, A. Increasing cyclic electron flow is related to Na+ sequestration into vacuoles for salt tolerance in soybean. J. Exp. Bot. 2015, 66, 6877–6889. [Google Scholar] [CrossRef]
- Jang, I.C.; Choi, W.B.; Lee, K.H.; Song, S.I.; Nahm, B.H.; Kim, J.K. High-level and ubiquitous expression of the rice cytochrome c gene OsCc1 and its promoter activity in transgenic plants provides a useful promoter for transgenesis of monocots. Plant Physiol. 2002, 129, 1473–1481. [Google Scholar] [CrossRef] [PubMed]
- Endo, T.; Shikanai, T.; Takabayashi, A.; Asada, K.; Sato, F. The role of chloroplastic NAD(P)H dehydrogenase in photoprotection. FEBS Lett. 1999, 457, 5–8. [Google Scholar] [CrossRef]
- Horváth, E.M.; Peter, S.O.; Joët, T.; Rumeau, D.; Cournac, L.; Horváth, G.V.; Kavanagh, T.A.; Schäfer, C.; Peltier, G.; Medgyesy, P. Targeted inactivation of the plastid ndhB gene in tobacco results in an enhanced sensitivity of photosynthesis to moderate stomatal closure. Plant Physiol. 2000, 123, 1337–1350. [Google Scholar] [CrossRef]
- Li, X.-G.; Duan, W.; Meng, Q.-W.; Zou, Q.; Zhao, S.-J. The function of chloroplastic NAD (P) H dehydrogenase in tobacco during chilling stress under low irradiance. Plant Cell Physiol. 2004, 45, 103–108. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Duan, W.; Takabayashi, A.; Endo, T.; Shikanai, T.; Ye, J.Y.; Mi, H. Chloroplastic NAD(P)H dehydrogenase in tobacco leaves functions in alleviation of oxidative damage caused by temperature stress. Plant Physiol. 2006, 141, 465–474. [Google Scholar] [CrossRef]
- Zhang, R.; Sharkey, T.D. Photosynthetic electron transport and proton flux under moderate heat stress. Photosynth. Res. 2009, 100, 29–43. [Google Scholar] [CrossRef]
- Rumeau, D.; Peltier, G.; Cournac, L. Chlororespiration and cyclic electron flow around PSI during photosynthesis and plant stress response. Plant Cell Environ. 2007, 30, 1041–1051. [Google Scholar] [CrossRef] [PubMed]
- Hajihashemi, S.; Noedoost, F.; Geuns, J.M.; Djalovic, I.; Siddique, K.H. Effect of cold stress on photosynthetic traits, carbohydrates, morphology, and anatomy in nine cultivars of stevia rebaudiana. Front. Plant Sci. 2018, 9, 1430. [Google Scholar] [CrossRef]
- Wang, S.; Bai, G.; Wang, S.; Yang, L.; Yang, F.; Wang, Y.; Zhu, J.-K.; Hua, J. Chloroplast RNA-binding protein RBD1 promotes chilling tolerance through 23S rRNA processing in Arabidopsis. PLoS Genet. 2016, 12, e1006027. [Google Scholar] [CrossRef]
- Hirose, T.; Sugiura, M. Involvement of a site-specific trans-acting factor and a common RNA-binding protein in the editing of chloroplast mRNAs: Development of a chloroplast in vitro RNA editing system. EMBO J. 2001, 20, 1144–1152. [Google Scholar] [CrossRef]
- Teubner, M.; Fuß, J.; Kühn, K.; Krause, K.; Schmitz-Linneweber, C. The RNA recognition motif protein CP33A is a global ligand of chloroplast mRNAs and is essential for plastid biogenesis and plant development. Plant J. 2017, 89, 472–485. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. Central role of cyclic electron transport around photosystem I in the regulation of photosynthesis. Curr. Opin. Biotechnol. 2014, 26, 25–30. [Google Scholar] [CrossRef] [PubMed]
- Shikanai, T. Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase. Biochim. Biophys. Acta. 2016, 7, 1015–1022. [Google Scholar] [CrossRef]
- Kofer, W.; Koop, H.U.; Wanner, G.; Steinmuller, K. Mutagenesis of the genes encoding subunits A, C, H, I, J and K of the plastid NAD(P)H-plastoquinone-oxidoreductase in tobacco by polyethylene glycol-mediated plastome transformation. Mol. Gen. Genet. 1998, 258, 166–173. [Google Scholar] [CrossRef] [PubMed]
- Rumeau, D.; Becuwe-Linkaa, N.; Beylya, A.; Louwagieb, M.; Garinb, J.; Peltiera, G. New subunits NDH-M, -N, and -O, encoded by nuclear genes, are essential for plastid Ndh complex functioning in higher plants. Plant Cell 2005, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Livingston, A.K.; Cruz, J.A.; Kohzuma, K.; Dhingra, A.; Kramer, D.M. An Arabidopsis mutant with high cyclic electron flow around photosystem I (hcef) involving the NADPH dehydrogenase complex. Plant Cell 2010, 22, 221–233. [Google Scholar] [CrossRef]
- Hou, Q.Z.; Sun, K.; Zhang, H.; Su, X.; Fan, B.Q.; Feng, H.Q. The responses of photosystem II and intracellularATP production of Arabidopsis leaves to salt stress are affected by extracellular ATP. J. Plant Res. 2018, 131, 331–339. [Google Scholar] [CrossRef]
- Lee, D.-K.; Jung, H.; Jang, G.; Jeong, J.S.; Kim, Y.S.; Ha, S.-H.; Choi, Y.D.; Kim, J.-K. Overexpression of the OsERF71 transcription factor alters rice root structure and drought resistance. Plant Physiol. 2016, 172, 575–588. [Google Scholar] [CrossRef]
- Hiei, Y.; Ohta, S.; Komari, T.; Kumashiro, T. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 1994, 6, 271–282. [Google Scholar] [CrossRef]
- Park, S.H.; Jeong, J.S.; Choi, Y.D.; Kim, J.K. Characterization of the rice RbcS3 promoter and its transitpeptide for use in chloroplast-targeted expression. Plant Biotechnol. Rep. 2015, 9, 395–403. [Google Scholar] [CrossRef]
- Bang, S.W.; Lee, D.-K.; Jung, H.; Chung, P.J.; Kim, Y.S.; Choi, Y.D.; Suh, J.-W.; Kim, J.-K. Overexpression of OsTF1L, a rice HD-Zip transcription factor, promotes lignin biosynthesis and stomatal closure that improves drought tolerance. Plant Biotechnol. J. 2019, 17, 118–131. [Google Scholar] [CrossRef] [PubMed]
- Keene, J.D.; Komisarow, J.M.; Friedersdorf, M.B. RIP-Chip: The isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat. Protoc. 2006, 1, 302–307. [Google Scholar] [CrossRef] [PubMed]
- Peritz, T.; Zeng, F.; Kannanayakal, T.J.; Kilk, K.; Eiriksdottir, E.; Langel, U.; Eberwine, J. Immunoprecipitation of mRNA-protein complexes. Nat. Protoc. 2006, 1, 577–580. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bang, S.W.; Lee, H.S.; Park, S.-H.; Lee, D.-K.; Seo, J.S.; Kim, Y.S.; Park, S.-C.; Kim, J.-K. OsCRP1, a Ribonucleoprotein Gene, Regulates Chloroplast mRNA Stability That Confers Drought and Cold Tolerance. Int. J. Mol. Sci. 2021, 22, 1673. https://doi.org/10.3390/ijms22041673
Bang SW, Lee HS, Park S-H, Lee D-K, Seo JS, Kim YS, Park S-C, Kim J-K. OsCRP1, a Ribonucleoprotein Gene, Regulates Chloroplast mRNA Stability That Confers Drought and Cold Tolerance. International Journal of Molecular Sciences. 2021; 22(4):1673. https://doi.org/10.3390/ijms22041673
Chicago/Turabian StyleBang, Seung Woon, Ho Suk Lee, Su-Hyun Park, Dong-Keun Lee, Jun Sung Seo, Youn Shic Kim, Soo-Chul Park, and Ju-Kon Kim. 2021. "OsCRP1, a Ribonucleoprotein Gene, Regulates Chloroplast mRNA Stability That Confers Drought and Cold Tolerance" International Journal of Molecular Sciences 22, no. 4: 1673. https://doi.org/10.3390/ijms22041673
APA StyleBang, S. W., Lee, H. S., Park, S.-H., Lee, D.-K., Seo, J. S., Kim, Y. S., Park, S.-C., & Kim, J.-K. (2021). OsCRP1, a Ribonucleoprotein Gene, Regulates Chloroplast mRNA Stability That Confers Drought and Cold Tolerance. International Journal of Molecular Sciences, 22(4), 1673. https://doi.org/10.3390/ijms22041673





