Abstract
Alzheimer’s disease (AD), an age-dependent, progressive neurodegenerative disorder, is the most common type of dementia, accounting for 50–70% of all dementia cases. Due to the increasing incidence and corresponding socioeconomic burden of dementia, it has rapidly emerged as a challenge to public health worldwide. The characteristics of AD include the development of extracellular amyloid-beta plaques and intracellular neurofibrillary tangles, vascular changes, neuronal inflammation, and progressive brain atrophy. However, the complexity of the biology of AD has hindered progress in elucidating the underlying pathophysiological mechanisms of AD, and the development of effective treatments. MicroRNAs (miRNAs, which are endogenous, noncoding RNAs of approximately 22 nucleotides that function as posttranscriptional regulators of various genes) are attracting attention as powerful tools for studying the mechanisms of diseases, as they are involved in several biological processes and diseases, including AD. AD is a multifactorial disease, and several reports have suggested that miRNAs play an important role in the pathological processes of AD. In this review, the basic biology of miRNAs is described, and the function and physiology of miRNAs in the pathological processes of AD are highlighted. In addition, the limitations of current pharmaceutical therapies for the treatment of AD and the development of miRNA-based next-generation therapies are discussed.
1. Introduction
Alzheimer’s disease (AD) is an irreversible and multifactorial neurodegenerative disorder, and the main cause of dementia in the global elderly population (>65 years of age) [1,2]. Approximately 10% of the elderly population is diagnosed with AD, and the age-related prevalence of this disease doubles every five years after the age of 65 [3,4]. AD is characterized by cognitive impairment and memory loss, which eventually result in a loss of the ability to independently perform daily activities. Therefore, as the aging population continues to increase globally, the increasing incidence and socioeconomic burden of AD continue to represent tremendous challenges to public health worldwide [5].
Although the cause of AD is unclear [6], several studies have reported that age, family history, the ε4 allele of apolipoprotein E (APOE), high cholesterol, type 2 diabetes, and cardiovascular disease are associated with the development of AD [7,8,9,10]. The main histopathological features of AD are amyloid-beta (Aβ) plaques and neurofibrillary tangles (NFTs) in the neocortex, hippocampus, and other subcortical brain regions [11,12,13], and these are believed to be causative factors of AD. Accumulated amyloid-beta plaques and NFTs in the brain tissue of Alzheimer’s patients cause neuroinflammation and apoptosis, leading to brain atrophy. However, no effective pharmacotherapies for the prevention and treatment of AD have been developed [6,14,15].
The currently developed drugs prescribed for AD patients are not fundamental treatment methods; rather, their purpose is to relieve symptoms or to delay disease progression. Therefore, the development of new therapeutic agents is required.
MicroRNAs (miRNAs) are composed of approximately 18–22 nucleotides, and are typically non-coding and single-stranded RNA sequences. miRNAs bind to complementary untranslated regions (3′-UTRs) of mRNAs to regulate target genes, resulting in translational repression or degradation [16,17]. miRNAs play an essential role in normal development, and are involved in various biological functions, including development, differentiation, proliferation, and apoptosis [18,19,20,21]. Therefore, the dysregulation of miRNAs is involved in many human diseases, including AD [22,23,24]. According to previous studies, dysregulation of some key factors, such as c-Myc and P53, increased the expression of the oncogenic miR-17-92 cluster in tumors [25]. In addition, miRNAs are closely implicated in the pathogenesis of major diseases, such as heart disease [26], hypertension [27], arthritis [28], diabetes [29], and obesity [30].
Several studies have reported that the miRNA profile changes in pathological conditions [31,32,33]. The relationships between diseases and miRNAs are currently being studied, including the ability of miRNAs to regulate multiple targets, which may be a promising tool for the study of multifactorial diseases, such as AD [34]. In addition, knockdown or overexpression of miRNAs can alter gene expression; thus, due to these properties, miRNAs have strong therapeutic potential to treat diseases [35].
In this review, a brief overview of the current therapeutic methods to treat AD, and the limitations of these methods, are provided. The role of miRNAs and the current trends of miRNA-based therapeutic strategies for AD are also discussed (Figure 1).
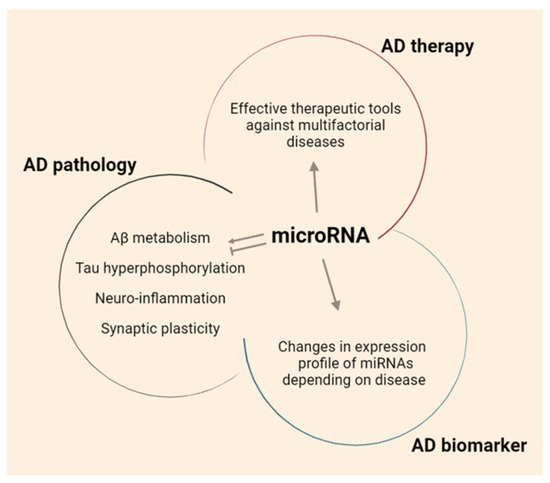
Figure 1.
The role of miRNAs in AD.
2. Current Treatment for AD
2.1. Pharmacotherapy for Alzheimer’s Disease
Though AD is the most common neurodegenerative dementia, no effective treatments have been developed. However, treatments that can alleviate symptoms are available for patients with AD. Until recently, five dementia treatments were approved by the Food and Drug Administration (FDA), including donepezil, rivastigmine, memantine, and aducanumab (Table 1).

Table 1.
FDA-approved drugs for AD.
Donepezil, the leading compound for the treatment of AD, rapidly and reversibly inhibits acetylcholinesterase (AChE), impeding the hydrolysis of the neurotransmitter acetylcholine, resulting in increased activity of acetylcholine [36]. Donepezil was approved by the FDA in 1996, and its side effects include nausea, difficulty sleeping, abnormal heart rhythms, and seizures [37]. In 1997 and 2001, rivastigmine and galantamine, two different types of cholinesterase inhibitor, were developed and approved for AD pharmacotherapy by the FDA. Like donepezil, rivastigmine and galantamine reversibly inhibit AChE to improve the intrinsic action of acetylcholine on cholinergic receptors [38]. Memantine is used to treat moderate-to-severe AD in the United States, Canada, Europe, China, and other countries, and was approved by the FDA in 2003. Memantine is an antagonist of the N-methyl-D-aspartate glutamate receptors (NMDAR) in the central nervous system [39].
Although these drugs have been used to treat AD, the cholinesterase inhibitors and memantine only treat patients’ symptoms, and do not prevent the progression of AD. Treatment with donepezil results in slightly improved cognitive function, but does not improve length of hospital stay, treatment costs, or caregiver burdens [40]. Memantine has been reported to safely and effectively improve cognitive function in patients with severe AD [41]. However, no clinical benefits have been reported for patients with early stage AD [41].
Aducanumab is a monoclonal IgG1 antibody that inhibits accumulation of extracellular Aβ plaques in the brain [42]. Although the drug received conditional approval from the FDA in June 2021, the history of aducanumab failure in phase 3 clinical trials remains controversial [43,44]. Nevertheless, aducanumab is the first drug to directly target Aβ plaques, which have been the focus of Alzheimer’s research and drug development for decades; aducanumab is being evaluated as an important therapeutic option for the treatment of AD.
2.2. Limitations of Drugs for AD
Various therapies have been developed to treat AD, though they each have limitations. The biggest obstacle to drug development is the lack of understanding of the pathophysiology of AD. As a result, existing AD treatments are being used for limited purposes, such as improving cognitive function or delaying memory decline. Therefore, no significant improvements in practical benefits, such as clinical improvement, reduced treatment costs, or shortened hospital stay, have been reported for patients receiving these therapies.
In addition, as AD is a multifactorial disease by, for example, accumulation of Aβ plaques and formation of NFTs, the effects of conventional pharmacotherapies with one target are limited. Although aducanumab reduces Aβ accumulation in the brain, no significant clinical effects were reported [42,43,45]. Therefore, an innovative approach that can efficiently control each risk factor of the multifactorial disease is required for the development of AD therapies. While multi-target therapeutics are attracting attention, their synergistic effects have not been proven, and more research is necessary.
Finally, a damaged brain has limited recovery abilities. An early diagnosis and regenerative medicine perspectives are required for the treatment of AD. As AD progresses, brain atrophy due to the loss of nerve cells cannot be reversed. Therefore, early treatment is important.
3. Pathological Characteristics of AD and the Role of miRNAs
miRNAs have recently been reported in many studies regarding neurodegenerative diseases, such as AD [46]. In addition, many studies reported that various miRNAs are involved in regulating Aβ, Tau and neuroinflammation in AD pathology (Table 2). In 2007, it was revealed that alterations in specific miRNAs, namely, miR-9, miR-125b, and miR-128, had been induced by the production of reactive oxygen species, likely generating neurotoxic metal sulfates in the hippocampi of patients with AD [47]. Subsequent studies have reported significant variations or decreased expression of miRNAs in different areas of the brain in patients with AD. The expressions of miR-106b, miR-29a/b/c, and miR-107 were decreased in the hippocampi of patients with AD, and miR-181c and miR-101 were deregulated in the cortices of patients with AD [48,49]. Several miRNAs, such as miR-212, are expressed at low levels in the white matter of the brain of patients with AD, and the expression of miR-15 and miR-107 is decreased in the grey matter [50]. In contrast, miR-125b, miR-128, and miR-146a have increased expression in the brains of these patients [51]. As such, several reports have revealed that miRNA is closely related to AD. In this review, we introduce the relationship between miRNAs and major factors involved in AD pathogenesis.
3.1. miRNAs Regulate Aβ
The amyloid hypothesis was first presented by John Hardy and David Allsop in 1991 [52]. Aβ is a transmembrane protein generated by the hydrolysis amyloid precursor protein (APP) via the amyloidogenic pathway. C-terminal fragments of APP are produced via hydrolysis by α-, β-, γ-, and η-secretase through three pathways [53]. Among them, Aβ1-42 is produced by the cleavage of APP by β-secretase and γ-secretase. Aβ deposited in the hippocampus and basal segment in the form of neurotoxic Aβ plaques recruits more Aβ, forming insoluble aggregates and inducing mitochondrial damage [54], disrupting homeostasis [55], and causing synaptic dysfunction [56].
Several studies have reported that various miRNAs (miR-20a, miR-106a, miR-106b, miR-17-5p, miR-16, miR-101, miR-147, miR-153, and miR-520c) directly regulate APP expression by targeting the 3′-UTR of APP mRNA [57,58,59,60]. Among these miRNAs, miR-106b, and miR-101 are downregulated in the brains of patients with AD, resulting in increased APP expression and Aβ production. Furthermore, AD-specific polymorphisms have been reported to influence the APP-modulating activity of miR-147 and miR-20a [61].
β-secretase cleaving enzyme 1 (BACE1) levels have been reported to be increased in patients with AD, increasing the risk of sporadic AD [62]. Several studies have reported a relationship between abnormal miRNA expression and increased levels of BACE1. miR-107, miR-298, miR-328, miR-15a, miR-15b, miR-195, miR-103, and miR-485 target the 3′-UTR of BACE1 [63,64,65,66,67]. These miRNAs are decreased in patients with AD [68,69,70,71]. In addition, significant reductions in miR-29a and miR-29b levels were reported to lead to an increased level of BACE1. Thus, miR-29 has been proposed as an endogenous BACE1 regulator [48,72]. These findings suggest that the miRNAs play important roles in regulating the amyloidogenic pathways in the brain of AD patients.
miRNAs can also affect AD pathology in other ways. Decreased monocyte lysosomal hydrolases, including the cathepsin B, D, and S enzymes, also play a role in Aβ accumulation [73]. Recent studies have shown that the upregulation of miR-128 is responsible for the downregulation of cathepsin B, D, and S. Furthermore, miR-128 inhibition in monocytes isolated from AD patients improved both lysosomal enzyme expression levels and Aβ degradation capacity [74]. These findings indicate that miRNAs are involved in the clearance of Aβ via the regulation of enzymatic activities in cleavage and degradation processes. Taken together, these findings suggest that miRNAs have the potential to be used as therapeutic agents or as mediators for regulation of Aβ in AD patients.
3.2. miRNAs Regulate Tau
Tau, a microtubule-associated protein generated by the alternative splicing of the MAPT gene, is mainly found in neuronal axons in the brain, where it maintains microtubule structure, cytoplasmic transport processes, and synaptic structure and function, and regulates neuronal signaling [75,76]. Tau is a phosphoprotein, and its phosphorylation is regulated by balanced protein kinase and protein phosphatase activities, and changes according to the stage of brain development. Under normal conditions, tau has few phosphorylation sites, and tau phosphorylation negatively regulates the binding of tau to microtubules. However, under pathological conditions, tau is hyperphosphorylated. Hyperphosphorylation of tau in the AD patient brain causes changes in microtubule configuration and the loss of tubulin polymerization, eventually resulting in defective microtubule function [77,78]. The elevated levels of cytosolic tau in AD lead to the formation of insoluble, paired helical filaments and straight filaments through tau–tau interactions and polymerization, resulting in the generation of intraneuronal fibrillar deposits [79]. These intraneuronal fibrillar deposits reduce the number of synapses, induce neurotoxicity, and eventually cause cell dysfunction [80,81,82].
Various studies reveled that miRNAs are closely involved in the regulation of tau protein homeostasis. Carrettiero et al. reported that miR-128a regulates the production of the auxiliary chaperone protein BAG2, which plays a role in the degradation and aggregation of tau [83]. Furthermore, miR-124, miR-132, and miR-9 can alter the accumulation of endogenous tau [84,85]. Both miR-34a and miR-26b can inhibit tau expression and affect NFT formation [86]. It is suggested that miR-9 expression increases concurrently with the decreased expression of sirtuin 1 (SIRT1), which participates in tau pathology as a deacetylase [85]. These findings suggest that miRNAs play crucial roles in maintaining microtubule structure and synaptic structure via regulation of tau protein homeostasis.
Phosphorylation of tau is very important in the progression of AD. The downregulation of miR-101 expression increases the levels of phosphorylated tau [87]. miR-922 binds the 3′-UTR of ubiquitin carboxyl-terminal hydrolase isozyme L1 (UCHL1), leading to a reduction in UCHL1 expression, which reduces the level of phosphorylated tau [88]. Wang et al. reported that miR-138 was increased in AD models, such as N2a/APP and HEK293/tau cell lines. In addition, miR-138 promotes tau phosphorylation by targeting the retinoic acid receptor alpha/GSK-3β pathway [89]. He et al. reported that miR-326 decreased tau phosphorylation. These findings suggest that miRNAs are involved in regulation of tau phosphorylation, which is closely related to tau pathology, such as defective microtubule configuration in AD patients. Therefore, it could be suggested that miRNAs are potential treatments that can achieve the suppression of tau-related pathological properties.
3.3. miRNAs Regulate Neuroinflammation
Several studies have suggested that AD is strongly related to immune mechanisms [90,91,92]. Protein accumulation, associated with the pathology of AD, stimulates receptors on microglia and astrocytes to trigger immune responses, including the release of inflammatory mediators [93,94,95]. A short-term inflammatory response removes the causative agent of inflammation and contributes to the recovery of the affected area, and a continuous inflammatory response causes irreversible tissue damage. Inflammatory responses in patients with AD are continuous, causing neuronal loss, promoting the progression of AD disease, or playing a significant role in exacerbations of AD.
Several studies have reported that the NLRP3-inflammasome is activated in microglia, increasing the expressions of inflammasome-forming factors, such as NLRP3, caspase-1, and ASC, leading to increased secretion of the pro-inflammatory cytokines interleukin (IL)-1 beta and IL-18, inducing neuronal death in patients with AD [95,96,97].
Zhou et al. reported that miR-7 inhibits the NLRP3-inflammasome activity of microglia, and anti-miR-7 activates the inflammasome. In addition, the inflammatory response was reduced by miR-7 in mouse models [98]. Cao et al. reported that the overexpression of miR-9 leads to suppression of NLRP1-inflammasome activation by regulating the expression of NLRP1 [99]. In addition, the levels of the pro-inflammatory cytokines IL-1 beta and IL-18 were also reduced. Thus, the overexpression of miR-9 may be an effective method to characterize brain damage after an ischemic stroke.
In the setting of neuroinflammation, activation of the inflammasome leads to pyroptotic cell death by cleaving the N-terminus of gasdermin D (GSDMD) and by the secretion of proinflammatory cytokines [100,101]. miR-22 targets GSDMD [102], and the expression of miR-22 was lower in patients with AD than in healthy adults. The overexpression of miR-22 in an APP/PS1 mouse model significantly improved memory and behavior, and suppressed the expression of proinflammatory cytokines, such as IL-1 beta and IL-18, by inhibiting GSDMD. This resulted in the prevention of pyroptotic cell death. Based on these findings, it could be suggested that miRNAs play an important role in controlling neuroinflammation; thus, miRNA could be an option for the treatment of degenerative brain diseases, such as AD.
3.4. miRNAs Regulate Synaptic Function
Increasing evidence suggests that miRNAs are involved in early synaptic alterations and synapse loss in patients with AD and in AD experimental models. For instance, miR-26b, miR-132, and miR-206 are able to target neurotrophic factors, such as insulin-like growth factor 1 (IGF1) and brain-derived neurotrophic factor (BDNF); these miRNAs regulate spine remodeling via dendrite maturation [103,104]. In addition, it is suggested that the dendritic miR-134 can facilitate homeostatic synaptic function in synapse formation through direct regulation of LIMK1 and PUM2 expression [105,106,107]. Moreover, miR-9 controls dendritic development by downregulation of the transcriptional repressor REST in primary culture and in the mouse brain [108]. These findings indicate that several miRNAs are involved in synaptic morphological changes, including dendritic formation and synapse maturation.
Several studies have reported that miRNAs also regulate the pre-synaptic function in AD [109,110,111]. For instance, miR-210 can bind its target, such as the synaptosomal-associated protein, Snap25 mRNA. In a hippocampal neuron culture, overexpression of miR-210 decreased the number of synapses. In an animal model, intracerebroventricular injection of miR-210 induced the phenotypes of synaptic loss in the brain and of cognitive impairment [109]. Furthermore, overexpression of miR-485 reduced the abundance and protein expression of synaptic vesicle glycoprotein 2A (SV2A) mRNA in hippocampal neurons, leading to a reduction of spontaneous neurotransmitter release [110]. Especially, Aβ-induced miR-34c alteration can decrease the expression of vesicle-associated membrane protein 2 (VAMP2), a protein component of the SNARE complex, which, in turn, causes learning and memory deficits with synaptic failure in AD models [111]. Based on these findings, it could be speculated that the miRNAs may play a critical role in vesicle trafficking and neurotransmitter releases, via the regulation of physiological conditions in pre-synaptic neurons in AD models.
Since glutamatergic neurotransmission plays a key role in synaptic function and synaptic plasticity processes, including long-term potentiation (LTP) and long-term depression (LTD), numerous studies of miRNAs’ targets have been conducted on glutamate receptors and excitatory synapses in AD. It is suggested that miR-9, miR-92, miR-137, and miR-501 selectively regulate GluA1 trafficking, resulting in a reduction of the α-amino-3-hydroxy-5-methylisoxazole-4-propionic acid glutamate receptor (AMPAR) insertion in the cell surface [112,113,114,115]. Additionally, the increased miR-181a, by accumulation of Aβ, significantly decreased the GluA2 subunit in AMPAR and other plasticity-related protein expression (i.e., scaffolding proteins and post-synaptic density 95) in mouse hippocampi, which, in turn, causes memory deficits [116]. Like AMPAR, surface expression of NMDAR is also regulated by several miRNAs, such as miR-125b and miR-34a [117,118]. In hippocampal culture, overexpression of miR-125b suppressed GluN2A expression. In addition, miR-125b prolonged excitatory postsynaptic currents by a half-width, which is consistent with the relative loss of synaptic GluN2A-containing NMDAR [117]. While miR-34a targets the GluN2B subunit of NMDAR, it also regulates synaptic plasticity modulation in the working memory [118]. Based on these findings, dysregulation of glutamate receptor internalization (AMPAR and NMDAR) causes an imbalance between LTP and LTD in AD, creating a functional deficit in glutamatergic neurotransmission.
Taken together, these studies showed that several miRNAs are able to regulate structural and morphological alterations in pre- and post-synaptic neurons, and they may play key roles in the maintenance of synaptic function on learning and memory in the brain of AD patients.

Table 2.
AD-related miRNAs and their targets.
Table 2.
AD-related miRNAs and their targets.
| AD Phenotype | miRNA | Target mRNA | Reference |
|---|---|---|---|
| Aβ | miR-16, miR-17, miR-20a, miR-101, miR-106a, miR-106b, miR-147, miR-153, miR-520c | APP | [57,58,59,60] |
| miR-15a, miR-15b, miR-29a, miR-29b, miR-29c, miR-103, miR-107, miR-298, miR-328, miR-195, miR-485 | BACE1 | [63,64,65,66,67] | |
| miR-128 | Cathepsin B, D, S | [74] | |
| Tau | miR-128 | BAG2 | [83] |
| miR-9 | SIRT1 | [85] | |
| miR-922, miR-181b | UCHL1 | [88,119] | |
| miR-124 | Caveolin-1 | [120] | |
| miR-132 | ITPKB | [121] | |
| miR-34a | Tau | [122] | |
| miR-26b | Rb1 | [86] | |
| Neuroinflammation | miR-7, miR-223 | NLRP3 | [98,123] |
| miR-9 | NLRP1 | [99] | |
| miR-22 | GSDMD | [102] | |
| let-7 family | TLR4 | [124] | |
| IL-6 | [125] | ||
| miR-485 | AKT3 | [126] | |
| Synaptic function | miR-26b, miR-206 | IGF1 | [103] |
| miR-132, miR-206 | BDNF | [104] | |
| miR-134 | LIMK1, PUM2 | [105] | |
| miR-9 | REST | [108] | |
| miR-210 | Snap25 | [109] | |
| miR-485 | SV2A | [110] | |
| miR-34c | VAMP2 | [111] | |
| miR-92, miR-137, miR-501 | GRIA1 | [113,114,115] | |
| miR-181a | GRIA2 | [116] | |
| miR-125b | GRIN2A | [117] | |
| miR-34a | GRIN2B | [118] |
4. miRNA as Diagnostic AD Biomarker
Early diagnosis is essential for the most effective treatment of AD. AD lesions cause irreversible damage to the brain, so if diagnosis is delayed, treatment options and recovery are very limited. A recent report suggests that early diagnosis can cut the risk of developing Alzheimer’s by one third [127]. Currently, abnormal accumulation of Aβ is visualized to diagnose AD through brain imaging, using the cerebrospinal fluid (CSF) test and positron emission tomography. However, these methods are either invasive to the patient or require a very expensive financial expenditure [128]. In addition, cognitive function evaluation performed for AD diagnosis is used only as reference data, due to individual differences and low accuracy [129].
As aforementioned, several miRNAs that regulate Aβ production or tau have been reported to have increased or decreased expression in AD lesions. These miRNA profile changes are observed not only in brain tissue, but also in blood. Based on this evidence, the development of an AD diagnostic method based on miRNA may improve some of the limitations of the current diagnostic method. As reported by Bhatnagar et al., in 2014, miR-34c was found to be elevated in plasma from patients with AD. miR-34c inhibits the expression of genes involved in survival and oxidative defense pathways, such as Bcl2 and SIRT1. Therefore, miR-34c shows potential as a biomarker for screening patients with AD [130]. In addition, according to Wang’s report, as a result of analyzing plasma from 120 control subjects, 120 PD patients, and 120 AD patients, miR-103 is promising as a biomarker for AD disease. miR-103 was decreased in AD patients, compared to controls, and was negatively correlated with dementia severity [131]. According to a recent report in 2020 by Souza et al., a sample of women aged 55 years and older, carrying the ε4 allele of APOE, found a 3-fold decrease in miR-9 expression levels, compared to the control group [132].
However, single-stranded miRNAs exhibit a short half-life in biofluids, such as CSF or blood. Nevertheless, the reason why miRNA-based diagnostic methods are attracting attention is that miRNAs encapsulated in exosomes and microvesicles, or bound to proteins, are stably present in biofluids. In addition, in order to increase the reliability of miRNA as a biomarker of diseases, such as AD, it is necessary to additionally standardize the sampling method and sample analysis method of biofluids.
5. Limitation of mRNAs
miRNAs are powerful tools to regulate gene expression. However, there are some obstacles to overcome for a clinical application. First, the relationship between miRNA and the target gene is not always a 1:1 match. Thus, it is difficult to define a suitable target as the reason for considering off-targets [133]. Therefore, the development of a sophisticated target gene prediction algorithm is required to minimize the side effects. Second, natural variations in the expression patterns of miRNAs must be considered, such as gender [134,135] and age [136,137]. Third, single-stranded miRNAs exhibit very fast decay kinetics in certain situations, such as in biofluids [138,139]. Therefore, additional means, such as a drug delivery system capable of delivering miRNA to the diseased site in a stable state, are required. Finally, results detecting expression of miRNAs may vary, depending on the detection platform, so that a careful and critical evaluation system is required when interpreting the data [140]. Despite the aforementioned limitations, miRNAs are attractive therapeutic tools to treat disease, such as neurodegenerative disease, especially AD. Thus, we suggest that these obstacles should be overcome, in order to treat disease using miRNAs as therapeutic drugs.
6. Conclusions and Prospects
miRNAs are closely related to the pathophysiology of various neurodegenerative diseases, including AD, and may also be key factors in the treatment of such diseases. The pathological functions of genes that are regulated by multiple miRNAs overlap and interact, and the effects of miRNA networks are better than those of individual miRNAs. The regulatory effects of miRNAs are sequence-specific, and each miRNA can regulate multiple genes; therefore, the use of miRNAs is promising for the treatment and regulation of complicated disease networks and pathways. Thus, these miRNA characteristics may allow for effective treatment of multifactorial disease.
Although several drugs have been studied comprehensively, a safe and effective drug for the treatment of AD has yet to be identified. Therefore, the development of successful miRNA-based drugs for the treatment of AD is needed; also required are extensive research and in-depth knowledge of the precise mechanism of miRNA-target interactions and regulation of the target by miRNAs. Studies regarding miRNA-based therapies for AD are in the early stages, though extensive research regarding the treatment of AD has been conducted. Therefore, effective miRNA therapies for AD are likely to be developed in the very near future.
Author Contributions
Conceptualization, C.Y.L.; writing—original draft preparation, C.Y.L.; writing—review and editing, C.Y.L., I.S.R. and H.-J.C.; supervision, J.-H.R.; project administration, H.-J.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Korea Ministry of Science, ICT and Future Planning (NRF-2017M3A9G2094069), and by grants from the Ministry of Health & Welfare, Republic of Korea (HI18C1671).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Ballard, C.; Gauthier, S.; Corbett, A.; Brayne, C.; Aarsland, D.; Jones, E. Alzheimer’s disease. Lancet 2011, 377, 1019–1031. [Google Scholar] [CrossRef]
- Apostolova, L.G. Alzheimer Disease. Continuum 2016, 22, 419–434. [Google Scholar] [CrossRef] [Green Version]
- Qiu, C.; Kivipelto, M.; von Strauss, E. Epidemiology of Alzheimer’s disease: Occurrence, determinants, and strategies toward intervention. Dialogues Clin. Neurosci. 2009, 11, 111–128. [Google Scholar]
- Prince, M.; Bryce, R.; Albanese, E.; Wimo, A.; Ribeiro, W.; Ferri, C.P. The global prevalence of dementia: A systematic review and metaanalysis. Alzheimer’s Dement. 2013, 9, 63–75.e2. [Google Scholar] [CrossRef]
- Fotenos, A.F.; Mintun, M.A.; Snyder, A.Z.; Morris, J.C.; Buckner, R.L. Brain volume decline in aging: Evidence for a relation between socioeconomic status, preclinical Alzheimer disease, and reserve. Arch. Neurol. 2008, 65, 113–120. [Google Scholar] [CrossRef]
- Camandola, S.; Mattson, M.P. Brain metabolism in health, aging, and neurodegeneration. EMBO J. 2017, 36, 1474–1492. [Google Scholar] [CrossRef]
- Coon, K.D.; Myers, A.J.; Craig, D.W.; Webster, J.A.; Pearson, J.V.; Lince, D.H.; Zismann, V.L.; Beach, T.G.; Leung, D.; Bryden, L.; et al. A high-density whole-genome association study reveals that APOE is the major susceptibility gene for sporadic late-onset Alzheimer’s disease. J. Clin. Psychiatry 2007, 68, 613–618. [Google Scholar] [CrossRef] [PubMed]
- Wijesekara, N.; Ahrens, R.; Sabale, M.; Wu, L.; Ha, K.; Verdile, G.; Fraser, P.E. Amyloid-beta and islet amyloid pathologies link Alzheimer’s disease and type 2 diabetes in a transgenic model. FASEB J. 2017, 31, 5409–5418. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chang, T.Y.; Yamauchi, Y.; Hasan, M.T.; Chang, C. Cellular cholesterol homeostasis and Alzheimer’s disease. J. Lipid Res. 2017, 58, 2239–2254. [Google Scholar] [CrossRef] [Green Version]
- Stampfer, M.J. Cardiovascular disease and Alzheimer’s disease: Common links. J. Intern. Med. 2006, 260, 211–223. [Google Scholar] [CrossRef] [PubMed]
- Serrano-Pozo, A.; Frosch, M.P.; Masliah, E.; Hyman, B.T. Neuropathological alterations in Alzheimer disease. Cold Spring Harb. Perspect. Med. 2011, 1, a006189. [Google Scholar] [CrossRef]
- Iqbal, K.; Alonso Adel, C.; Chen, S.; Chohan, M.O.; El-Akkad, E.; Gong, C.X.; Khatoon, S.; Li, B.; Liu, F.; Rahman, A.; et al. Tau pathology in Alzheimer disease and other tauopathies. Biochim. Biophys. Acta 2005, 1739, 198–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesulam, M.M. Neuroplasticity failure in Alzheimer’s disease: Bridging the gap between plaques and tangles. Neuron 1999, 24, 521–529. [Google Scholar] [CrossRef] [Green Version]
- Idda, M.L.; Munk, R.; Abdelmohsen, K.; Gorospe, M. Noncoding RNAs in Alzheimer’s disease. Wiley Interdiscip. Rev. RNA 2018, 9, e1463. [Google Scholar] [CrossRef]
- Chin-Chan, M.; Navarro-Yepes, J.; Quintanilla-Vega, B. Environmental pollutants as risk factors for neurodegenerative disorders: Alzheimer and Parkinson diseases. Front. Cell Neurosci. 2015, 9, 124. [Google Scholar] [CrossRef] [Green Version]
- Eddy, S.R. Non-coding RNA genes and the modern RNA world. Nat. Rev. Genet. 2001, 2, 919–929. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Kong, W.; He, L.; Zhao, J.J.; O’Donnell, J.D.; Wang, J.; Wenham, R.M.; Coppola, D.; Kruk, P.A.; Nicosia, S.V.; et al. MicroRNA expression profiling in human ovarian cancer: miR-214 induces cell survival and cisplatin resistance by targeting PTEN. Cancer Res. 2008, 68, 425–433. [Google Scholar] [CrossRef] [Green Version]
- Chang, W.; Lee, C.Y.; Park, J.H.; Park, M.S.; Maeng, L.S.; Yoon, C.S.; Lee, M.Y.; Hwang, K.C.; Chung, Y.A. Survival of hypoxic human mesenchymal stem cells is enhanced by a positive feedback loop involving miR-210 and hypoxia-inducible factor 1. J. Vet. Sci. 2013, 14, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Fu, G.; Brkic, J.; Hayder, H.; Peng, C. MicroRNAs in Human Placental Development and Pregnancy Complications. Int. J. Mol. Sci. 2013, 14, 5519–5544. [Google Scholar] [CrossRef] [Green Version]
- Lee, C.Y.; Shin, S.; Lee, J.; Seo, H.H.; Lim, K.H.; Kim, H.; Choi, J.W.; Kim, S.W.; Lee, S.; Lim, S.; et al. MicroRNA-Mediated Down-Regulation of Apoptosis Signal-Regulating Kinase 1 (ASK1) Attenuates the Apoptosis of Human Mesenchymal Stem Cells (MSCs) Transplanted into Infarcted Heart. Int. J. Mol. Sci. 2016, 17, 1752. [Google Scholar] [CrossRef] [Green Version]
- Paul, P.; Chakraborty, A.; Sarkar, D.; Langthasa, M.; Rahman, M.; Bari, M.; Singha, R.S.; Malakar, A.K.; Chakraborty, S. Interplay between miRNAs and human diseases. J. Cell Physiol. 2018, 233, 2007–2018. [Google Scholar] [CrossRef] [PubMed]
- Tufekci, K.U.; Oner, M.G.; Meuwissen, R.L.; Genc, S. The role of microRNAs in human diseases. Methods Mol. Biol. 2014, 1107, 33–50. [Google Scholar]
- Wang, M.; Qin, L.; Tang, B. MicroRNAs in Alzheimer’s Disease. Front. Genet. 2019, 10, 153. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal. Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [Green Version]
- Dong, S.; Cheng, Y.; Yang, J.; Li, J.; Liu, X.; Wang, X.; Wang, D.; Krall, T.J.; Delphin, E.S.; Zhang, C. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J. Biol. Chem. 2009, 284, 29514–29525. [Google Scholar] [CrossRef] [Green Version]
- Kontaraki, J.E.; Marketou, M.E.; Zacharis, E.A.; Parthenakis, F.I.; Vardas, P.E. MicroRNA-9 and microRNA-126 expression levels in patients with essential hypertension: Potential markers of target-organ damage. J. Am. Soc. Hypertens. 2014, 8, 368–375. [Google Scholar] [CrossRef]
- Anaparti, V.; Smolik, I.; Meng, X.; Spicer, V.; Mookherjee, N.; El-Gabalawy, H. Whole blood microRNA expression pattern differentiates patients with rheumatoid arthritis, their seropositive first-degree relatives, and healthy unrelated control subjects. Arthritis Res. Ther. 2017, 19, 249. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.H.; Gao, Y.; Ren, A.J.; Zhao, S.H.; Zhong, M.; Peng, Y.J.; Shen, W.; Jing, M.; Liu, L. Altered microRNA expression profiles in retinas with diabetic retinopathy. Ophthalmic Res. 2012, 47, 195–201. [Google Scholar] [CrossRef] [PubMed]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA expression profile of human subcutaneous adipose and during adipocyte differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [Green Version]
- Fehlmann, T.; Lehallier, B.; Schaum, N.; Hahn, O.; Kahraman, M.; Li, Y.; Grammes, N.; Geffers, L.; Backes, C.; Balling, R.; et al. Common diseases alter the physiological age-related blood microRNA profile. Nat. Commun. 2020, 11, 5958. [Google Scholar] [CrossRef]
- Gui, Y.; Liu, H.; Zhang, L.; Lv, W.; Hu, X. Altered microRNA profiles in cerebrospinal fluid exosome in Parkinson disease and Alzheimer disease. Oncotarget 2015, 6, 37043–37053. [Google Scholar] [CrossRef] [Green Version]
- Sproviero, D.; Gagliardi, S.; Zucca, S.; Arigoni, M.; Giannini, M.; Garofalo, M.; Olivero, M.; Dell’Orco, M.; Pansarasa, O.; Bernuzzi, S.; et al. Different miRNA Profiles in Plasma Derived Small and Large Extracellular Vesicles from Patients with Neurodegenerative Diseases. Int. J. Mol. Sci. 2021, 22, 2737. [Google Scholar] [CrossRef]
- Shamsuzzama; Kumar, L.; Haque, R.; Nazir, A. Role of MicroRNA Let-7 in Modulating Multifactorial Aspect of Neurodegenerative Diseases: An Overview. Mol. Neurobiol. 2016, 53, 2787–2793. [Google Scholar] [CrossRef]
- Christopher, A.F.; Kaur, R.P.; Kaur, G.; Kaur, A.; Gupta, V.; Bansal, P. MicroRNA therapeutics: Discovering novel targets and developing specific therapy. Perspect. Clin. Res. 2016, 7, 68–74. [Google Scholar]
- Birks, J.S.; Harvey, R.J. Donepezil for dementia due to Alzheimer’s disease. Cochrane Database Syst. Rev. 2018, 6, CD001190. [Google Scholar] [CrossRef]
- Shintani, E.Y.; Uchida, K.M. Donepezil: An anticholinesterase inhibitor for Alzheimer’s disease. Am. J. Health Syst. Pharm. 1997, 54, 2805–2810. [Google Scholar] [CrossRef]
- McShane, R.; Westby, M.J.; Roberts, E.; Minakaran, N.; Schneider, L.; Farrimond, L.E.; Maayan, N.; Ware, J.; Debarros, J. Memantine for dementia. Cochrane Database Syst. Rev. 2019, 3, CD003154. [Google Scholar] [CrossRef]
- Popik, P.; Wrobel, M.; Rygula, R.; Bisaga, A.; Bespalov, A.Y. Effects of memantine, an NMDA receptor antagonist, on place preference conditioned with drug and nondrug reinforcers in mice. Behav. Pharmacol. 2003, 14, 237–244. [Google Scholar] [CrossRef]
- Knowles, J. Donepezil in Alzheimer’s disease: An evidence-based review of its impact on clinical and economic outcomes. Core Evid. 2006, 1, 195–219. [Google Scholar]
- Tampi, R.R.; van Dyck, C.H. Memantine: Efficacy and safety in mild-to-severe Alzheimer’s disease. Neuropsychiatr. Dis. Treat. 2007, 3, 245–258. [Google Scholar] [CrossRef]
- Sevigny, J.; Chiao, P.; Bussiere, T.; Weinreb, P.H.; Williams, L.; Maier, M.; Dunstan, R.; Salloway, S.; Chen, T.; Ling, Y.; et al. The antibody aducanumab reduces Abeta plaques in Alzheimer’s disease. Nature 2016, 537, 50–56. [Google Scholar] [CrossRef]
- Liu, K.Y.; Schneider, L.S.; Howard, R. The need to show minimum clinically important differences in Alzheimer’s disease trials. Lancet Psychiatry 2021, 8, 1013–1016. [Google Scholar] [CrossRef]
- Dunn, B.; Stein, P.; Cavazzoni, P. Approval of Aducanumab for Alzheimer Disease-The FDA’s Perspective. JAMA Intern. Med. 2021, 181, 1276–1278. [Google Scholar] [CrossRef]
- Mullard, A. Landmark Alzheimer’s drug approval confounds research community. Nature 2021, 594, 309–310. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Yu, J.T.; Hu, N.; Tan, L. Non-coding RNAs in Alzheimer’s disease. Mol. Neurobiol. 2013, 47, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Lukiw, W.J. Micro-RNA speciation in fetal, adult and Alzheimer’s disease hippocampus. Neuroreport 2007, 18, 297–300. [Google Scholar] [CrossRef] [PubMed]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Papadopoulou, A.S.; Mandemakers, W.; Silahtaroglu, A.N.; Kauppinen, S.; Delacourte, A.; De Strooper, B. Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer’s disease correlates with increased BACE1/beta-secretase expression. Proc. Natl. Acad. Sci. USA 2008, 105, 6415–6420. [Google Scholar] [CrossRef] [Green Version]
- Miya Shaik, M.; Tamargo, I.A.; Abubakar, M.B.; Kamal, M.A.; Greig, N.H.; Gan, S.H. The Role of microRNAs in Alzheimer’s Disease and Their Therapeutic Potentials. Genes 2018, 9, 174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarbrick, S.; Wragg, N.; Ghosh, S.; Stolzing, A. Systematic Review of miRNA as Biomarkers in Alzheimer’s Disease. Mol. Neurobiol. 2019, 56, 6156–6167. [Google Scholar] [CrossRef] [Green Version]
- Schipper, H.M.; Maes, O.C.; Chertkow, H.M.; Wang, E. MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Biol. 2007, 1, 263–274. [Google Scholar] [CrossRef]
- Hardy, J.; Allsop, D. Amyloid deposition as the central event in the aetiology of Alzheimer’s disease. Trends Pharmacol. Sci. 1991, 12, 383–388. [Google Scholar] [CrossRef]
- Coronel, R.; Bernabeu-Zornoza, A.; Palmer, C.; Muniz-Moreno, M.; Zambrano, A.; Cano, E.; Liste, I. Role of Amyloid Precursor Protein (APP) and Its Derivatives in the Biology and Cell Fate Specification of Neural Stem Cells. Mol. Neurobiol. 2018, 55, 7107–7117. [Google Scholar] [CrossRef] [PubMed]
- Lustbader, J.W.; Cirilli, M.; Lin, C.; Xu, H.W.; Takuma, K.; Wang, N.; Caspersen, C.; Chen, X.; Pollak, S.; Chaney, M.; et al. ABAD directly links Abeta to mitochondrial toxicity in Alzheimer’s disease. Science 2004, 304, 448–452. [Google Scholar] [CrossRef] [Green Version]
- Kuchibhotla, K.V.; Goldman, S.T.; Lattarulo, C.R.; Wu, H.Y.; Hyman, B.T.; Bacskai, B.J. Abeta plaques lead to aberrant regulation of calcium homeostasis in vivo resulting in structural and functional disruption of neuronal networks. Neuron 2008, 59, 214–225. [Google Scholar] [CrossRef] [Green Version]
- Hunt, D.L.; Castillo, P.E. Synaptic plasticity of NMDA receptors: Mechanisms and functional implications. Curr. Opin. Neurobiol. 2012, 22, 496–508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hebert, S.S.; Horre, K.; Nicolai, L.; Bergmans, B.; Papadopoulou, A.S.; Delacourte, A.; De Strooper, B. MicroRNA regulation of Alzheimer’s Amyloid precursor protein expression. Neurobiol. Dis. 2009, 33, 422–428. [Google Scholar] [CrossRef]
- Patel, N.; Hoang, D.; Miller, N.; Ansaloni, S.; Huang, Q.; Rogers, J.T.; Lee, J.C.; Saunders, A.J. MicroRNAs can regulate human APP levels. Mol. Neurodegener. 2008, 3, 10. [Google Scholar] [CrossRef] [Green Version]
- Vilardo, E.; Barbato, C.; Ciotti, M.; Cogoni, C.; Ruberti, F. MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 2010, 285, 18344–18351. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.; Zhu, H.; Xu, Y.; Huang, L.; Ma, C.; Deng, W.; Liu, Y.; Qin, C. MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 2012, 1455, 103–113. [Google Scholar] [CrossRef]
- Delay, C.; Calon, F.; Mathews, P.; Hebert, S.S. Alzheimer-specific variants in the 3’UTR of Amyloid precursor protein affect microRNA function. Mol. Neurodegener. 2011, 6, 70. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; He, P.; Yao, H.; Dong, Q.; Li, R.; Shen, Y. Occludin deficiency with BACE1 elevation in cerebral amyloid angiopathy. Neurology 2014, 82, 1707–1715. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, F.; Gong, G.; Wang, Y.; Bian, M.; Yu, L.; Wei, C. MiR-124 acts as a target for Alzheimer’s disease by regulating BACE1. Oncotarget 2017, 8, 114065–114071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chopra, N.; Wang, R.; Maloney, B.; Nho, K.; Beck, J.S.; Pourshafie, N.; Niculescu, A.; Saykin, A.J.; Rinaldi, C.; Counts, S.E.; et al. MicroRNA-298 reduces levels of human amyloid-beta precursor protein (APP), beta-site APP-converting enzyme 1 (BACE1) and specific tau protein moieties. Mol. Psychiatry 2020, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Ding, Y.; Hou, D. Research status of the regulation of miRNA on BACE1. Int. J. Neurosci. 2014, 124, 474–477. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Zhao, Y.; Zhou, Y.; Liu, L.; Liu, Y.; Wang, D.; Zhang, S.; Yang, M. MiR-9 Regulates the Expression of BACE1 in Dementia Induced by Chronic Brain Hypoperfusion in Rats. Cell Physiol. Biochem. 2017, 42, 1213–1226. [Google Scholar] [CrossRef] [Green Version]
- Faghihi, M.A.; Zhang, M.; Huang, J.; Modarresi, F.; Van der Brug, M.P.; Nalls, M.A.; Cookson, M.R.; St-Laurent, G., 3rd; Wahlestedt, C. Evidence for natural antisense transcript-mediated inhibition of microRNA function. Genome Biol. 2010, 11, R56. [Google Scholar] [CrossRef] [Green Version]
- Boissonneault, V.; Plante, I.; Rivest, S.; Provost, P. MicroRNA-298 and microRNA-328 regulate expression of mouse beta-amyloid precursor protein-converting enzyme 1. J. Biol. Chem. 2009, 284, 1971–1981. [Google Scholar] [CrossRef] [Green Version]
- Gong, G.; An, F.; Wang, Y.; Bian, M.; Yu, L.J.; Wei, C. miR-15b represses BACE1 expression in sporadic Alzheimer’s disease. Oncotarget 2017, 8, 91551–91557. [Google Scholar] [CrossRef] [Green Version]
- Moncini, S.; Lunghi, M.; Valmadre, A.; Grasso, M.; Del Vescovo, V.; Riva, P.; Denti, M.A.; Venturin, M. The miR-15/107 Family of microRNA Genes Regulates CDK5R1/p35 with Implications for Alzheimer’s Disease Pathogenesis. Mol. Neurobiol. 2017, 54, 4329–4342. [Google Scholar] [CrossRef]
- Zhu, H.C.; Wang, L.M.; Wang, M.; Song, B.; Tan, S.; Teng, J.F.; Duan, D.X. MicroRNA-195 downregulates Alzheimer’s disease amyloid-beta production by targeting BACE1. Brain Res. Bull. 2012, 88, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Lei, X.; Lei, L.; Zhang, Z.; Zhang, Z.; Cheng, Y. Downregulated miR-29c correlates with increased BACE1 expression in sporadic Alzheimer’s disease. Int. J. Clin. Exp. Pathol. 2015, 8, 1565–1574. [Google Scholar] [PubMed]
- Sun, B.; Zhou, Y.; Halabisky, B.; Lo, I.; Cho, S.H.; Mueller-Steiner, S.; Devidze, N.; Wang, X.; Grubb, A.; Gan, L. Cystatin C-cathepsin B axis regulates amyloid beta levels and associated neuronal deficits in an animal model of Alzheimer’s disease. Neuron 2008, 60, 247–257. [Google Scholar] [CrossRef] [Green Version]
- Tiribuzi, R.; Crispoltoni, L.; Porcellati, S.; Di Lullo, M.; Florenzano, F.; Pirro, M.; Bagaglia, F.; Kawarai, T.; Zampolini, M.; Orlacchio, A.; et al. miR128 up-regulation correlates with impaired amyloid beta(1-42) degradation in monocytes from patients with sporadic Alzheimer’s disease. Neurobiol. Aging 2014, 35, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Dixit, R.; Ross, J.L.; Goldman, Y.E.; Holzbaur, E.L. Differential regulation of dynein and kinesin motor proteins by tau. Science 2008, 319, 1086–1089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kimura, T.; Ishiguro, K.; Hisanaga, S. Physiological and pathological phosphorylation of tau by Cdk5. Front. Mol. Neurosci. 2014, 7, 65. [Google Scholar] [CrossRef] [Green Version]
- Jara, C.; Aranguiz, A.; Cerpa, W.; Tapia-Rojas, C.; Quintanilla, R.A. Genetic ablation of tau improves mitochondrial function and cognitive abilities in the hippocampus. Redox Biol. 2018, 18, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Wilson, D.M.; Binder, L.I. Polymerization of microtubule-associated protein tau under near-physiological conditions. J. Biol. Chem. 1995, 270, 24306–24314. [Google Scholar] [CrossRef] [Green Version]
- Grundke-Iqbal, I.; Iqbal, K.; Tung, Y.C.; Quinlan, M.; Wisniewski, H.M.; Binder, L.I. Abnormal phosphorylation of the microtubule-associated protein tau (tau) in Alzheimer cytoskeletal pathology. Proc. Natl. Acad. Sci. USA 1986, 83, 4913–4917. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Spires-Jones, T.L.; Hyman, B.T. The intersection of amyloid beta and tau at synapses in Alzheimer’s disease. Neuron 2014, 82, 756–771. [Google Scholar] [CrossRef] [Green Version]
- Callahan, L.M.; Vaules, W.A.; Coleman, P.D. Quantitative decrease in synaptophysin message expression and increase in cathepsin D message expression in Alzheimer disease neurons containing neurofibrillary tangles. J. Neuropathol. Exp. Neurol. 1999, 58, 275–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, S.S.; Bloom, G.S. Tau: The Center of a Signaling Nexus in Alzheimer’s Disease. Front. Neurosci. 2016, 10, 31. [Google Scholar] [CrossRef] [Green Version]
- Carrettiero, D.C.; Hernandez, I.; Neveu, P.; Papagiannakopoulos, T.; Kosik, K.S. The cochaperone BAG2 sweeps paired helical filament- insoluble tau from the microtubule. J. Neurosci. 2009, 29, 2151–2161. [Google Scholar] [CrossRef]
- Hebert, S.S.; Sergeant, N.; Buee, L. MicroRNAs and the Regulation of Tau Metabolism. Int. J. Alzheimer’s Dis. 2012, 2012, 406561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramachandran, D.; Roy, U.; Garg, S.; Ghosh, S.; Pathak, S.; Kolthur-Seetharam, U. Sirt1 and mir-9 expression is regulated during glucose-stimulated insulin secretion in pancreatic beta-islets. FEBS J. 2011, 278, 1167–1174. [Google Scholar] [CrossRef] [PubMed]
- Absalon, S.; Kochanek, D.M.; Raghavan, V.; Krichevsky, A.M. MiR-26b, upregulated in Alzheimer’s disease, activates cell cycle entry, tau-phosphorylation, and apoptosis in postmitotic neurons. J. Neurosci. 2013, 33, 14645–14659. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Tang, H.; Li, X.Y.; Deng, M.F.; Wei, N.; Wang, X.; Zhou, Y.F.; Wang, D.Q.; Fu, P.; Wang, J.Z.; et al. Targeting the HDAC2/HNF-4A/miR-101b/AMPK Pathway Rescues Tauopathy and Dendritic Abnormalities in Alzheimer’s Disease. Mol. Ther. 2017, 25, 752–764. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Z.B.; Wu, L.; Xiong, R.; Wang, L.L.; Zhang, B.; Wang, C.; Li, H.; Liang, L.; Chen, S.D. MicroRNA-922 promotes tau phosphorylation by downregulating ubiquitin carboxy-terminal hydrolase L1 (UCHL1) expression in the pathogenesis of Alzheimer’s disease. Neuroscience 2014, 275, 232–237. [Google Scholar] [CrossRef]
- Wang, X.; Tan, L.; Lu, Y.; Peng, J.; Zhu, Y.; Zhang, Y.; Sun, Z. MicroRNA-138 promotes tau phosphorylation by targeting retinoic acid receptor alpha. FEBS Lett. 2015, 589, 726–729. [Google Scholar] [CrossRef] [Green Version]
- Craft, J.M.; Watterson, D.M.; Van Eldik, L.J. Human amyloid beta-induced neuroinflammation is an early event in neurodegeneration. Glia 2006, 53, 484–490. [Google Scholar] [CrossRef]
- Pizza, V.; Agresta, A.; D’Acunto, C.W.; Festa, M.; Capasso, A. Neuroinflamm-aging and neurodegenerative diseases: An overview. CNS Neurol. Disord Drug Targets 2011, 10, 621–634. [Google Scholar] [CrossRef]
- Varnum, M.M.; Ikezu, T. The classification of microglial activation phenotypes on neurodegeneration and regeneration in Alzheimer’s disease brain. Arch. Immunol. Ther. Exp. 2012, 60, 251–266. [Google Scholar] [CrossRef] [PubMed]
- Cagnin, A.; Brooks, D.J.; Kennedy, A.M.; Gunn, R.N.; Myers, R.; Turkheimer, F.E.; Jones, T.; Banati, R.B. In-vivo measurement of activated microglia in dementia. Lancet 2001, 358, 461–467. [Google Scholar] [CrossRef]
- Fillit, H.; Ding, W.H.; Buee, L.; Kalman, J.; Altstiel, L.; Lawlor, B.; Wolf-Klein, G. Elevated circulating tumor necrosis factor levels in Alzheimer’s disease. Neurosci. Lett. 1991, 129, 318–320. [Google Scholar] [CrossRef]
- Tan, M.S.; Yu, J.T.; Jiang, T.; Zhu, X.C.; Tan, L. The NLRP3 inflammasome in Alzheimer’s disease. Mol. Neurobiol. 2013, 48, 875–882. [Google Scholar] [CrossRef]
- Hanslik, K.L.; Ulland, T.K. The Role of Microglia and the Nlrp3 Inflammasome in Alzheimer’s Disease. Front. Neurol. 2020, 11, 570711. [Google Scholar] [CrossRef]
- Gustin, A.; Kirchmeyer, M.; Koncina, E.; Felten, P.; Losciuto, S.; Heurtaux, T.; Tardivel, A.; Heuschling, P.; Dostert, C. NLRP3 Inflammasome Is Expressed and Functional in Mouse Brain Microglia but Not in Astrocytes. PLoS ONE 2015, 10, e0130624. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Lu, M.; Du, R.H.; Qiao, C.; Jiang, C.Y.; Zhang, K.Z.; Ding, J.H.; Hu, G. MicroRNA-7 targets Nod-like receptor protein 3 inflammasome to modulate neuroinflammation in the pathogenesis of Parkinson’s disease. Mol. Neurodegener. 2016, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhang, H.; Lu, X.; Wang, J.; Zhang, X.; Sun, S.; Bao, Z.; Tian, W.; Ning, S.; Wang, L.; et al. Overexpression of MicroRNA-9a-5p Ameliorates NLRP1 Inflammasome-mediated Ischemic Injury in Rats Following Ischemic Stroke. Neuroscience 2020, 444, 106–117. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Voet, S.; Srinivasan, S.; Lamkanfi, M.; van Loo, G. Inflammasomes in neuroinflammatory and neurodegenerative diseases. EMBO Mol. Med. 2019, 11, e10248. [Google Scholar] [CrossRef] [PubMed]
- Han, C.; Guo, L.; Yang, Y.; Guan, Q.; Shen, H.; Sheng, Y.; Jiao, Q. Mechanism of microRNA-22 in regulating neuroinflammation in Alzheimer’s disease. Brain Behav. 2020, 10, e01627. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Chu, W.; Gong, L.; Gao, X.; Wang, W. MicroRNA-26b is upregulated in a double transgenic mouse model of Alzheimer’s disease and promotes the expression of amyloid-beta by targeting insulin-like growth factor 1. Mol. Med. Rep. 2016, 13, 2809–2814. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.T.; Chu, K.; Jung, K.H.; Kim, J.H.; Huh, J.Y.; Yoon, H.; Park, D.K.; Lim, J.Y.; Kim, J.M.; Jeon, D.; et al. miR-206 regulates brain-derived neurotrophic factor in Alzheimer disease model. Ann. Neurol. 2012, 72, 269–277. [Google Scholar] [CrossRef]
- Park, I.; Kim, H.J.; Kim, Y.; Hwang, H.S.; Kasai, H.; Kim, J.H.; Park, J.W. Nanoscale imaging reveals miRNA-mediated control of functional states of dendritic spines. Proc. Natl. Acad. Sci. USA 2019, 116, 9616–9621. [Google Scholar] [CrossRef] [Green Version]
- Schratt, G.M.; Tuebing, F.; Nigh, E.A.; Kane, C.G.; Sabatini, M.E.; Kiebler, M.; Greenberg, M.E. A brain-specific microRNA regulates dendritic spine development. Nature 2006, 439, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Fiore, R.; Rajman, M.; Schwale, C.; Bicker, S.; Antoniou, A.; Bruehl, C.; Draguhn, A.; Schratt, G. MiR-134-dependent regulation of Pumilio-2 is necessary for homeostatic synaptic depression. EMBO J. 2014, 33, 2231–2246. [Google Scholar] [CrossRef] [Green Version]
- Giusti, S.A.; Vogl, A.M.; Brockmann, M.M.; Vercelli, C.A.; Rein, M.L.; Trumbach, D.; Wurst, W.; Cazalla, D.; Stein, V.; Deussing, J.M.; et al. MicroRNA-9 controls dendritic development by targeting REST. Elife 2014, 3, e02755. [Google Scholar] [CrossRef]
- Ren, Z.; Yu, J.; Wu, Z.; Si, W.; Li, X.; Liu, Y.; Zhou, J.; Deng, R.; Chen, D. MicroRNA-210-5p Contributes to Cognitive Impairment in Early Vascular Dementia Rat Model Through Targeting Snap25. Front. Mol. Neurosci. 2018, 11, 388. [Google Scholar] [CrossRef]
- Cohen, J.E.; Lee, P.R.; Chen, S.; Li, W.; Fields, R.D. MicroRNA regulation of homeostatic synaptic plasticity. Proc. Natl. Acad. Sci. USA 2011, 108, 11650–11655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, S.; Wang, H.; Chen, K.; Cheng, P.; Gao, S.; Liu, J.; Li, X.; Sun, X. MicroRNA-34c Downregulation Ameliorates Amyloid-beta-Induced Synaptic Failure and Memory Deficits by Targeting VAMP2. J. Alzheimer’s Dis. 2015, 48, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Sim, S.E.; Lim, C.S.; Kim, J.I.; Seo, D.; Chun, H.; Yu, N.K.; Lee, J.; Kang, S.J.; Ko, H.G.; Choi, J.H.; et al. The Brain-Enriched MicroRNA miR-9-3p Regulates Synaptic Plasticity and Memory. J. Neurosci. 2016, 36, 8641–8652. [Google Scholar] [CrossRef] [PubMed]
- Letellier, M.; Elramah, S.; Mondin, M.; Soula, A.; Penn, A.; Choquet, D.; Landry, M.; Thoumine, O.; Favereaux, A. miR-92a regulates expression of synaptic GluA1-containing AMPA receptors during homeostatic scaling. Nat. Neurosci. 2014, 17, 1040–1042. [Google Scholar] [CrossRef]
- Hu, Z.; Zhao, J.; Hu, T.; Luo, Y.; Zhu, J.; Li, Z. miR-501-3p mediates the activity-dependent regulation of the expression of AMPA receptor subunit GluA1. J. Cell Biol. 2015, 208, 949–959. [Google Scholar] [CrossRef] [Green Version]
- Olde Loohuis, N.F.; Ba, W.; Stoerchel, P.H.; Kos, A.; Jager, A.; Schratt, G.; Martens, G.J.; van Bokhoven, H.; Nadif Kasri, N.; Aschrafi, A. MicroRNA-137 Controls AMPA-Receptor-Mediated Transmission and mGluR-Dependent LTD. Cell Rep. 2015, 11, 1876–1884. [Google Scholar] [CrossRef] [Green Version]
- Baby, N.; Alagappan, N.; Dheen, S.T.; Sajikumar, S. MicroRNA-134-5p inhibition rescues long-term plasticity and synaptic tagging/capture in an Abeta(1-42)-induced model of Alzheimer’s disease. Aging Cell 2020, 19, e13046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edbauer, D.; Neilson, J.R.; Foster, K.A.; Wang, C.F.; Seeburg, D.P.; Batterton, M.N.; Tada, T.; Dolan, B.M.; Sharp, P.A.; Sheng, M. Regulation of synaptic structure and function by FMRP-associated microRNAs miR-125b and miR-132. Neuron 2010, 65, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Engler-Chiurazzi, E.B.; Cavendish, J.Z.; Povroznik, J.M.; Russell, A.E.; Quintana, D.D.; Mathers, P.H.; Simpkins, J.W. Over-expression of miR-34a induces rapid cognitive impairment and Alzheimer’s disease-like pathology. Brain Res. 2019, 1721, 146327. [Google Scholar] [CrossRef]
- Peng, Z.; Li, J.; Li, Y.; Yang, X.; Feng, S.; Han, S.; Li, J. Downregulation of miR-181b in mouse brain following ischemic stroke induces neuroprotection against ischemic injury through targeting heat shock protein A5 and ubiquitin carboxyl-terminal hydrolase isozyme L1. J. Neurosci. Res. 2013, 91, 1349–1362. [Google Scholar] [CrossRef]
- Kang, Q.; Xiang, Y.; Li, D.; Liang, J.; Zhang, X.; Zhou, F.; Qiao, M.; Nie, Y.; He, Y.; Cheng, J.; et al. MiR-124-3p attenuates hyperphosphorylation of Tau protein-induced apoptosis via caveolin-1-PI3K/Akt/GSK3beta pathway in N2a/APP695swe cells. Oncotarget 2017, 8, 24314–24326. [Google Scholar] [CrossRef]
- Salta, E.; Sierksma, A.; Vanden Eynden, E.; De Strooper, B. miR-132 loss de-represses ITPKB and aggravates amyloid and TAU pathology in Alzheimer’s brain. EMBO Mol. Med. 2016, 8, 1005–1018. [Google Scholar] [CrossRef]
- Dickson, J.R.; Kruse, C.; Montagna, D.R.; Finsen, B.; Wolfe, M.S. Alternative polyadenylation and miR-34 family members regulate tau expression. J. Neurochem. 2013, 127, 739–749. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bauernfeind, F.; Rieger, A.; Schildberg, F.A.; Knolle, P.A.; Schmid-Burgk, J.L.; Hornung, V. NLRP3 inflammasome activity is negatively controlled by miR-223. J. Immunol. 2012, 189, 4175–4181. [Google Scholar] [CrossRef] [Green Version]
- Teng, G.G.; Wang, W.H.; Dai, Y.; Wang, S.J.; Chu, Y.X.; Li, J. Let-7b is involved in the inflammation and immune responses associated with Helicobacter pylori infection by targeting Toll-like receptor 4. PLoS ONE 2013, 8, e56709. [Google Scholar]
- Schulte, L.N.; Eulalio, A.; Mollenkopf, H.J.; Reinhardt, R.; Vogel, J. Analysis of the host microRNA response to Salmonella uncovers the control of major cytokines by the let-7 family. EMBO J. 2011, 30, 1977–1989. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, L.; Li, H.; Liu, W.; Zhang, L.; Tian, Q.; Li, H.; Li, M. MiR-485-3p serves as a biomarker and therapeutic target of Alzheimer’s disease via regulating neuronal cell viability and neuroinflammation by targeting AKT3. Mol. Genet. Genom. Med. 2021, 9, e1548. [Google Scholar] [CrossRef]
- Norton, S.; Matthews, F.E.; Barnes, D.E.; Yaffe, K.; Brayne, C. Potential for primary prevention of Alzheimer’s disease: An analysis of population-based data. Lancet Neurol. 2014, 13, 788–794. [Google Scholar] [CrossRef] [Green Version]
- Dolgin, E. Alzheimer’s disease is getting easier to spot. Nature 2018, 559, S10–S12. [Google Scholar] [CrossRef]
- McKhann, G.M.; Knopman, D.S.; Chertkow, H.; Hyman, B.T.; Jack, C.R., Jr.; Kawas, C.H.; Klunk, W.E.; Koroshetz, W.J.; Manly, J.J.; Mayeux, R.; et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dement. 2011, 7, 263–269. [Google Scholar] [CrossRef] [Green Version]
- Bhatnagar, S.; Chertkow, H.; Schipper, H.M.; Yuan, Z.; Shetty, V.; Jenkins, S.; Jones, T.; Wang, E. Increased microRNA-34c abundance in Alzheimer’s disease circulating blood plasma. Front. Mol. Neurosci. 2014, 7, 2. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, C.; Zhang, Y. An investigation of microRNA-103 and microRNA-107 as potential blood-based biomarkers for disease risk and progression of Alzheimer’s disease. J. Clin. Lab. Anal. 2020, 34, e23006. [Google Scholar] [CrossRef] [Green Version]
- Souza, V.C.; Morais, G.S., Jr.; Henriques, A.D.; Machado-Silva, W.; Perez, D.I.V.; Brito, C.J.; Camargos, E.F.; Moraes, C.F.; Nobrega, O.T. Whole-Blood Levels of MicroRNA-9 Are Decreased in Patients with Late-Onset Alzheimer Disease. Am. J. Alzheimer’s Dis. Other Dement. 2020, 35, 1533317520911573. [Google Scholar] [CrossRef] [PubMed]
- Loganantharaj, R.; Randall, T.A. The Limitations of Existing Approaches in Improving MicroRNA Target Prediction Accuracy. Methods Mol. Biol. 2017, 1617, 133–158. [Google Scholar]
- Guo, L.; Zhang, Q.; Ma, X.; Wang, J.; Liang, T. miRNA and mRNA expression analysis reveals potential sex-biased miRNA expression. Sci. Rep. 2017, 7, 39812. [Google Scholar] [CrossRef] [Green Version]
- Cui, C.; Yang, W.; Shi, J.; Zhou, Y.; Yang, J.; Cui, Q.; Zhou, Y. Identification and Analysis of Human Sex-biased MicroRNAs. Genom. Proteom. Bioinform. 2018, 16, 200–211. [Google Scholar] [CrossRef]
- Huan, T.; Chen, G.; Liu, C.; Bhattacharya, A.; Rong, J.; Chen, B.H.; Seshadri, S.; Tanriverdi, K.; Freedman, J.E.; Larson, M.G.; et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 2018, 17, e12687. [Google Scholar] [CrossRef]
- Noren Hooten, N.; Abdelmohsen, K.; Gorospe, M.; Ejiogu, N.; Zonderman, A.B.; Evans, M.K. microRNA expression patterns reveal differential expression of target genes with age. PLoS ONE 2010, 5, e10724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Qin, Y.W.; Brewer, G.; Jing, Q. MicroRNA degradation and turnover: Regulating the regulators. Wiley Interdiscip. Rev. RNA 2012, 3, 593–600. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.M.; Bhakta, N.R.; Barczak, A.J.; Cakmak, H.; Fisher, S.; MacKenzie, T.C.; Patel, T.; Price, R.W.; Smith, J.F.; Woodruff, P.G.; et al. Large Differences in Small RNA Composition Between Human Biofluids. Cell Rep. 2018, 25, 1346–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koshiol, J.; Wang, E.; Zhao, Y.; Marincola, F.; Landi, M.T. Strengths and limitations of laboratory procedures for microRNA detection. Cancer Epidemiol. Biomark. Prev. 2010, 19, 907–911. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).