Abstract
Hirschsprung disease is a congenital malformation where ganglia of the neural crest-derived enteric nervous system are missing over varying lengths of the distal gastrointestinal tract. This complex genetic condition involves both rare and common variants in dozens of genes, many of which have been functionally validated in animal models. Modifier loci present in the genetic background are also believed to influence disease penetrance and severity, but this has not been frequently tested in animal models. Here, we addressed this question using Holstein mice in which aganglionosis is due to excessive deposition of collagen VI around the developing enteric nervous system, thereby allowing us to model trisomy 21-associated Hirschsprung disease. We also asked whether the genetic background might influence the response of Holstein mice to GDNF enemas, which we recently showed to have regenerative properties for the missing enteric nervous system. Compared to Holstein mice in their original FVB/N genetic background, Holstein mice maintained in a C57BL/6N background were found to have a less severe enteric nervous system defect and to be more responsive to GDNF enemas. This change of genetic background had a positive impact on the enteric nervous system only, leaving the neural crest-related pigmentation phenotype of Holstein mice unaffected. Taken together with other similar studies, these results are thus consistent with the notion that the enteric nervous system is more sensitive to genetic background changes than other neural crest derivatives.
1. Introduction
Hirschsprung disease (HSCR) is a birth defect of the neurocristopathy class characterized by the lack of enteric neural ganglia (aganglionosis) in the distal bowel [1,2]. This occurs because neural crest-derived progenitors of the enteric nervous system (ENS) have failed to complete their colonization of the gastrointestinal tract during prenatal development [3]. In absence of ENS innervation, smooth muscles from the affected distal bowel segment remain tonically contracted, causing a functional intestinal obstruction (megacolon). Because the ENS influences epithelial barrier function as well, children with HSCR also have a high risk of enterocolitis leading to sepsis and premature death [4,5].
HSCR is a complex genetic condition characterized by non-Mendelian inheritance [1,6,7], as best exemplified by variation in disease penetrance and severity between family members carrying a similar set of mutations [8,9,10]. Current knowledge suggests that most cases are due to different combinations of rare coding variants, common regulatory variants, and/or copy-number variants in many genes important for proper colonization of the developing gastrointestinal tract by ENS progenitors [11,12]. Dozens of such genes have been identified so far [11,12,13,14,15,16,17,18,19]. The major HSCR-associated gene is RET (REarranged during Transfection) [20,21], which codes for a transmembrane tyrosine kinase that is activated by GDNF (Glial cell line-derived Neurotrophic Factor) upon binding to the co-receptor GFRα1 (GDNF family receptor alpha-1). Other genes that influence HSCR risk include transcription factors (SOX10, ZFHX1B, PHOX2B), EDNRB signaling pathway molecules (EDNRB, EDN3, ECE1), cell adhesion proteins (L1CAM), guidance molecules (SEMA3D), molecules needed for cell-extracellular matrix interactions (ITGB1 and COL6), and diverse additional genes (BACE2, NRG1, ERBB2, ADAMTS17, ACSS2) [11,12]. Male sex and trisomy 21 are other contributing factors, increasing HSCR risk ~4-fold [7] and ~130-fold [22], respectively.
Influence of the genetic background on the expressivity of ENS defects has been reported not only for mouse models of HSCR (with loss-of-function mutation of Sox10, Ednrb, or Ret) [23,24,25] but also for other ENS disorders like intestinal neuronal dysplasia (with loss-of-function mutation of Tlx2 or Kif26a) [26,27]. Almost all these studies tested the C57BL/6 background (either 6N or 6J substrains), which was compared to other strains—like BALB/c, C3Fe, and 129S—that differed as a function of the study. The consensus emanating from these studies is that the C57BL/6 background is the most susceptible to develop ENS defects. Yet, these studies also revealed that this is not the case for all neural crest derivatives. The opposite was seen for neural crest-derived melanocytes in Sox10-mutant mice, which presented a more severe pigmentation defect in the C3Fe background compared to the C57BL/6 background [25]. A similar disconnection between ENS and melanocyte defects was also reported in a study investigating the impact of the genetic background on the phenotype of Ednrb-mutant rats [28].
The Holstein mouse model of HSCR is issued from a pigmentation-based forward genetic screen aimed at identifying neurocristopathy-associated loci in FVB/N mice [29]. This screen was based on the transgenic rescue of the albino mutation in the tyrosinase gene (Tyr) [30,31,32], an approach that can result in non-uniform patterns of pigmentation when the rescuing Tyr minigene is inserted in a neural crest-relevant locus [29]. Holstein mice display a fully penetrant recessive phenotype combining unpigmented fur with aganglionic megacolon [29,33]. This phenotype is due to insertional mutation of a CTCF insulator element upstream of Collagen-6 alpha-4 (Col6a4) on Chr.9, leading to neural crest-specific upregulation of this gene [33]. The resulting increase of total collagen VI protein levels interferes with ENS progenitor migration in homozygous Holstein embryos (HolTg/Tg) [33]. Our analysis of the most distal ENS-containing colon of patients with short-segment HSCR further revealed that collagen VI protein levels are especially elevated in children also having trisomy 21 [33], consistent with the presence of two collagen VI genes (COL6A1 and COL6A2) in a region of Chr.21 that increases HSCR risk when present in 3 copies [34].
Using this model of trisomy 21-associated HSCR (HolTg/Tg) and other models for male-biased (TashTTg/Tg) [35,36] and EDNRB mutation-associated (Ednrbs-l/s-l) [37] HSCR, we recently developed a new therapy for HSCR allowing us to generate a new ENS from tissue-resident ENS progenitors in the otherwise aganglionic colon [38]. Acute post-natal administration of the potent neurotrophic factor GDNF via rectal enemas was found to be sufficient not only for inducing ENS ganglia with both neurons and glia but also for globally improving colon structure and function, thereby preventing premature megacolon-associated death of a significant number of treated animals [38]. When exposed to GDNF in vitro, cultured explants of aganglionic bowel from children with HSCR also developed new neurons [38]. While the origin of GDNF-induced ENS ganglia is not yet fully understood, about a third of induced neural cells were found to arise from Schwann cells in extrinsic nerves [38], which are more abundant than normal in the aganglionic colon of both mice [24,33,35] and humans [39].
In the current study, we verified if (1) the C57BL/6N background could specifically worsen the ENS defect of HolTg/Tg mice as previously reported for other mouse models of HSCR and if (2) this could also impact response to GDNF enemas. Surprisingly, the ENS phenotype of HolTg/Tg mice maintained in the C57BL/6N background (hereafter referred to as HolTg/Tg[BL6]) was found to be less severe than for HolTg/Tg mice maintained in their original FVB/N background (hereafter referred to as HolTg/Tg[FVB]). Accordingly, the outcome of GDNF enemas in terms of resulting ENS density appeared better for HolTg/Tg[BL6] than for HolTg/Tg[FVB] mice. The change of genetic background specifically impacted the ENS, leaving the melanocyte defect of HolTg/Tg unaffected.
2. Results
2.1. Increased Survival of HolTg/Tg[BL6] Mice Compared to HolTg/Tg[FVB] Mice
To determine if changing the genetic background from FVB/N to C57BL/6N could influence the phenotype of Holstein mice, heterozygous HolTg/+[FVB] mice were backcrossed with C57BL/6N mice for nine generations. As previously reported for HolTg/+[FVB] mice [33], the resulting HolTg/+[BL6] presented large areas of unpigmented fur covering about half of their body (44.2 ± 2.2%; Figure 1a,b). Intercrosses of HolTg/+[BL6] mice yielded the expected Mendelian ratio of homozygous HolTg/Tg[BL6] mice, which were easily recognizable by their almost entirely unpigmented fur (Figure 1a,b) as previously described for HolTg/Tg[FVB] mice [33].
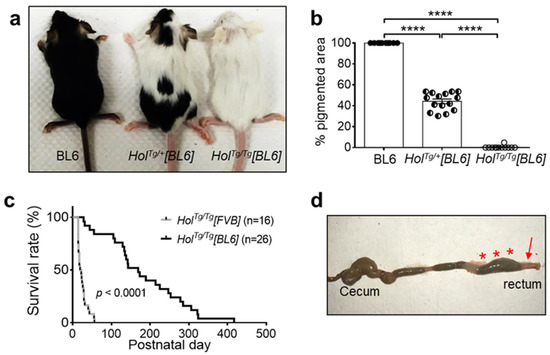
Figure 1.
The C57BL/6N genetic background delays megacolon-associated death in HolTg/Tg mice. (a,b) Allele dosage-dependent decrease of pigmentation in P20 HolTg/Tg[BL6] mice. **** p < 0.0001, one-way ANOVA with post-hoc Sidak’s test. (c) Comparison of survival rates between HolTg/Tg[BL6] and HolTg/Tg[FVB] mice. **** p < 0.0001, Mantel–Cox test. (d) HolTg/Tg[BL6] mice die from complications of megacolon, as evidenced in this euthanized P31 animal by the blockage in the distal colon and rectum (arrow) and accumulation of fecal material in more proximal regions (asterisks).
Yet, HolTg/Tg[BL6] mice markedly differed from HolTg/Tg[FVB] mice in terms of survival rate. Close monitoring of both colonies during the same period revealed an 8.2-fold increase in median survival age of HolTg/Tg[BL6] mice (169 days; interval of 28–417 days) compared to HolTg/Tg[FVB] mice (20.5 days; interval of 14–56 days) (Figure 1c). Regardless of genetic background, all dying HolTg/Tg mice exhibited typical megacolon-associated symptoms including abdominal distention, growth delay/weight loss, hunched posture, and ruffled fur. Accordingly, all of these mice had a narrow distal colon and enlarged mid/proximal colon (Figure 1d).
The increased survival of HolTg/Tg[BL6] mice further allowed us to test their fertility. All tested breeding pairs (n = 21) were fertile, producing an average of 6.2 ± 2.1 pups per litter. However, 43% (9/21) of reproductive HolTg/Tg[BL6] females eventually died of dystocia. At necropsy, all these females displayed fecal impaction, which appeared to compress the vagina. Altogether, these observations are consistent with the notion that the C57BL/6N genetic background specifically decreases the severity of the ENS defect of HolTg/Tg mice, without influencing their pigmentation defect.
2.2. Short-Segment Aganglionosis and Colonic Dysmotility in HolTg/Tg[BL6] Mice
To confirm that aganglionosis was the underlying cause of megacolon/fecal impaction in HolTg/Tg[BL6] mice, we examined their myenteric plexus at P20 via staining for acetylcholinesterase (AchE) activity (Figure 2a). As previously observed in HolTg/Tg[FVB] mice [33], the rectum of HolTg/Tg[BL6] mice is completely devoid of myenteric ganglia which are instead replaced by hypertrophic extrinsic nerve fibers (Figure 2a,b). This aganglionic segment is preceded by a transition zone with markedly decreased ENS density (25.4 ± 3.4% in mutants vs. 38.7 ± 1.1% in controls), while no overt differences are noted in the proximal colon (Figure 2a,b). In accordance with the increased survival in the C57BL/6N background (Figure 1c), we further found that the length of ENS-covered colon is longer for HolTg/Tg[BL6] mice (86.6 ± 1.2%; Figure 2c) than in HolTg/Tg[FVB] mice (74.2 ± 2.2%, [33]). Interestingly, this analysis also revealed that the majority of HolTg/Tg[BL6] mice had reached what we previously described as the minimal length of ENS innervation necessary to avoid blockage in the FVB/N background (between 78–84%, [35,36]; see dashed lines in Figure 1c), suggesting that the same threshold level applies in the C57BL/6N background.
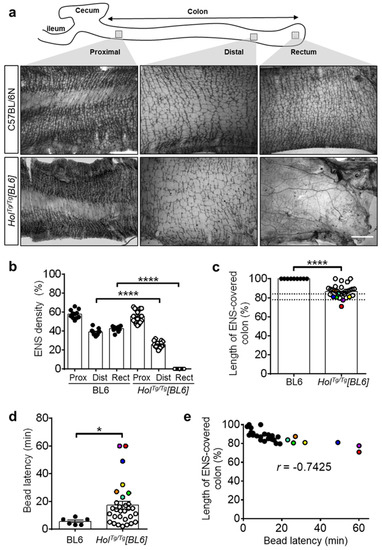
Figure 2.
HolTg/Tg[BL6] mice display short-segment aganglionosis and colonic dysmotility. (a) Staining of AChE activity in muscle strips from P20 mice, evidencing hypoganglionosis in the distal colon and aganglionosis combined to an overabundance of extrinsic nerve fibers in the rectum of HolTg/Tg[BL6]. Scale bar, 1 mm. (b) Quantitative analysis of the area occupied by the myenteric plexus (expressed in % of the image area), using images such as those displayed in (a). Each value is a field a view, for a minimum of 3 fields of view per region (n = 3–5 animals per group). Values for the aganglionic rectum of HolTg/Tg[BL6] mice were set at 0% by default, to avoid the confounder extrinsic nerves. **** p < 0.0001, one-way ANOVA with post-hoc Sidak’s test. (c) Quantitative analysis of the length of ENS-covered colon (in % of total colon length) in P20 mice. The dashed lines refer to the previously described threshold level interval beyond which megacolon is less likely to occur in FVB/N mice [35,36]. **** p < 0.0001, two-tailed Student’s t-test. (d) In vivo analysis of colonic motility in P20 mice using the bead latency test. Time to expel the glass bead after rectal insertion was capped at 60 min to simplify the analysis without impacting statistical significance. * p < 0.05, two-tailed Mann–Whitney U test. (e) Correlation between length of ENS-covered colon and time to expel the bead. Colored symbols were attributed to all HolTg/Tg[BL6] mice with latency time above average in panel d and used to highlight the same mice in panels c and e. r, Pearson’s correlation coefficient.
To verify the functional impact of short-segment aganglionosis in HolTg/Tg[BL6] mice, we analyzed colonic motility at P20 via the bead latency test (Figure 2d). Although most HolTg/Tg[BL6] mice were able to expel a rectally inserted glass bead, it took them much longer than for control C57BL/6N mice (17.1 ± 2.8 min vs. 5.5 ± 1.0 min, respectively). HolTg/Tg[BL6] mice that have not expelled the bead during the 60-min test were only rarely observed, whereas this was the norm for HolTg/Tg[FVB] mice [38]. Direct comparison with the length of the ENS-covered colon revealed a robust inverse correlation between time to expel the bead and the extent of aganglionosis (Figure 2e). All these data confirm that the ENS defect is less severe in HolTg/Tg[BL6] than in HolTg/Tg[FVB] mice.
2.3. Col6a4 Overexpression Is Less Extensive in HolTg/Tg[BL6] than in HolTg/Tg[FVB] Mice
Aganglionosis in HolTg/Tg[FVB] mice is due to transgene insertion-induced upregulation of Col6a4 and accompanying excessive, anti-migratory, secretion of collagen VI microfibrils by neural crest-derived ENS progenitors [33]. To determine the impact of the C57BL/6N background on this pathological mechanism, we first analyzed the extent of colonization by ENS progenitors via whole-mount immunofluorescence staining of βIII-tubulin. As previously reported for the HolTg/Tg[FVB] line [33], colonization by ENS progenitors was found to be less extensive in the colon of HolTg/Tg[BL6] embryos in comparison to WT controls (Figure 3). However, at both analyzed stages (e12.5 and e15.5), this phenotype again appears less severe in the C57BL/6N background (83% and 77% of WT levels at e12.5 and e15.5, respectively; Figure 3) compared to the FVB/N background (52% and 62% of WT levels at e12.5 and e15.5, respectively [33]).
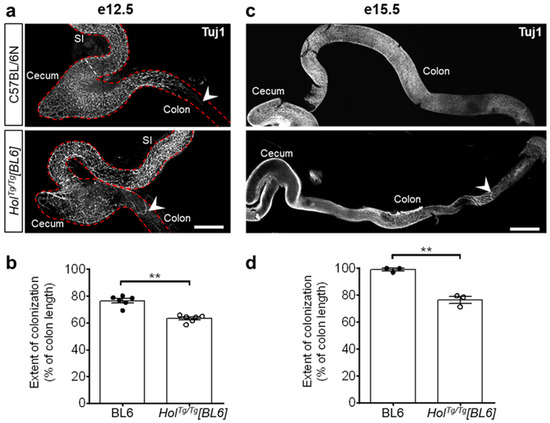
Figure 3.
Colonization by ENS progenitors has delayed in the colon of HolTg/Tg[BL6] embryos. (a,c) Immunofluorescence staining of βIII-Tubulin (Tuj1) in whole-mount preparations of embryonic guts at e12.5 (a) and e15.5 (c), with migratory front indicated by arrowheads. Scale bar, 400 µm (a) and 700 µm (c). (b,d) Quantification of extent of colonization by ENS progenitors (expressed in % of total colon length) at e12.5 (b) and e15.5 (d), using images such as those displayed in panels a and c. ** p < 0.01, two-tailed Student’s t-test.
We next sought to analyze Col6a4 expression levels by RT-qPCR, specifically in ENS progenitors. For this analysis, we thus introduced the Gata4p[5kb]-RFP (G4-RFP) transgene [40] in the HolTg/Tg[BL6] background by breeding—this transgene allowing to recover of RFP-positive ENS progenitors by fluorescence-activated cell sorting (FACS) [33,41]. Then, using e12.5 ENS progenitors recovered from these HolTg/Tg[BL6]; G4-RFP embryos as well as from HolTg/Tg[FVB]; G4-RFP embryos [33], we measured mRNA levels of Col6a4 and both of the collagen VI gene isoforms required for the production of collagen VI microfibrils (Col6a1 and Col6a2) [42]. In line with all other phenotypic data, our comparative analysis revealed a specific 2-fold decrease of Col6a4 expression levels in the C57BL/6N background compared to the FVB/N background, without any impact on Col6a1 and Col6a2 levels (Figure 4a). Using immunofluorescence, we further confirmed that this specific variation of Col6a4 expression levels similarly impacted the overall production of collagen VI microfibrils in e12.5 guts. Compared to respective WT controls, the immunofluorescence signal of collagen VI was found to be either doubled or unaffected in small intestines and cecum from HolTg/Tg[BL6] embryos (Figure 4b–e), whereas it was previously found to be tripled in all corresponding regions from HolTg/Tg[FVB] embryos [33]. These results strongly suggest that the Holstein ENS defect differs in FVB/N and C57BL/6N backgrounds because associated genetic differences are modulating the impact of the insertional mutation on Col6a4 gene expression.
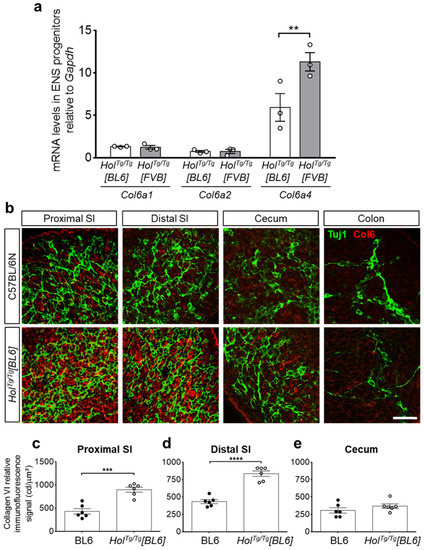
Figure 4.
The increased gene expression of Col6a4 and secretion of collagen VI microfibrils are both lower in HolTg/Tg[BL6] embryos than in HolTg/Tg[FVB] embryos. (a) RT-qPCR analysis of Col6a1, Col6a2, and Col6a4 in e12.5 ENS progenitors recovered by FACS from HolTg/Tg[BL6]; G4-RFP and HolTg/Tg[FVB]; G4-RFP embryos. ** p < 0.01, two-way ANOVA with post-hoc Sidak’s test. (b) Double immunofluorescence staining of collagen VI microfibrils (Col6, red) and βIII-tubulin+ neuronal progenitors (Tuj1, green) in e12.5 embryonic guts from control C57BL/6 and mutant HolTg/Tg[BL6] embryos. SI, small intestine. Scale bar, 20 µm. (c–e) Quantitative analysis of collagen VI immunofluorescence signal (in candella [cd] per µm2) in e12.5 C57BL/6 and HolTg/Tg[BL6] guts, using images such as those displayed in panel b. *** p < 0.001, **** p < 0.0001, two-way ANOVA with post-hoc Sidak’s test.
2.4. GDNF Enemas Restore Nearly Normal ENS Density and Function in HolTg/Tg[BL6] Mice
Having recently discovered that GDNF enemas can induce a new ENS in the otherwise aganglionic colon of several mouse models of HSCR (including HolTg/Tg[FVB]) [38], we wondered if the genetic background might influence the response to this treatment. To address this question, we used the exact same experimental approach as before and administered GDNF enemas (10 µL at 1 µg/µL) to HolTg/Tg[BL6] pups once daily between P4 and P8. Then, we analyzed the response to GDNF treatment via whole-mount immunofluorescence staining of HuC/D+ myenteric neurons at P20. As previously observed in HolTg/Tg[FVB] animals [38], GDNF treatment markedly increased the number of HuC/D+ myenteric neurons in HolTg/Tg[BL6] mice, reaching WT-like levels in ENS-containing colon (both proximal and distal) and about two-thirds of WT levels in the otherwise aganglionic rectum (Figure 5a and Figure S1). The magnitude of the response to GDNF enemas thus appears greater in HolTg/Tg[BL6] mice than in HolTg/Tg[FVB] mice, in which density of GDNF-induced ENS only reached about a third of WT levels in otherwise aganglionic distal colon [38].
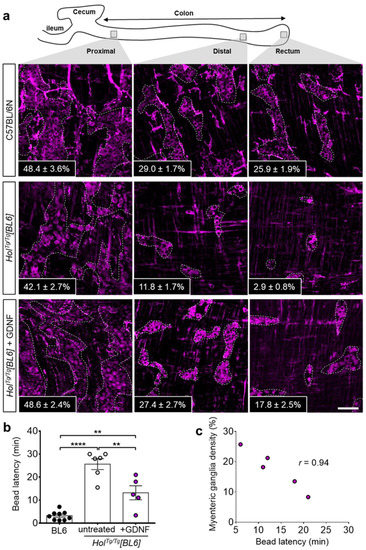
Figure 5.
GDNF-induced neurogenesis in the colon of HolTg/Tg[BL6] pups. (a) Immunofluorescence staining of HuC/D+ neurons in the myenteric plexus of P20 C57BL/6 controls and HolTg/Tg[BL6] mutants treated or not with GDNF between P4–P8. Insets refer to the average percentage of surface area occupied by HuC/D+ myenteric ganglia in 3–4 fields of view per corresponding region (n = 3–5 mice per group, as detailed in Figure S1). Scale bar, 100 µm. (b) In vivo analysis of colonic motility in P20 mice using the bead latency test. Time to expel the glass bead after rectal insertion was capped at 30 min to simplify the analysis without impacting statistical significance. ** p < 0.01, **** p < 0.0001, two-way ANOVA with post-hoc Sidak’s test. (c) Correlation between myenteric ganglia density and time to expel the bead for P20 HolTg/Tg[BL6] mice treated with GDNF between P4–P8.
To confirm the functionality of GDNF-induced myenteric ganglia, we analyzed colonic motility using the bead latency test. In contrast to untreated HolTg/Tg[BL6] mice for which one third (2/6) did not expel the bead before the end of the assay (average of 25.6 ± 2.4 min for the entire group), all GDNF-treated HolTg/Tg[BL6] mice did expel the bead (average of 13.2 ± 3.0 min) although not as quick as for control C57BL/6N mice (average of 3.2 ± 0.6 min) (Figure 5b). Strikingly, the time needed for GDNF-treated HolTg/Tg[BL6] mice to expel the bead was also found to be inversely correlated with the percentage of ENS density in the rectum (Figure 5c). This set of data suggests that GDNF-based correction of Holstein-associated ENS defect is more efficient in the C57BL/6N background than in the FVB/N background.
3. Discussion
In the current study, we first wanted to determine if the genetic background could influence the expressivity of aganglionic megacolon in mice bearing the Holstein insertional mutation, as observed for human HSCR. Transfer of the Holstein mutation from the FVB/N to the C57BL/6N background specifically decreased the severity of the collagen VI-dependent ENS defect of HolTg/Tg mice, without influencing its associated pigmentation defect. On one hand, this finding thus rules out the possibility that the C57BL/6 background is a general sensitized background for ENS defects, as previously suggested by similar analyses of mice mutated for Sox10, Ednrb, Tlx2, or Kif26a [23,25,26,27]. On the other hand, the observed genetic background-induced disconnection in severity between these two neural crest-related phenotypes replicates prior findings made in rodents bearing mutation of Sox10 or Ednrb [25,28]. These observations are consistent with the notion that the formation of the ENS is more sensitive to genetic variations than the melanocyte lineage. This most likely has something to do with the much greater number of genes required for building a complex structure like the ENS as opposed to the generation of a single cell lineage (melanocyte in this case). The corollary of this is that the greater genetic complexity associated with ENS formation obviously provides a richer source of phenotypically impactful variations.
Our Col6a4 RT-qPCR and collagen VI immunofluorescence data suggest that local genomic changes around the Holstein transgene insertion site might be one reason for the observed genetic background effect. The Holstein transgenic insertion is believed to perturb the insulation activity of a region enriched in CTCF binding motifs separating Col6a4 from a neighbor region highly transcribed in neural crest cells [33]. In agreement with this possibility, close examination of this CTCF-enriched region using the Ensembl Genome Browser revealed the presence of an indel variant immediately downstream of a CTCF ChIP-seq peak (Figure S2a). As confirmed by Sanger sequencing of DNA amplicons from our Holstein mouse colonies, the corresponding 12-bp sequence is present in the FVB/N background but absent in the C57BL/6N background (Figure S2b). The functional impact of this particular indel variant is currently unknown. However, previous studies in other pathological contexts suggest that variations flanking CTCF binding sites can influence the binding of CTCF proteins and thereby insulation activity of corresponding regions [43,44].
Variation in the pathological secretion of collagen VI also provides a plausible explanation as to why GDNF treatments appear more efficient in HolTg/Tg[BL6] mice compared to HolTg/Tg[FVB] mice. Indeed, the extracellular matrix is a major determinant of GDNF diffusion and activity [45,46]. Accordingly, levels of collagen VI in the diseased segment might affect both the distribution of rectally administered GDNF and the subsequent response of targeted ENS progenitors. Although the impact on GDNF diffusion in the colon is currently unknown, we do know that GDNF-induced migration of ENS progenitors from gut explants is more efficient when collagen VI levels are reduced [33]. Our data thus add credence to the growing belief that modulation of the extracellular matrix around ENS progenitors might have therapeutic value for enteric neuropathies [2,47].
4. Materials and Methods
4.1. Animals
Holstein (HolTg/Tg[FVB]) and Gata4p[5kb]-RFP (G4-RFP) single and double transgenics in the FVB/N background were as previously described [33,40]. Both transgenic alleles were transferred on the C57BL/6N background via consecutive backcrossing over 5 (for G4-RFP) to 9 (for Holstein) generations and resulting single transgenic lines were then intercrossed to generate double transgenics. Genotyping of adult HolTg/Tg[BL6] animals was made by visual inspection of coat color whereas genotyping of Holstein and G4-RFP alleles in embryonic tissues was performed by standard PCR using primers listed in Table S1. For embryo analyses, mice were mated during the night and noon on the day of vaginal plug detection was designated as e0.5. Euthanasia of pregnant dams or mice used for postnatal analyses was performed by CO2 inhalation following isoflurane-mediated anesthesia. Clinical-grade GDNF (Medgenesis Therapeutix Inc, Victoria, BC, Canada) was administered to some HolTg/Tg[BL6] pups via rectal enemas (10 µL of a 1 µg/µL solution in phosphate-buffered saline) once a day for 5 consecutive days from P4 to P8, as previously described [38].
4.2. Tissue Staining and Imaging
For postnatal tissues, whole colons were dissected from P20 mice, cut longitudinally along the mesentery, and fixed in 4% paraformaldehyde (PFA) overnight at 4 °C. Muscle layers and associated myenteric plexus were then microdissected from mucosa/submucosa and subsequently stained for acetylcholinesterase activity [33,48] or via immunofluorescence [38], as previously described. For embryonic tissues, whole intestines were dissected from e12.5 and e15.5 embryos, fixed in 4% PFA for 1 h at room temperature, and then processed for immunofluorescence staining, as previously described [33]. Details about all antibodies used in this study are listed in Table S2. All immunofluorescence images were acquired with 20×/60× objectives on a Nikon A1R confocal unit. To quantify immunofluorescence signal, relevant images were acquired using the exact same settings, and mean fluorescence intensity in candela/μm2 was determined within 20× fields of view using the Image J software, as previously described [33]. Images of acetylcholinesterase staining were acquired with a Leica DFC 495 camera mounted on a Leica M205 FA stereomicroscope (Leica Microsystems Canada, Vaughan, ON, Canada).
4.3. Bead Latency Test
Distal colonic motility was analyzed using an in vivo bead expulsion assay, as previously described [38,49]. Briefly, a 2-mm glass bead (Sigma-Aldrich, St. Louis, MO, USA) was inserted using a probe over a distance of 0.5 cm from the anus of overnight-fasted mice, under isoflurane anesthesia. Mice were then individually isolated in their cage without access to food and water, and colonic transit was determined by monitoring the time required for bead expulsion after insertion. The maximum time allowed for bead expulsion was set at either 30 or 60 min.
4.4. Fluorescence-Activated Cell Sorting (FACS) and RT-qPCR
Whole intestines were dissected from e12.5 embryos obtained from HolTg/+[FVB]; G4-RFP or HolTg/+[BL6]; G4-RFP intercrosses and individually dissociated at 37 °C using a cocktail of collagenase (0.4 mg/mL; Sigma C2674), dispase II (1.3 mg/mL; Life Technologies 17105-041, Carlsbad, CA, USA) and DNAse I (0.5 mg/mL; Sigma DN25), as previously described [33]. Single RFP-positive cells (~10,000 per intestine) were recovered from each preparation using a FACSJazz cell sorter (BD Biosciences, San Jose, CA, USA) and then kept frozen at −80 °C. Following PCR-based genotyping of embryo heads, relevant samples were individually processed for RNA extraction using the RNAeasy Plus purification mini kit (Qiagen, Germantown, MD, USA) and reverse transcription (with 50 ng of total RNA) using the Superscript III kit (Invitrogen, St. Louis, MO, USA), in accordance with manufacturers’ instructions. RT-qPCR analyses were performed with the Ssofast EvaGreen Supermix and C1000 Touch thermal cycler (BioRad, Hercules, CA, USA), in accordance with the manufacturer’s protocol. PCR consisted of 35 cycles of 20 s at 95 °C, 40 s at 60 °C, and 60 s at 72 °C. Quantitative gene expression relative to Gapdh was calculated by the 2−ΔΔCt method [50]. All primers used for RT-qPCR are described in Table S1.
4.5. Statistical Analysis
For all quantitative analyses, data are expressed as the mean ± standard error of the mean (SEM). The number of independent biological replicates (n) is indicated either in the figure or accompanying legend. The significance of differences was determined using relevant tests in GraphPad Prism, as indicated in figure legends. p values below 0.05 were considered statistically significant.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222313140/s1.
Author Contributions
Conceptualization, R.S. and N.P.; methodology, R.S.; formal analysis, R.S. and N.P.; investigation, R.S., N.L., G.B. (Grégoire Bonnamour), G.B. (Guillaume Bernas), A.B., M.P. and M.A.; writing—original draft preparation, R.S. and N.P.; writing—review and editing, N.P.; supervision, R.S. and N.P.; project administration, N.P.; funding acquisition, N.P. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Natural Science and Engineering Research Council of Canada (grant number RGPIN-2019-07076) and the Canadian Institutes of Health Research (grant number PJT-153264).
Institutional Review Board Statement
Experiments were performed according to the guidelines of the Canadian Council on Animal Care (CCAC) and approved by the relevant institutional review board (Comité institutionnel de protection des animaux; CIPA reference #878) of University of Quebec at Montreal (UQAM).
Data Availability Statement
All data are included in the current article and associated supplementary materials.
Acknowledgments
The authors thank Denis Flipo (Cell Analysis and Imaging research core, CERMO-FC) for help with confocal imaging, and MedGenesis Therapeutix for having generously provided clinical-grade human recombinant GDNF.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Heuckeroth, R.O. Hirschsprung disease—Integrating basic science and clinical medicine to improve outcomes. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Pilon, N. Treatment and Prevention of Neurocristopathies. Trends Mol. Med. 2021, 27, 451–468. [Google Scholar] [CrossRef]
- Bergeron, K.F.; Silversides, D.W.; Pilon, N. The developmental genetics of Hirschsprung’s disease. Clin. Genet. 2013, 83, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Gosain, A.; Brinkman, A.S. Hirschsprung’s associated enterocolitis. Curr Opin Pediatr. 2015, 27, 364–369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleinhaus, S.; Boley, S.J.; Sheran, M.; Sieber, W.K. Hirschsprung’s disease—A survey of the members of the Surgical Section of the American Academy of Pediatrics. J. Pediatr. Surg. 1979, 14, 588–597. [Google Scholar] [CrossRef]
- Alves, M.M.; Sribudiani, Y.; Brouwer, R.W.; Amiel, J.; Antinolo, G.; Borrego, S.; Ceccherini, I.; Chakravarti, A.; Fernandez, R.M.; Garcia-Barcelo, M.M.; et al. Contribution of rare and common variants determine complex diseases-Hirschsprung disease as a model. Dev. Biol. 2013, 382, 320–329. [Google Scholar] [CrossRef]
- Amiel, J.; Sproat-Emison, E.; Garcia-Barcelo, M.; Lantieri, F.; Burzynski, G.; Borrego, S.; Pelet, A.; Arnold, S.; Miao, X.; Griseri, P.; et al. Hirschsprung disease, associated syndromes and genetics: A review. J. Med. Genet. 2008, 45, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Attie, T.; Pelet, A.; Edery, P.; Eng, C.; Mulligan, L.M.; Amiel, J.; Boutrand, L.; Beldjord, C.; Nihoul-Fekete, C.; Munnich, A.; et al. Diversity of RET proto-oncogene mutations in familial and sporadic Hirschsprung disease. Hum. Mol. Genet. 1995, 4, 1381–1386. [Google Scholar] [CrossRef]
- Bolk, S.; Pelet, A.; Hofstra, R.M.; Angrist, M.; Salomon, R.; Croaker, D.; Buys, C.H.; Lyonnet, S.; Chakravarti, A. A human model for multigenic inheritance: Phenotypic expression in Hirschsprung disease requires both the RET gene and a new 9q31 locus. Proc. Natl. Acad. Sci. USA 2000, 97, 268–273. [Google Scholar] [CrossRef] [Green Version]
- Pingault, V.; Bondurand, N.; Kuhlbrodt, K.; Goerich, D.E.; Prehu, M.O.; Puliti, A.; Herbarth, B.; Hermans-Borgmeyer, I.; Legius, E.; Matthijs, G.; et al. SOX10 mutations in patients with Waardenburg-Hirschsprung disease. Nat. Genet. 1998, 18, 171–173. [Google Scholar] [CrossRef]
- Karim, A.; Tang, C.S.; Tam, P.K. The Emerging Genetic Landscape of Hirschsprung Disease and Its Potential Clinical Applications. Front. Pediatr. 2021, 9, 638093. [Google Scholar] [CrossRef]
- Luzon-Toro, B.; Villalba-Benito, L.; Torroglosa, A.; Fernandez, R.M.; Antinolo, G.; Borrego, S. What is new about the genetic background of Hirschsprung disease? Clin. Genet. 2020, 97, 114–124. [Google Scholar] [CrossRef] [Green Version]
- Gui, H.; Schriemer, D.; Cheng, W.W.; Chauhan, R.K.; Antinolo, G.; Berrios, C.; Bleda, M.; Brooks, A.S.; Brouwer, R.W.; Burns, A.J.; et al. Whole exome sequencing coupled with unbiased functional analysis reveals new Hirschsprung disease genes. Genome Biol. 2017, 18, 48. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, A.; Nandakumar, P.; Auer, D.R.; Sosa, M.X.; Ross, H.; Bollinger, J.; Yan, J.; Berrios, C.; Hirschsprung Disease Research, C.; Chakravarti, A. Multiple, independent, common variants at RET, SEMA3 and NRG1 gut enhancers specify Hirschsprung disease risk in European ancestry subjects. J. Pediatr. Surg. 2021. [Google Scholar] [CrossRef] [PubMed]
- Kuil, L.E.; MacKenzie, K.C.; Tang, C.S.; Windster, J.D.; Le, T.L.; Karim, A.; de Graaf, B.M.; van der Helm, R.; van Bever, Y.; Sloots, C.E.J.; et al. Size matters: Large copy number losses in Hirschsprung disease patients reveal genes involved in enteric nervous system development. PLoS Genet. 2021, 17, e1009698. [Google Scholar] [CrossRef]
- Mederer, T.; Schmitteckert, S.; Volz, J.; Martinez, C.; Roth, R.; Thumberger, T.; Eckstein, V.; Scheuerer, J.; Thoni, C.; Lasitschka, F.; et al. A complementary study approach unravels novel players in the pathoetiology of Hirschsprung disease. PLoS Genet. 2020, 16, e1009106. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.S.; Li, P.; Lai, F.P.; Fu, A.X.; Lau, S.T.; So, M.T.; Lui, K.N.; Li, Z.; Zhuang, X.; Yu, M.; et al. Identification of Genes Associated With Hirschsprung Disease, Based on Whole-Genome Sequence Analysis, and Potential Effects on Enteric Nervous System Development. Gastroenterology 2018, 155, 1908–1922. [Google Scholar] [CrossRef] [PubMed]
- Tang, C.S.; Zhuang, X.; Lam, W.Y.; Ngan, E.S.; Hsu, J.S.; Michelle, Y.U.; Man-Ting, S.O.; Cherny, S.S.; Ngo, N.D.; Sham, P.C.; et al. Uncovering the genetic lesions underlying the most severe form of Hirschsprung disease by whole-genome sequencing. Eur. J. Hum. Genet. 2018, 26, 818–826. [Google Scholar] [CrossRef] [Green Version]
- Tilghman, J.M.; Ling, A.Y.; Turner, T.N.; Sosa, M.X.; Krumm, N.; Chatterjee, S.; Kapoor, A.; Coe, B.P.; Nguyen, K.H.; Gupta, N.; et al. Molecular Genetic Anatomy and Risk Profile of Hirschsprung’s Disease. N. Engl. J. Med. 2019, 380, 1421–1432. [Google Scholar] [CrossRef]
- Edery, P.; Lyonnet, S.; Mulligan, L.M.; Pelet, A.; Dow, E.; Abel, L.; Holder, S.; Nihoul-Fekete, C.; Ponder, B.A.; Munnich, A. Mutations of the RET proto-oncogene in Hirschsprung’s disease. Nature 1994, 367, 378–380. [Google Scholar] [CrossRef]
- Romeo, G.; Ronchetto, P.; Luo, Y.; Barone, V.; Seri, M.; Ceccherini, I.; Pasini, B.; Bocciardi, R.; Lerone, M.; Kaariainen, H.; et al. Point mutations affecting the tyrosine kinase domain of the RET proto-oncogene in Hirschsprung’s disease. Nature 1994, 367, 377–378. [Google Scholar] [CrossRef] [PubMed]
- Friedmacher, F.; Puri, P. Hirschsprung’s disease associated with Down syndrome: A meta-analysis of incidence, functional outcomes and mortality. Pediatric Surg. Int. 2013, 29, 937–946. [Google Scholar] [CrossRef]
- Cantrell, V.A.; Owens, S.E.; Chandler, R.L.; Airey, D.C.; Bradley, K.M.; Smith, J.R.; Southard-Smith, E.M. Interactions between Sox10 and EdnrB modulate penetrance and severity of aganglionosis in the Sox10Dom mouse model of Hirschsprung disease. Hum. Mol. Genet. 2004, 13, 2289–2301. [Google Scholar] [CrossRef]
- McCallion, A.S.; Stames, E.; Conlon, R.A.; Chakravarti, A. Phenotype variation in two-locus mouse models of Hirschsprung disease: Tissue-specific interaction between Ret and Ednrb. Proc. Natl. Acad. Sci. USA 2003, 100, 1826–1831. [Google Scholar] [CrossRef] [Green Version]
- Southard-Smith, E.M.; Angrist, M.; Ellison, J.S.; Agarwala, R.; Baxevanis, A.D.; Chakravarti, A.; Pavan, W.J. The Sox10(Dom) mouse: Modeling the genetic variation of Waardenburg-Shah (WS4) syndrome. Genome Res. 1999, 9, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Ohara, Y.; Fujimura, L.; Sakamoto, A.; Teratake, Y.; Hiraoka, S.; Koseki, H.; Saito, T.; Terui, K.; Mitsunaga, T.; Nakata, M.; et al. Genetic background-dependent abnormalities of the enteric nervous system and intestinal function in Kif26a-deficient mice. Sci. Rep. 2021, 11, 3191. [Google Scholar] [CrossRef]
- Parisi, M.A.; Baldessari, A.E.; Iida, M.H.; Clarke, C.M.; Doggett, B.; Shirasawa, S.; Kapur, R.P. Genetic background modifies intestinal pseudo-obstruction and the expression of a reporter gene in Hox11L1-/- mice. Gastroenterology 2003, 125, 1428–1440. [Google Scholar] [CrossRef]
- Dang, R.; Torigoe, D.; Suzuki, S.; Kikkawa, Y.; Moritoh, K.; Sasaki, N.; Agui, T. Genetic background strongly modifies the severity of symptoms of Hirschsprung disease, but not hearing loss in rats carrying Ednrb(sl) mutations. PLoS ONE 2011, 6, e24086. [Google Scholar] [CrossRef]
- Pilon, N. Pigmentation-based insertional mutagenesis is a simple and potent screening approach for identifying neurocristopathy-associated genes in mice. Rare Dis. 2016, 4, e1156287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Methot, D.; Reudelhuber, T.L.; Silversides, D.W. Evaluation of tyrosinase minigene co-injection as a marker for genetic manipulations in transgenic mice. Nucleic Acids Res. 1995, 23, 4551–4556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Overbeek, P.A.; Aguilar-Cordova, E.; Hanten, G.; Schaffner, D.L.; Patel, P.; Lebovitz, R.M.; Lieberman, M.W. Coinjection strategy for visual identification of transgenic mice. Transgenic Res. 1991, 1, 31–37. [Google Scholar] [CrossRef]
- Yokoyama, T.; Silversides, D.W.; Waymire, K.G.; Kwon, B.S.; Takeuchi, T.; Overbeek, P.A. Conserved cysteine to serine mutation in tyrosinase is responsible for the classical albino mutation in laboratory mice. Nucleic Acids Res. 1990, 18, 7293–7298. [Google Scholar] [CrossRef] [Green Version]
- Soret, R.; Mennetrey, M.; Bergeron, K.F.; Dariel, A.; Neunlist, M.; Grunder, F.; Faure, C.; Silversides, D.W.; Pilon, N. A collagen VI-dependent pathogenic mechanism for Hirschsprung’s disease. J. Clin. Investig. 2015, 125, 4483–4496. [Google Scholar] [CrossRef] [Green Version]
- Korbel, J.O.; Tirosh-Wagner, T.; Urban, A.E.; Chen, X.N.; Kasowski, M.; Dai, L.; Grubert, F.; Erdman, C.; Gao, M.C.; Lange, K.; et al. The genetic architecture of Down syndrome phenotypes revealed by high-resolution analysis of human segmental trisomies. Proc. Natl. Acad. Sci. USA 2009, 106, 12031–12036. [Google Scholar] [CrossRef] [Green Version]
- Bergeron, K.F.; Cardinal, T.; Toure, A.M.; Beland, M.; Raiwet, D.L.; Silversides, D.W.; Pilon, N. Male-Biased Aganglionic Megacolon in the TashT Mouse Line Due to Perturbation of Silencer Elements in a Large Gene Desert of Chromosome 10. PLoS Genet. 2015, 11, e1005093. [Google Scholar] [CrossRef] [Green Version]
- Cardinal, T.; Bergeron, K.F.; Soret, R.; Souchkova, O.; Faure, C.; Guillon, A.; Pilon, N. Male-biased aganglionic megacolon in the TashT mouse model of Hirschsprung disease involves upregulation of p53 protein activity and Ddx3y gene expression. PLoS Genet. 2020, 16, e1009008. [Google Scholar] [CrossRef] [PubMed]
- Hosoda, K.; Hammer, R.E.; Richardson, J.A.; Baynash, A.G.; Cheung, J.C.; Giaid, A.; Yanagisawa, M. Targeted and natural (piebald-lethal) mutations of endothelin-B receptor gene produce megacolon associated with spotted coat color in mice. Cell 1994, 79, 1267–1276. [Google Scholar] [CrossRef]
- Soret, R.; Schneider, S.; Bernas, G.; Christophers, B.; Souchkova, O.; Charrier, B.; Righini-Grunder, F.; Aspirot, A.; Landry, M.; Kembel, S.W.; et al. Glial Cell Derived Neurotrophic Factor Induces Enteric Neurogenesis and Improves Colon Structure and Function in Mouse Models of Hirschsprung Disease. Gastroenterology 2020, 159, 1824–1838. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, Y.; Ito, F.; Ando, H.; Seo, T.; Harada, T.; Kaneko, K.; Ishiguro, Y.; Kobayashi, S. Extrinsic nerve strands in the aganglionic segment of Hirschsprung’s disease. J. Pediatr. Surg. 1998, 33, 1233–1237. [Google Scholar] [CrossRef]
- Pilon, N.; Raiwet, D.; Viger, R.S.; Silversides, D.W. Novel pre- and post-gastrulation expression of Gata4 within cells of the inner cell mass and migratory neural crest cells. Dev. Dyn. 2008, 237, 1133–1143. [Google Scholar] [CrossRef]
- Bergeron, K.F.; Nguyen, C.M.; Cardinal, T.; Charrier, B.; Silversides, D.W.; Pilon, N. Upregulation of the Nr2f1-A830082K12Rik gene pair in murine neural crest cells results in a complex phenotype reminiscent of waardenburg syndrome type 4. Dis. Models Mech. 2016, 9, 1283–1293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gara, S.K.; Grumati, P.; Urciuolo, A.; Bonaldo, P.; Kobbe, B.; Koch, M.; Paulsson, M.; Wagener, R. Three novel collagen VI chains with high homology to the alpha3 chain. J. Biol. Chem. 2008, 283, 10658–10670. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.A.; Chang, M.M.; Huang, W.; Ooi, W.F.; Xing, M.; Tan, P.; Skanderup, A.J. Mutation hotspots at CTCF binding sites coupled to chromosomal instability in gastrointestinal cancers. Nat. Commun. 2018, 9, 1520. [Google Scholar] [CrossRef] [Green Version]
- Essien, K.; Vigneau, S.; Apreleva, S.; Singh, L.N.; Bartolomei, M.S.; Hannenhalli, S. CTCF binding site classes exhibit distinct evolutionary, genomic, epigenomic and transcriptomic features. Genome Biol. 2009, 10, R131. [Google Scholar] [CrossRef]
- Bespalov, M.M.; Sidorova, Y.A.; Tumova, S.; Ahonen-Bishopp, A.; Magalhaes, A.C.; Kulesskiy, E.; Paveliev, M.; Rivera, C.; Rauvala, H.; Saarma, M. Heparan sulfate proteoglycan syndecan-3 is a novel receptor for GDNF, neurturin, and artemin. J. Cell Biol. 2011, 192, 153–169. [Google Scholar] [CrossRef] [Green Version]
- Bondarenko, O.; Saarma, M. Neurotrophic Factors in Parkinson’s Disease: Clinical Trials, Open Challenges and Nanoparticle-Mediated Delivery to the Brain. Front. Cell. Neurosci. 2021, 15, 682597. [Google Scholar] [CrossRef]
- Ji, Y.; Tam, P.K.; Tang, C.S. Roles of Enteric Neural Stem Cell Niche and Enteric Nervous System Development in Hirschsprung Disease. Int. J. Mol. Sci. 2021, 22, 9659. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, H.; Araki, T.; Jackman, A.; Heuckeroth, R.O.; Snider, W.D.; Johnson, E.M., Jr.; Milbrandt, J. GFR alpha1-deficient mice have deficits in the enteric nervous system and kidneys. Neuron 1998, 21, 317–324. [Google Scholar] [CrossRef] [Green Version]
- Toure, A.M.; Charrier, B.; Pilon, N. Male-specific colon motility dysfunction in the TashT mouse line. Neurogastroenterol. Motil. 2016, 28, 1494–1507. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).