Sequence Does Not Matter: The Biomedical Applications of DNA-Based Coatings and Cores
Abstract
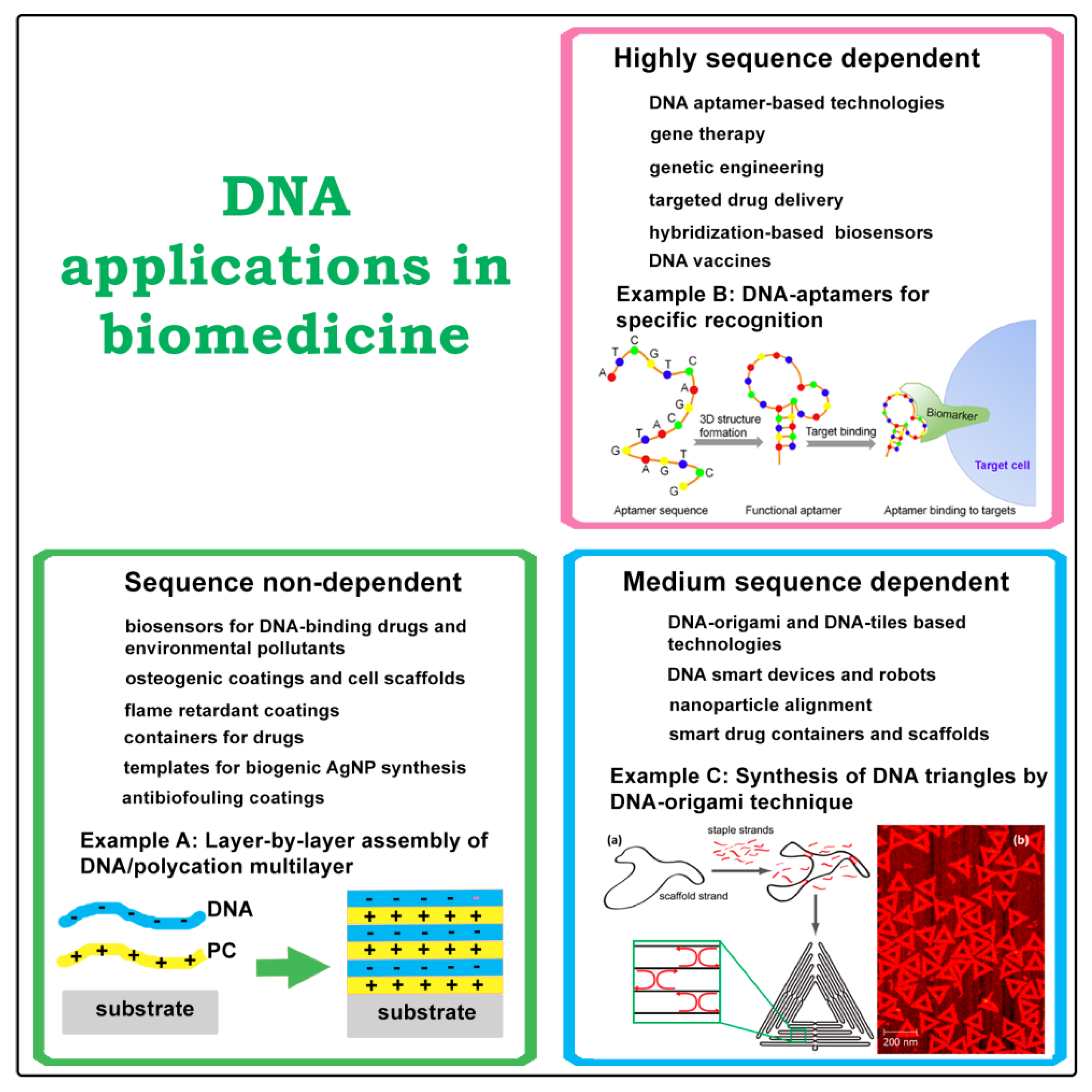
:1. Introduction
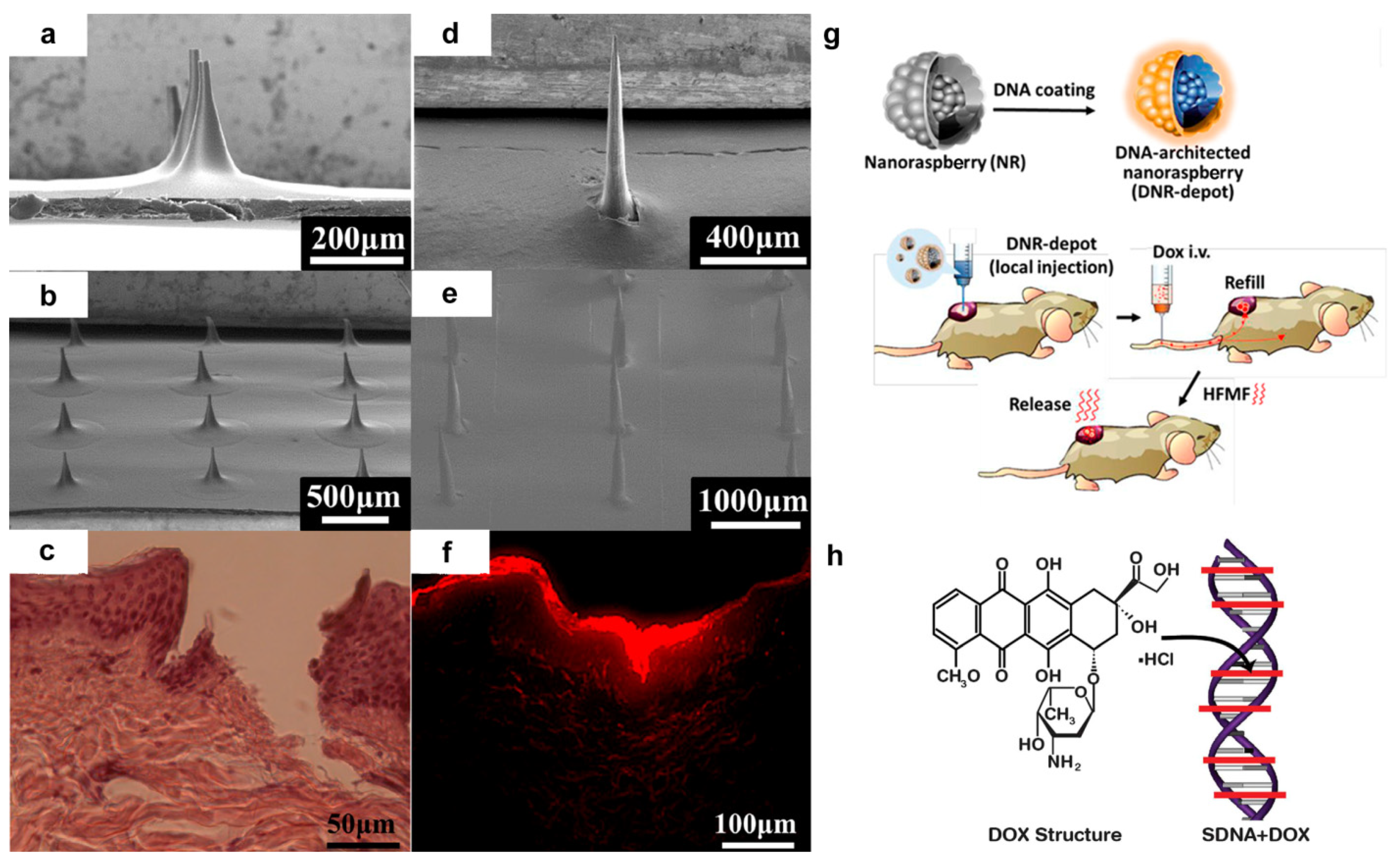
2. DNA Based Drug Delivery Vehicles
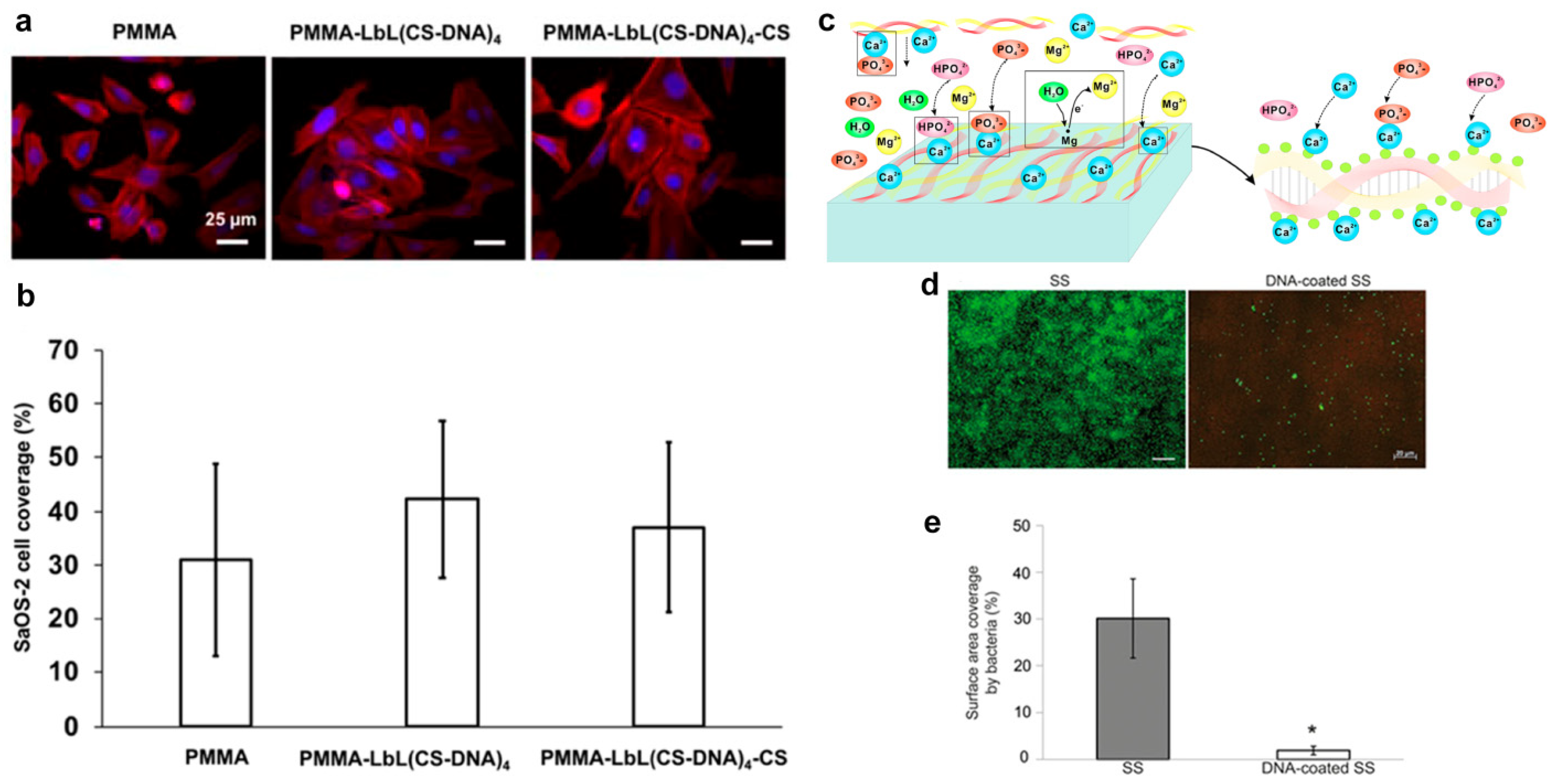
3. Cytocompatible and Antibiofouling DNA Coatings
4. Antibacterial and Flame-Retardant Coatings for Fabrics Based on DNA
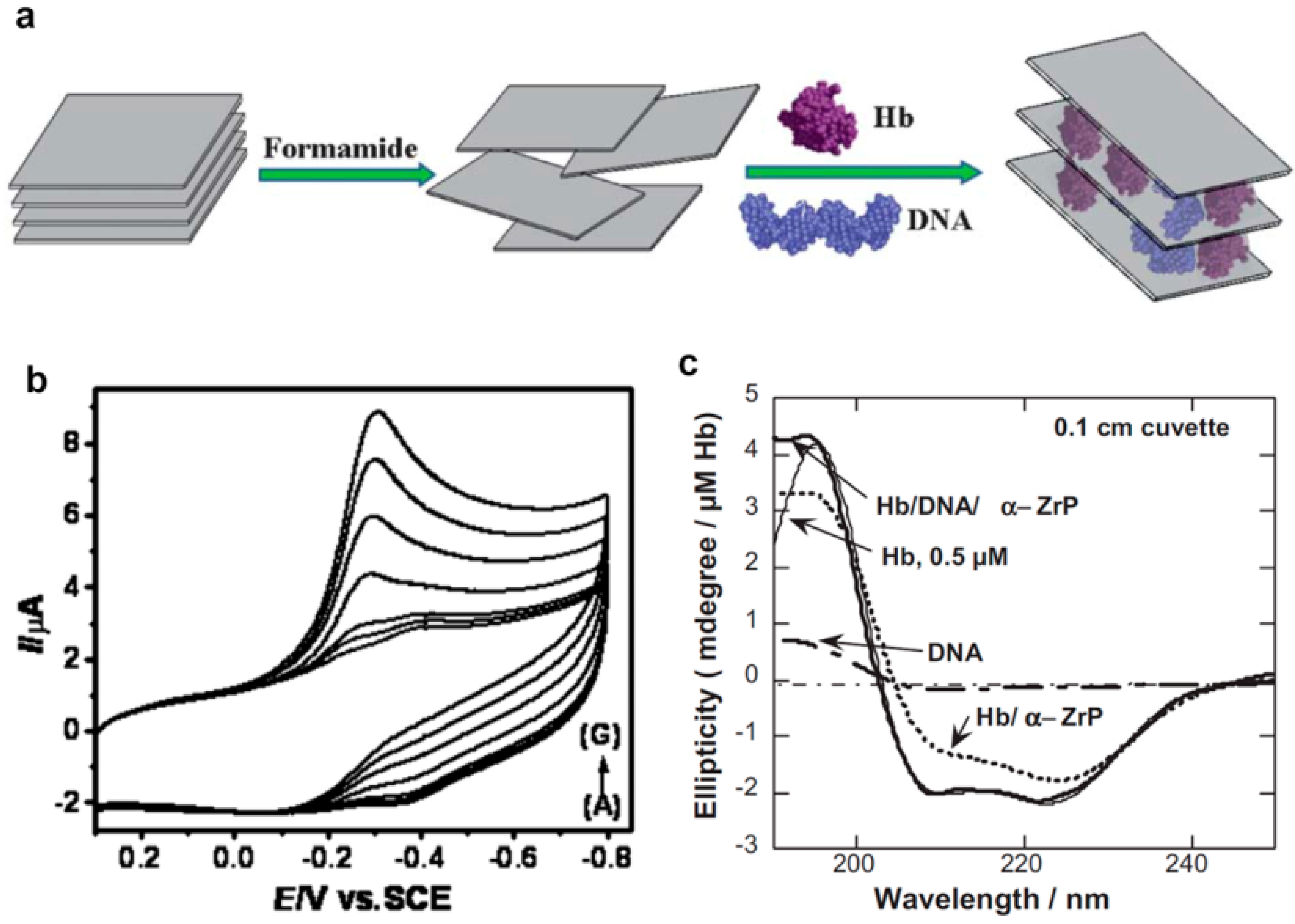
5. DNA as a Part of Electrochemical Biosensors
6. The Limitations, Challenges and Future Prospects of Raw DNA Applications in Biomedicine
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Linnik, D.S.; Tarakanchikova, Y.V.; Zyuzin, M.V.; Lepik, K.V.; Aerts, J.L.; Sukhorukov, G.; Timin, A.S. Layer-by-Layer technique as a versatile tool for gene delivery applications. Expert Opin. Drug Deliv. 2021, 18, 1047–1066. [Google Scholar] [CrossRef] [PubMed]
- Franck, C.O.; Fanslau, L.; Bistrovic Popov, A.; Tyagi, P.; Fruk, L. Biopolymer-based carriers for DNA vaccine design. Angew. Chem. Int. Ed. 2021, 60, 13225–13243. [Google Scholar] [CrossRef] [PubMed]
- Joshi, J.; Rubart, M.; Zhu, W. Optogenetics: Background, methodological advances and potential applications for cardiovascular research and medicine. Front. Bioeng. Biotechnol. 2020, 7, 466. [Google Scholar] [CrossRef] [Green Version]
- Mackelprang, R.; Lemaux, P.G. Genetic engineering and editing of plants: An analysis of new and persisting questions. Annu. Rev. Plant Biol. 2020, 71, 659–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, L.; Zhang, Q.; Deng, W.; Liu, H. DNA-based nanofabrication: Pathway to applications in surface engineering. Small 2019, 15, 1805428. [Google Scholar] [CrossRef]
- Ariga, K.; Jia, X.; Song, J.; Hill, J.P.; Leong, D.T.; Jia, Y.; Li, J. Nanoarchitectonics beyond self-assembly: Challenges to create bio-like hierarchic organization. Angew. Chem. Int. Ed. 2020, 59, 15424–15446. [Google Scholar] [CrossRef]
- Ghosh, D.; Datta, L.P.; Govindaraju, T. Molecular architectonics of DNA for functional nanoarchitectures. Beilstein J. Nanotechnol. 2020, 11, 124–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, S.; Wang, D.; Kenaan, A.; Cheng, J.; Cui, D.; Song, J. Create nanoscale patterns with DNA origami. Small 2019, 15, 1805554. [Google Scholar] [CrossRef]
- Zhang, Y.; Tu, J.; Wang, D.; Zhu, H.; Maity, S.K.; Qu, X.; Bogaert, B.; Pei, H.; Zhang, H. Programmable and multifunctional DNA-based materials for biomedical applications. Adv. Mater. 2018, 30, 1703658. [Google Scholar] [CrossRef]
- Yang, W.; Shen, Y.; Zhang, D.; Li, C.; Yuan, R.; Xu, W. Programmed dual-functional DNA tweezer for simultaneous and recognizable fluorescence detection of microRNA and protein. Anal. Chem. 2019, 91, 7782–7789. [Google Scholar] [CrossRef]
- Kolpashchikov, D.M. Evolution of hybridization probes to DNA machines and robots. Acc. Chem. Res. 2019, 52, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Molden, T.A.; Niccum, C.T.; Kolpashchikov, D.M. Cut and Paste for Cancer Treatment: A DNA nanodevice that cuts out an RNA marker sequence to activate a therapeutic function. Angew. Chem. Int. Ed. 2020, 59, 21190–21194. [Google Scholar] [CrossRef] [PubMed]
- Tang, J.; Yao, C.; Gu, Z.; Jung, S.; Luo, D.; Yang, D. Super-soft and super-elastic DNA robot with magnetically driven navigational locomotion for cell delivery in confined space. Angew. Chem. Int. Ed. 2020, 132, 2511–2516. [Google Scholar] [CrossRef]
- Wang, K.; He, M.Q.; Zhai, F.H.; Wang, J.; He, R.H.; Yu, Y.L. Autonomous DNA nanomachine based on cascade amplification of strand displacement and DNA walker for detection of multiple DNAs. Biosens. Bioelectron. 2018, 105, 159–165. [Google Scholar] [CrossRef]
- Chen, J.; Luo, Z.; Sun, C.; Huang, Z.; Zhou, C.; Yin, S.; Duan, Y.; Li, Y. Research progress of DNA walker and its recent applications in biosensor. Trends Anal. Chem. 2019, 120, 115626. [Google Scholar] [CrossRef]
- Thubagere, A.J.; Li, W.; Johnson, R.F.; Chen, Z.; Doroudi, S.; Lee, Y.L.; Izatt, G.; Wittman, S.; Srinivas, N.; Woods, D.; et al. A cargo-sorting DNA robot. Science 2017, 357, eaan6558. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Jiang, Q.; Wang, Y.; Ding, B. Biomedical applications of DNA-based molecular devices. Adv. Healthc. Mater. 2019, 8, 1801658. [Google Scholar] [CrossRef]
- Jahanban-Esfahlan, A.; Seidi, K.; Jaymand, M.; Schmidt, T.L.; Majdi, H.; Javaheri, T.; Jahanban-Esfahlan, R.; Zare, P. Dynamic DNA nanostructures in biomedicine: Beauty, utility and limits. J. Control. Release 2019, 315, 166–185. [Google Scholar] [CrossRef] [PubMed]
- Ramezani, H.; Dietz, H. Building machines with DNA molecules. Nat. Rev. Genet. 2020, 21, 5–26. [Google Scholar] [CrossRef]
- Nummelin, S.; Shen, B.; Piskunen, P.; Liu, Q.; Kostiainen, M.A.; Linko, V. Robotic DNA nanostructures. ACS Synth. Biol. 2020, 9, 1923–1940. [Google Scholar] [CrossRef] [PubMed]
- Scharnweber, D.; Bierbaum, S.; Wolf-Brandstetter, C. Utilizing DNA for functionalization of biomaterial surfaces. FEBS Lett. 2018, 592, 2181–2196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, K.; Zheng, L.; Ma, C.; Gösti, R.; Herrmann, A. DNA–surfactant complexes: Self-assembly properties and applications. Chem. Soc. Rev. 2017, 46, 5147–5172. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Li, Q.; Aldalbahi, A.; Wang, L.; Fan, C.; Liu, X. DNA-based fabrication for nanoelectronics. Nano Lett. 2020, 20, 5604–5615. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Li, H.; Xu, L.; Deng, Z.; Han, W.; Liu, Y.; Jiang, W.; Zu, Y. Oligonucleotide aptamer-mediated precision therapy of hematological malignancies. Mol. Ther. Nucleic Acids 2018, 7, 164–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bald, I.; Keller, A. Molecular processes studied at a single-molecule level using DNA origami nanostructures and atomic force microscopy. Molecules 2014, 19, 13803–13823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decher, G.; Hong, J.D.; Schmitt, J. Buildup of ultrathin multilayer films by a self-assembly process: III. Consecutively alternating adsorption of anionic and cationic polyelectrolytes on charged surfaces. Thin Solid Films 1992, 210, 831–835. [Google Scholar] [CrossRef]
- Sukhorukov, G.B.; Möhwald, H.; Decher, G.; Lvov, Y.M. Assembly of polyelectrolyte multilayer films by consecutively alternating adsorption of polynucleotides and polycations. Thin Solid Films 1996, 284, 220–223. [Google Scholar] [CrossRef]
- Borges, J.; Mano, J.F. Molecular interactions driving the layer-by-layer assembly of multilayers. Chem. Rev. 2014, 114, 8883–8942. [Google Scholar] [CrossRef]
- Lee, Y.; Dugansani, S.R.; Jeon, S.H.; Hwang, S.H.; Kim, J.H.; Park, S.H.; Jeong, J.H. Drug-delivery system based on salmon DNA nano-and micro-scale structures. Sci. Rep. 2017, 7, 9724. [Google Scholar] [CrossRef] [Green Version]
- Doberenz, F.; Zeng, K.; Willems, C.; Zhang, K.; Groth, T. Thermoresponsive polymers and their biomedical application in tissue engineering—A review. J. Mater. Chem. B 2020, 8, 607–628. [Google Scholar] [CrossRef]
- Hsu, R.S.; Fang, J.H.; Shen, W.T.; Sheu, Y.C.; Su, C.K.; Chiang, W.H.; Hu, S.H. Injectable DNA-architected nanoraspberry depot-mediated on-demand programmable refilling and release drug delivery. Nanoscale 2020, 12, 11153–11164. [Google Scholar] [CrossRef]
- Badran, M.M.; Kuntsche, J.; Fahr, A. Skin penetration enhancement by a microneedle device (Dermaroller®) in vitro: Dependency on needle size and applied formulation. Eur. J. Pharm. Sci. 2009, 36, 511–523. [Google Scholar] [CrossRef]
- Gnapareddy, B.; Dugasani, S.R.; Ha, T.; Paulson, B.; Hwang, T.; Kim, T.; Kim, J.H.; Oh, K.; Park, S.H. Chemical and physical characteristics of doxorubicin hydrochloride drug-doped salmon DNA thin films. Sci. Rep. 2015, 5, 12722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schüler, C.; Caruso, F. Decomposable hollow biopolymer-based capsules. Biomacromolecules 2001, 2, 921–926. [Google Scholar] [CrossRef]
- Yoshida, K.; Ono, T.; Dairaku, T.; Kashiwagi, Y.; Sato, K. Preparation of hydrogen peroxide sensitive nanofilms by a layer-by-layer technique. Nanomaterials 2018, 8, 941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, K.; Kashimura, Y.; Kamijo, T.; Ono, T.; Dairaku, T.; Sato, T.; Kashiwagi, Y.; Sato, K. Decomposition of glucose-sensitive layer-by-layer films using hemin, DNA, and glucose oxidase. Polymers 2020, 12, 319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nie, X.; Zhang, Z.; Wang, C.H.; Fan, Y.S.; Meng, Q.Y.; You, Y.Z. Interactions in DNA condensation: An important factor for improving the efficacy of gene transfection. Bioconj. Chem. 2018, 30, 284–292. [Google Scholar] [CrossRef]
- Kim, Y.H.; Lee, K.; Li, S. Nucleic acids based polyelectrolyte complexes: Their complexation mechanism, morphology, and stability. Chem. Mater. 2021, 33, 7923–7943. [Google Scholar] [CrossRef]
- Huo, S.; Li, H.; Boersma, A.J.; Herrmann, A. DNA nanotechnology enters cell membranes. Adv. Sci. 2019, 6, 1900043. [Google Scholar] [CrossRef]
- Qian, B.; Shi, S.; Wang, H.; Russell, T.P. Reconfigurable liquids stabilized by DNA surfactants. ACS App. Mater. Interfaces 2020, 12, 13551–13557. [Google Scholar] [CrossRef]
- Yamamoto, S.; Yamamoto, T.; Shimada, S.; Kuramoto, E.; Yano, O.; Kataoka, T.; Tokunaga, T. DNA from bacteria, but not from vertebrates, induces interferons, activates natural killer cells and inhibits tumor growth. Microbiol. Immunol. 1992, 36, 983–997. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Wang, X.M. Spectroscopic studies on the interaction of β-cyclodextrin-8-Hydroxyquiuoline inclusion complex with herring sperm DNA. J. Mol. Struct. 2013, 1036, 51–55. [Google Scholar] [CrossRef]
- Hill, I.R.; Garnett, M.C.; Bignotti, F.; Davis, S.S. Determination of protection from serum nuclease activity by DNA–polyelectrolyte complexes using an electrophoretic method. Anal. Biochem. 2001, 291, 62–68. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Ohno, J.; Sakagami, R.; Hayakawa, T.; Fukushima, T. Cell viabilities and biodegradation rates of DNA/protamine complexes with two different molecular weights of DNA. J. Biomed. Mater. Res. B Appl. Biomater. 2013, 101, 743–751. [Google Scholar] [CrossRef]
- Van den Beucken, J.J.; Vos, M.R.; Thüne, P.C.; Hayakawa, T.; Fukushima, T.; Okahata, Y.; Walboomers, X.F.; Sommerdijk, N.A.J.M.; Nolte, R.J.M.; Jansen, J.A. Fabrication, characterization, and biological assessment of multilayered DNA coatings for biomaterial purposes. Biomaterials 2006, 27, 691–701. [Google Scholar] [CrossRef]
- Van den Beucken, J.J.; Walboomers, X.F.; Boerman, O.C.; Vos, M.R.; Sommerdijk, N.A.; Hayakawa, T.; Fukushima, T.; Okahata, Y.; Nolte, R.J.; Jansen, J.A. Functionalization of multilayered DNA-coatings with bone morphogenetic protein 2. J. Control. Release 2006, 113, 63–72. [Google Scholar] [CrossRef] [PubMed]
- van den Beucken, J.J.; Walboomers, X.F.; Nillesen, S.T.; Vos, M.R.; Sommerdijk, N.A.; Van Kuppevelt, T.H.; Nolte, R.J.; Jansen, J.A. In vitro and in vivo effects of deoxyribonucleic acid–based coatings funtionalized with vascular endothelial growth factor. Tissue Eng. 2007, 13, 711–720. [Google Scholar] [CrossRef] [Green Version]
- van den Beucken, J.J.J.P.; Walboomers, X.F.; Leeuwenburgh, S.C.G.; Vos, M.R.J.; Sommerdijk, N.A.J.M.; Nolte, R.J.M.; Jansen, J.A. Multilayered DNA coatings: In vitro bioactivity studies and effects on osteoblast-like cell behavior. Acta Biomater. 2007, 3, 587–596. [Google Scholar] [CrossRef] [PubMed]
- Gao, W.; Feng, B.; Ni, Y.; Yang, Y.; Lu, X.; Weng, J. Protein adsorption and biomimetic mineralization behaviors of PLL-DNA multilayered films assembled onto titanium. Appl. Surf. Sci. 2010, 257, 538–546. [Google Scholar] [CrossRef]
- Liu, P.; Wang, J.M.; Yu, X.T.; Chen, X.B.; Li, S.Q.; Chen, D.C.; Guan, S.K.; Zeng, R.C.; Cui, L.Y. Corrosion resistance of bioinspired DNA-induced Ca–P coating on biodegradable magnesium alloy. J. Magnesium Alloys 2019, 7, 144–154. [Google Scholar] [CrossRef]
- Cui, L.Y.; Fang, X.H.; Cao, W.; Zeng, R.C.; Li, S.Q.; Chen, X.B.; Zou, Y.H.; Guan, S.K.; Han, E.H. In vitro corrosion resistance of a layer-by-layer assembled DNA coating on magnesium alloy. Appl. Surf. Sci. 2018, 457, 49–58. [Google Scholar] [CrossRef]
- Fukushima, T.; Hayakawa, T.; Okamura, K.; Takeda, S.; Inoue, Y.; Miyazaki, K.; Okahata, Y. Buffer solution can control the porosity of DNA/chitosan complexes. J. Biomed. Mater. Res. B Appl. Biomater. 2006, 76, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Fukushima, T.; Kawaguchi, M.; Hayakawa, T.; Ohno, J.; Iwahashi, T.; Taniguchi, K.; Inoue, Y.; Takeda, S. Complexation of DNA with cationic polyamino acid for biomaterial purposes. J. Oral Tissue Eng. 2008, 6, 24–32. [Google Scholar]
- Fukushima, T.; Ohno, J.; Hayakawa, T.; Kawaguchi, M.; Inoue, Y.; Takeda, S.; Toyoda, M.; Okahata, Y. Mold fabrication and biological assessment of porous DNA-chitosan complexes. J. Biomed. Mater. Res. B Appl. Biomater. 2009, 91, 746–754. [Google Scholar] [CrossRef]
- Fukushima, T.; Ohno, J.; Hayakawa, T.; Imayoshi, R.; Kawaguchi, M.; Doi, Y.; Kanaya, K.; Mitarai, M. Polycation protamine for waterinsoluble complex formation with DNA. Dent. Mater. J. 2010, 29, 529–535. [Google Scholar] [CrossRef] [Green Version]
- Fukushima, T.; Ohno, J.; Hayakawa, T.; Imayoshi, R.; Mori, N.; Sakagami, R.; Mitarai, M.; Hayakawa, T. DNA/protamine complex paste for an injectable dental material. J. Mater. Sci. Mater. Med. 2011, 22, 2607–2615. [Google Scholar] [CrossRef]
- Katsumata, Y.; Kajiya, H.; Okabe, K.; Fukushima, T.; Ikebe, T. A salmon DNA scaffold promotes osteogenesis through activation of sodium-dependent phosphate cotransporters. Biochem. Biophys. Res. Commun. 2015, 468, 622–628. [Google Scholar] [CrossRef]
- Ouni, O.A.; Subbiahdoss, G.; Scheberl, A.; Reimhult, E. DNA polyelectrolyte multilayer coatings are antifouling and promote mammalian cell adhesion. Materials 2021, 14, 4596. [Google Scholar] [CrossRef]
- Subbiahdoss, G.; Zeng, G.; Aslan, H.; Ege-Friis, J.; Iruthayaraj, J.; Zelikin, A.N.; Meyer, R.L. Antifouling properties of layer by layer DNA coatings. Biofouling 2019, 35, 75–88. [Google Scholar] [CrossRef]
- Zhang, C.; Lin, J.; Nguyen, N.T.; Guo, Y.; Xu, C.; Seo, C.; Villafana, E.; Jimenez, H.; Chai, Y.; Guan, R.; et al. Antimicrobial bioresorbable Mg–Zn–Ca alloy for bone repair in a comparison study with Mg–Zn–Sr alloy and pure Mg. ACS Biomater. Sci. Eng. 2020, 6, 517–538. [Google Scholar] [CrossRef]
- Zhang, J.; Tang, L.; Qi, H.; Zhao, Q.; Liu, Y.; Zhang, Y. Dual function of magnesium in bone biomineralization. Adv. Healthc. Mater. 2019, 8, 1901030. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.Q.; Wang, L.; Zeng, M.Q.; Zheng, R.C.; Kannan, M.B.; Lin, C.G.; Zheng, Y.F. Biodegradation behavior of micro-arc oxidation coating on magnesium alloy—From a protein perspective. Bioact. Mater. 2020, 5, 398–409. [Google Scholar] [CrossRef] [PubMed]
- Chandra, G.; Pandey, A. Preparation strategies for Mg-alloys for biodegradable orthopaedic implants and other biomedical applications: A review. IRBM 2020, in press. [Google Scholar] [CrossRef]
- Li, C.Y.; Yu, C.; Zeng, R.C.; Zhang, B.C.; Cui, L.Y.; Wan, J.; Xia, Y. In vitro corrosion resistance of a Ta2O5 nanofilm on MAO coated magnesium alloy AZ31 by atomic layer deposition. Bioact. Mater. 2020, 5, 34–43. [Google Scholar] [CrossRef]
- Cui, L.Y.; Qin, P.H.; Huang, X.L.; Yin, Z.Z.; Zeng, R.C.; Li, S.Q.; Han, E.H.; Wang, Z.L. Electrodeposition of TiO2 layer-by-layer assembled composite coating and silane treatment on Mg alloy for corrosion resistance. Surf. Coat. Technol. 2017, 324, 560–568. [Google Scholar] [CrossRef]
- Cui, L.Y.; Liu, H.P.; Xue, K.; Zhang, W.L.; Zeng, R.C.; Li, S.Q.; Xu, D.; Han, E.H.; Guan, S.K. In vitro corrosion and antibacterial performance of micro-arc oxidation coating on AZ31 magnesium alloy: Effects of tannic acid. J. Electrochem. Soc. 2018, 165, C821–C829. [Google Scholar] [CrossRef]
- Cui, L.Y.; Wei, G.B.; Han, Z.Z.; Zeng, R.C.; Wang, L.; Zou, Y.-H.; Li, S.Q.; Xu, D.K.; Guan, S.K. In vitro corrosion resistance and antibacterial performance of novel tin dioxide-doped calcium phosphate coating on degradable Mg–1Li–1Ca alloy. J. Mater. Sci. Technol. 2019, 35, 254–265. [Google Scholar] [CrossRef]
- Cui, L.Y.; Gao, L.; Zhang, J.C.; Tang, Z.; Fan, X.L.; Liu, J.C.; Chen, D.C.; Zeng, R.C.; Li, S.Q.; Zhi, K.Q. In vitro corrosion resistance, antibacterial activity and cytocompatibility of a layer-by-layer assembled DNA coating on magnesium alloy. J. Magnes. Alloys 2021, 9, 266–280. [Google Scholar] [CrossRef]
- Zhang, Z.Y.; Wang, D.; Liang, L.X.; Cheng, S.C.; Cui, L.Y.; Li, S.Q.; Wang, Z.L.; Zeng, R.C. Corrosion resistance of Ca-P coating induced by layer-by-layer assembled polyvinylpyrrolidone/DNA multilayer on magnesium AZ31 alloy. Front. Mater. Sci. 2021, 15, 391–405. [Google Scholar] [CrossRef]
- Johansen, C.; Gill, T.; Gram, L. Antibacterial effect of protamine assayed by impedimetry. J. Appl. Bacteriol. 1995, 78, 297–303. [Google Scholar] [CrossRef]
- Shinozaki, Y.; Toda, M.; Ohno, J.; Kawaguchi, M.; Kido, H.; Fukushima, T. Evaluation of bone formation guided by DNA/protamine complex with FGF-2 in an adult rat calvarial defect model. J. Biomed. Mater. Res. B Appl. Biomater. 2014, 102, 1669–1676. [Google Scholar] [CrossRef]
- Toda, M.; Ohno, J.; Shinozaki, Y.; Ozaki, M.; Fukushima, T. Osteogenic potential for replacing cells in rat cranial defects implanted with a DNA/protamine complex paste. Bone 2014, 67, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, Y.; Yanagi, T.; Yamaguchi, Y.; Kido, H.; Fukushima, T. Osteogenic evaluation of DNA/Protamine complex paste in rat cranial defects. J. Hard Tissue Biol. 2013, 22, 401–408. [Google Scholar] [CrossRef] [Green Version]
- Sakurai, T.; Yoshinari, M.; Toyama, T.; Hayakawa, T.; Ohkubo, C. Effects of a multilayered DNA/protamine coating on titanium implants on bone responses. J. Biomed. Mater. Res. A 2016, 104, 1500–1509. [Google Scholar] [CrossRef]
- Miyamoto, N.; Yamachika, R.; Sakurai, T.; Hayakawa, T.; Hosoya, N. Bone response to titanium implants coated with double-or single-stranded DNA. BioMed Res. Int. 2018, 2018, 9204391. [Google Scholar] [CrossRef] [Green Version]
- Yamachika, R.; Miyamoto, N.; Mishima, H.; Hayakawa, T.; Hosoya, N. Effects of DNA/protamine and DNA/gelatin paste on bone formation at tooth extraction wound sites. J. Hard Tissue Biol. 2019, 28, 191–198. [Google Scholar] [CrossRef]
- Naumenko, E.; Akhatova, F.; Rozhina, E.; Fakhrullin, R. Revisiting the cytotoxicity of cationic polyelectrolytes as a principal component in layer-by-layer assembly fabrication. Pharmaceutics 2021, 13, 1230. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Meng, Q.; Li, Q.; Liu, J.; Zhou, M.; Jin, Z.; Zhao, K. Chitosan derivatives and their application in biomedicine. Int. J. Mol. Sci. 2020, 21, 487. [Google Scholar] [CrossRef] [Green Version]
- Hu, B.; Guo, Y.; Li, H.; Liu, X.; Fu, Y.; Ding, F. Recent advances in chitosan-based layer-by-layer biomaterials and their biomedical applications. Carbohydr. Polym. 2021, 271, 118427. [Google Scholar] [CrossRef]
- Ogata, N.; Yamaoka, K.; Yoshida, J. Progress of DNA biotronics and other applications. In Proceedings of the SPIE 7765, Nanobiosystems: Processing, Characterization, and Applications III, San Diego, CA, USA, 24 August 2010; p. 776508. [Google Scholar]
- Jiang, J.; Oberdörster, G.; Biswas, P. Characterization of size, surface charge, and agglomeration state of nanoparticle dispersions for toxicological studies. J. Nanopart. Res. 2009, 11, 77–89. [Google Scholar] [CrossRef]
- Shamsi, M.H.; Geckeler, K.E. The first biopolymer-wrapped non-carbon nanotubes. Nanotechnology 2008, 19, 075604. [Google Scholar] [CrossRef] [PubMed]
- Nepal, D.; Sohn, J.I.; Aicher, W.K.; Lee, S.; Geckeler, K.E. Supramolecular conjugates of carbon nanotubes and DNA by a solid-state reaction. Biomacromolecules 2005, 6, 2919–2922. [Google Scholar] [CrossRef] [PubMed]
- Batasheva, S.; Kryuchkova, M.; Fakhrullin, R.; Cavallaro, G.; Lazzara, G.; Akhatova, F.; Nigamatzyanova, L.; Evtugin, V.; Rozhina, E.; Fakhrullin, R. Facile fabrication of natural polyelectrolyte-nanoclay composites: Halloysite nanotubes, nucleotides and DNA study. Molecules 2020, 25, 3557. [Google Scholar] [CrossRef]
- Zhao, Y.; Cavallaro, G.; Lvov, Y. Orientation of charged clay nanotubes in evaporating droplet meniscus. J. Colloid Interface Sci. 2015, 440, 68–77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kryuchkova, M.; Batasheva, S.; Naumenko, E.; Rozhina, E.; Akhatova, F.; Panchal, A.; Lvov, Y.; Fakhrullin, R. Self-assembly of concentric microrings of tubule and platy nanoclays for cell patterning and capturing. Appl. Clay Sci. 2020, 195, 105707. [Google Scholar] [CrossRef]
- Verderosa, A.D.; Totsika, M.; Fairfull-Smith, K.E. Bacterial biofilm eradication agents: A current review. Front. Chem. 2019, 7, 824. [Google Scholar] [CrossRef] [Green Version]
- Borges, A.; J Saavedra, M.; Simoes, M. Insights on antimicrobial resistance, biofilms and the use of phytochemicals as new antimicrobial agents. Curr. Med. Chem. 2015, 22, 2590–2614. [Google Scholar] [CrossRef] [Green Version]
- Chua, S.; Liu, Y.; Yam, J.; Chen, Y.; Vejborg, R.M.; Tan, B.G.C.; Kjelleberg, S.; Tolker-Nielsen, Y.; Givskov, M.; Yang, L. Dispersed cells represent a distinct stage in the transition from bacterial biofilm to planktonic lifestyles. Nat. Commun. 2014, 5, 4462. [Google Scholar] [CrossRef]
- Pingle, H.; Wang, P.Y.; Cavaliere, R.; Whitchurch, C.B.; Thissen, H.; Kingshott, P. Minimal attachment of Pseudomonas aeruginosa to DNA modified surfaces. Biointerphases 2018, 13, 06E405. [Google Scholar] [CrossRef] [Green Version]
- Carneiro, K.M.; Avakyan, N.; Sleiman, H.F. Long-range assembly of DNA into nanofibers and highly ordered networks. WIREs Nanomed. Nanobiotechnol. 2013, 5, 266–285. [Google Scholar] [CrossRef]
- Quartinello, F.; Kremser, K.; Vecchiato, S.; Schoen, H.; Vielnascher, R.; PLoszczanski, L.; Pellis, A.; Guebitz, G.M. Increased flame retardancy of enzymatic functionalized PET and Nylon fabrics via DNA immobilization. Front. Chem. 2019, 7, 685. [Google Scholar] [CrossRef] [PubMed]
- Camino, G.; Costa, L.; Martinasso, G. Intumescent fire-retardant systems. Polym. Degrad. Stab. 1989, 23, 359–376. [Google Scholar] [CrossRef]
- Alongi, J.; Di Blasio, A.; Milnes, J.; Malucelli, G.; Bourbigot, S.; Kandola, B.; Camino, G. Thermal degradation of DNA, an all-in-one natural intumescent flame retardant. Polym. Degrad. Stab. 2015, 113, 110–118. [Google Scholar] [CrossRef]
- Alongi, J.; Carletto, R.A.; Di Blasio, A.; Carosio, F.; Bosco, F.; Malucelli, G. DNA: A novel, green, natural flame retardant and suppressant for cotton. J. Mater. Chem. A 2013, 1, 4779–4785. [Google Scholar] [CrossRef]
- Alongi, J.; Carletto, R.A.; Di Blasio, A.; Cuttica, F.; Carosio, F.; Bosco, F.; Malucelli, G. Intrinsic intumescent-like flame retardant properties of DNA-treated cotton fabrics. Carbohydr. Polym. 2013, 96, 296–304. [Google Scholar] [CrossRef] [PubMed]
- Bosco, F.; Casale, A.; Mollea, C.; Terlizzi, M.E.; Gribaudo, G.; Alongi, J.; Malucelli, G. DNA coatings on cotton fabrics: Effect of molecular size and pH on flame retardancy. Surf. Coat. Technol. 2015, 272, 86–95. [Google Scholar] [CrossRef] [Green Version]
- Luo, Q.; Gao, P.; Zhou, J.; Zhang, J.; Wu, W.; Cao, J.; Reddy, N.; Ma, H. Imparting flame resistance to citric acid–modified cotton fabrics using DNA. J. Eng. Fibers Fabr. 2020, 15, 1–10. [Google Scholar] [CrossRef]
- Carosio, F.; Di Blasio, A.; Alongi, J.; Malucelli, G. Green DNA-based flame retardant coatings assembled through layer by layer. Polymer 2013, 54, 5148–5153. [Google Scholar] [CrossRef]
- Annalisa, C.; Francesca, B.; Giulio, M.; Chiara, M.; Monica, P. DNA-chitosan cross-linking and photografting to cotton fabrics to improve washing fastness of the fire-resistant finishing. Cellulose 2016, 23, 3963–3984. [Google Scholar] [CrossRef]
- Suryaprabha, T.; Sethuraman, M.G. Fabrication of a superhydrophobic and flame-retardant cotton fabric using a DNA-based coating. J. Mater. Sci. 2020, 55, 11959–11969. [Google Scholar] [CrossRef]
- Carosio, F.; Alongi, J.; Paravidino, C.; Frache, A. Improving the flame retardant efficiency of layer by layer coatings containing deoxyribonucleic acid by post-diffusion of hydrotalcite nanoparticles. Materials 2017, 10, 709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zabihi, O.; Ahmadi, M.; Khayyam, H.; Naebe, M. Fish DNA-modified clays: Towards highly flame retardant polymer nanocomposite with improved interfacial and mechanical performance. Sci. Rep. 2016, 6, 38194. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortelli, S.; Malucelli, G.; Blosi, M.; Zanoni, I.; Costa, A.L. NanoTiO2@ DNA complex: A novel eco, durable, fire retardant design strategy for cotton textiles. J. Colloid Interface Sci. 2019, 546, 174–183. [Google Scholar] [CrossRef] [PubMed]
- Alongi, J.; Di Blasio, A.; Cuttica, F.; Carosio, F.; Malucelli, G. Flame retardant properties of ethylene vinyl acetate copolymers meltcompounded with deoxyribonucleic acid in the presence of acellulose or b-cyclodextrins. Curr. Org. Chem. 2014, 18, 1651–1660. [Google Scholar] [CrossRef]
- Alongi, J.; Di Blasio, A.; Cuttica, F.; Carosio, F.; Malucelli, G. Bulk or surface treatments of ethylene vinyl acetate copolymers with DNA: Investigation on the flame retardant properties. Eur. Polym. J. 2014, 51, 112–119. [Google Scholar] [CrossRef]
- Malucelli, G.; Barbalini, M. UV-curable acrylic coatings containing biomacromolecules: A new fire retardant strategy for ethylene-vinyl acetate copolymers. Prog. Org. Coat. 2019, 127, 330–337. [Google Scholar] [CrossRef]
- Rajczak, E.; Arrigo, R.; Malucelli, G. Thermal stability and flame retardance of EVA containing DNA-modified clays. Thermochim. Acta 2020, 686, 178546. [Google Scholar] [CrossRef]
- Zhang, H.; Wu, W.; Zhou, J.; Cao, J.; Guo, X.; Kong, L.; Ma, H. Preparation and properties of DNA/PLLA, whey protein/PLLA and collagen/PLLA composites. Plast. Rubber Compos. 2018, 47, 87–93. [Google Scholar] [CrossRef]
- Tang, S.; Zheng, J. Antibacterial activity of silver nanoparticles: Structural effects. Adv. Healthc. Mater. 2018, 7, 1701503. [Google Scholar] [CrossRef]
- Takeshima, T.; Tada, Y.; Sakaguchi, N.; Watari, F.; Fugetsu, B. DNA/Ag nanoparticles as antibacterial agents against gram-negative bacteria. Nanomaterials 2015, 5, 284–297. [Google Scholar] [CrossRef]
- Takeshima, T.; Sun, L.; Wang, Y.; Yamada, Y.; Nishi, N.; Yonezawa, T.; Fugetsu, B. Salmon milt DNA as a template for the mass production of Ag nanoparticles. Polym. J. 2014, 46, 36–41. [Google Scholar] [CrossRef]
- Chumpol, J.; Siri, S. Light-mediated green synthesis of DNA-capped silver nanoparticles and their antibacterial activity. J. Nanosci. Nanotechnol. 2020, 20, 1678–1684. [Google Scholar] [CrossRef]
- Sritong, N.; Chumsook, S.; Siri, S. Light emitting diode irradiation induced shape conversion of DNA-capped silver nanoparticles and their antioxidant and antibacterial activities. Artif. Cells Nanomed. Biotechnol. 2018, 46, 955–963. [Google Scholar] [CrossRef] [Green Version]
- Storhoff, J.J.; Mirkin, C.A. Programmed materials synthesis with DNA. Chem. Rev. 1999, 99, 1849–1862. [Google Scholar] [CrossRef]
- Cavallaro, G.; Milioto, S.; Konnova, S.; Fakhrullina, G.; Akhatova, F.; Lazzara, G.; Fakhrullin, R.; Lvov, Y. Halloysite/keratin nanocomposite for human hair photoprotection coating. ACS Appl. Mater. Interfaces 2020, 12, 24348–24362. [Google Scholar] [CrossRef]
- Rahman, N.; Scott, F.H.; Lvov, Y.; Stavitskaya, A.; Akhatova, F.; Konnova, S.; Fakhrullina, G.; Fakhrullin, R. Clay nanotube immobilization on animal hair for sustained anti-lice protection. Pharmaceutics 2021, 13, 1477. [Google Scholar] [CrossRef] [PubMed]
- Drummond, T.G.; Hill, M.G.; Barton, J.K. Electrochemical DNA sensors. Nat. Biotechnol. 2003, 21, 1192–1199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, A.R.; Grodick, M.A.; Barton, J.K. DNA charge transport: From chemical principles to the cell. Cell Chem. Biol. 2016, 23, 183–197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, J.; Bai, J.; Dong, L.; Yang, M.; Hu, Y.; Gao, L.; Qian, H. A novel electrochemical biosensor based on layered hydroxide nanosheets/DNA composite for the determination of phenformin hydrochloride. Int. J. Electrochem. Sci. 2021, 16, 210237. [Google Scholar] [CrossRef]
- Dauphin-Ducharme, P.; Arroyo-Currás, N.; Plaxco, K.W. High-precision electrochemical measurements of the guanine-, mismatch-, and length-dependence of electron transfer from electrode-bound DNA are consistent with a contact-mediated mechanism. J. Am. Chem. Soc. 2019, 141, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Nano, A.; Furst, A.L.; Hill, M.G.; Barton, J.K. DNA Electrochemistry: Charge-transport pathways through DNA films on gold. J. Am. Chem. Soc. 2021, 143, 11631–11640. [Google Scholar] [CrossRef]
- Genereux, J.C.; Barton, J.K. Mechanisms for DNA charge transport. Chem. Rev. 2010, 110, 1642–1662. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, C.Z.; Liu, Y.; Luong, J.H. Impedance sensing of DNA binding drugs using gold substrates modified with gold nanoparticles. Anal. Chem. 2005, 77, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Pingarrón, J.M.; Yañez-Sedeño, P.; González-Cortés, A. Gold nanoparticle-based electrochemical biosensors. Electrochim. Acta 2008, 53, 5848–5866. [Google Scholar] [CrossRef]
- Benvidi, A.; Firouzabadi, A.D.; Moshtaghiun, S.M.; Mazloum-Ardakani, M.; Tezerjani, M.D. Ultrasensitive DNA sensor based on gold nanoparticles/reduced graphene oxide/glassy carbon electrode. Anal. Biochem. 2015, 484, 24–30. [Google Scholar] [CrossRef]
- Mello, L.D.; Hernandez, S.; Marrazza, G.; Mascini, M.; Kubota, L.T. Investigations of the antioxidant properties of plant extracts using a DNA-electrochemical biosensor. Biosens. Bioelectron. 2006, 21, 1374–1382. [Google Scholar] [CrossRef]
- Bučková, M.; Labuda, J.; Šandula, J.; Križková, L.; Štěpánek, I.; Ďuračková, Z. Detection of damage to DNA and antioxidative activity of yeast polysaccharides at the DNA-modified screen-printed electrode. Talanta 2002, 56, 939–947. [Google Scholar] [CrossRef]
- Qiu, Y.; Qu, X.; Dong, J.; Ai, S.; Han, R. Electrochemical detection of DNA damage induced by acrylamide and its metabolite at the graphene-ionic liquid-Nafion modified pyrolytic graphite electrode. J. Hazard. Mater. 2011, 190, 480–485. [Google Scholar] [CrossRef]
- Xiong, H.; Chen, Y.; Zhang, X.; Gu, H.; Wang, S. An electrochemical biosensor for the rapid detection of DNA damage induced by xanthine oxidase-catalyzed Fenton reaction. Sens. Actuator B Chem. 2013, 181, 85–91. [Google Scholar] [CrossRef]
- Hájková, A.; Barek, J.; Vyskočil, V. Electrochemical DNA biosensor for detection of DNA damage induced by hydroxyl radicals. Bioelectrochemistry 2017, 116, 1–9. [Google Scholar] [CrossRef]
- Mousavisani, S.Z.; Raoof, J.B.; Ojani, R.; Bagheryan, Z. An impedimetric biosensor for DNA damage detection and study of the protective effect of deferoxamine against DNA damage. Bioelectrochemistry 2018, 122, 142–148. [Google Scholar] [CrossRef]
- Evtugyn, G.A.; Stepanova, V.B.; Porfireva, A.V.; Zamaleeva, A.I.; Fakhrullin, R.R. Electrochemical DNA sensors based on nanostructured organic dyes/DNA/polyelectrolyte complexes. J. Nanosci. Nanotechnol. 2014, 14, 6738–6747. [Google Scholar] [CrossRef]
- Sun, Y.; Wang, J.; Jin, L.; Chang, Y.; Duan, J.; Lu, Y. A new conjugated poly(pyridinium salt) derived from phenanthridine diamine: Its synthesis, optical properties and interaction with calf thymus DNA. Polym. J. 2015, 47, 753–759. [Google Scholar] [CrossRef]
- Ensafi, A.A.; Amini, M.; Rezaei, B. Detection of DNA damage induced by chromium/glutathione/H2O2 system at MWCNTs–poly(diallyldimethylammonium chloride) modified pencil graphite electrode using methylene blue as an electroactive probe. Sens. Actuators B Chem. 2013, 177, 862–870. [Google Scholar] [CrossRef]
- Evtugyn, G.A.; Porfireva, A.V.; Stepanova, V.B.; Budnikov, H.C. Electrochemical biosensors based on native DNA and nanosized mediator for the detection of anthracycline preparations. Electroanalysis 2015, 27, 629–637. [Google Scholar] [CrossRef]
- Evtugyn, G.; Hianik, T. Electrochemical DNA sensors and aptasensors based on electropolymerized materials and polyelectrolyte complexes. Trends Analyt. Chem. 2016, 79, 168–178. [Google Scholar] [CrossRef]
- Shamagsumova, R.; Porfireva, A.; Stepanova, V.; Osin, Y.; Evtugyn, G.; Hianik, T. Polyaniline–DNA based sensor for the detection of anthracycline drugs. Sens. Actuators B Chem. 2015, 220, 573–582. [Google Scholar] [CrossRef]
- Porfir’eva, A.V.; Shibaeva, K.S.; Evtyugin, V.G.; Yakimova, L.S.; Stoikov, I.I.; Evtyugin, G.A. An electrochemical DNA sensor for doxorubicin based on a polyelectrolyte complex and aminated thiacalix[4]arene. J. Anal. Chem. 2019, 74, 707–714. [Google Scholar] [CrossRef]
- Ivanov, A.N.; Kuzin, Y.I.; Evtugyn, G.A. SPR sensor based on polyelectrolyte complexes with DNA inclusion. Sens. Actuators B Chem. 2019, 281, 574–581. [Google Scholar] [CrossRef]
- Banitaba, M.H.; Davarani, S.S.H.; Mehdinia, A. Study of interactions between DNA and aflatoxin B1 using electrochemical and fluorescence methods. Anal. Biochem. 2011, 411, 218–222. [Google Scholar] [CrossRef]
- Fotouhi, L.; Atoofi, Z.; Heravi, M.M. Interaction of ciprofloxacin with DNA studied by spectroscopy and voltammetry at MWCNT/DNA modified glassy carbon electrode. Talanta 2013, 103, 194–200. [Google Scholar] [CrossRef]
- Fotouhi, L.; Hashkavayi, A.B.; Heravi, M.M. Interaction of sulfadiazine with DNA on a MWCNT modified glassy carbon electrode: Determination of DNA. Int. J. Biol. Macromol. 2013, 53, 101–106. [Google Scholar] [CrossRef] [PubMed]
- Zhong, J.; Qi, Z.; Dai, H.; Fan, C.; Li, G.; Matsuda, N. Sensing phenothiazine drugs at a gold electrode co-modified with DNA and gold nanoparticles. Anal. Sci. 2003, 19, 653–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fotouhi, L.; Tabatabaee, R. A study of the interaction tyrosine and DNA using voltammetry and spectroscopy methods. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2014, 121, 152–156. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Hong, H.; Fen-Yue, Z.; Zhong, J.C.; Jian-Ping, L. Electrochemical luminescent DNA sensor based on polymerase-assisted signal amplification. Chin. J. Anal. Chem. 2018, 46, 203–209. [Google Scholar] [CrossRef]
- Zhu, L.; Zhao, R.; Wang, K.; Xiang, H.; Shang, Z.; Sun, W. Electrochemical behaviors of methylene blue on DNA modified electrode and its application to the detection of PCR product from NOS sequence. Sensors 2008, 8, 5649–5660. [Google Scholar] [CrossRef]
- Pang, D.W.; Zhang, M.; Wang, Z.L.; Qi, Y.P.; Cheng, J.K.; Liu, Z.Y. Modification of glassy carbon and gold electrodes with DNA. J. Electroanal. Chem. 1996, 403, 183–188. [Google Scholar] [CrossRef]
- Zhao, Z.; Qi, Y.; Wei, M.; Zhang, F.; Xu, S. Layer-by-layer assembly and morphological characterizations of DNA/layered double hydroxide thin films. Mater. Lett. 2012, 78, 62–65. [Google Scholar] [CrossRef]
- Liu, L.M.; Jiang, L.P.; Liu, F.; Lu, G.Y.; Abdel-Halim, E.S.; Zhu, J.J. Hemoglobin/DNA/layered double hydroxide composites for biosensing applications. Anal. Methods 2013, 5, 3565–3571. [Google Scholar] [CrossRef]
- Bhambhani, A.; Kumar, C.V. Protein/DNA/Inorganic Materials: DNA binding to layered α-zirconium phosphate enhances bound protein structure and activity. Adv. Mater. 2006, 18, 939–942. [Google Scholar] [CrossRef]
- Bohlin, J.; Pettersson, J.H.O. Evolution of genomic base composition: From single cell microbes to multicellular animals. Comput. Struct. Biotechnol. J. 2019, 17, 362–370. [Google Scholar] [CrossRef]
- Yamada, M.; Kato, K.; Nomizu, M.; Sakairi, N.; Ohkawa, K.; Yamamoto, H.; Nishi, N. Preparation and characterization of DNA films induced by UV irradiation. Chem. Eur. J. 2002, 8, 1407–1412. [Google Scholar] [CrossRef]
- Okholm, A.H.; Kjems, J. The utility of DNA nanostructures for drug delivery in vivo. Exp. Opin. Drug Deliv. 2017, 14, 137–139. [Google Scholar] [CrossRef] [Green Version]
- Green, C.M.; Mathur, D.; Medintz, I.L. Understanding the fate of DNA nanostructures inside the cell. J. Mater. Chem. B 2020, 8, 6170–6178. [Google Scholar] [CrossRef] [PubMed]
- Surana, S.; Bhatia, D.; Krishnan, Y. A method to study in vivo stability of DNA nanostructures. Methods 2013, 64, 94–100. [Google Scholar] [CrossRef]
- Lee, D.S.; Qian, H.; Tay, C.Y.; Leong, D.T. Cellular processing and destinies of artificial DNA nanostructures. Chem. Soc. Rev. 2016, 45, 4199–4225. [Google Scholar] [CrossRef] [PubMed]
- Surana, S.; Shenoy, A.R.; Krishnan, Y. Designing DNA nanodevices for compatibility with the immune system of higher organisms. Nat. Nanotechnol. 2015, 10, 741–747. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.B.; Cheng, W.; Webber, A.; Bhambhani, A.; Duff, M.R.; Kumar, C.V.; McLendon, G.L. Endonuclease-like activity of heme proteins. J. Biol. Inorg. Chem. 2005, 10, 790–799. [Google Scholar] [CrossRef]
- Bosco, F.; Casale, A.; Gribaudo, G.; Mollea, C.; Malucelli, G. Nucleic acids from agro-industrial wastes: A green recovery method for fire retardant applications. Ind. Crops Prod. 2017, 108, 208–218. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Batasheva, S.; Fakhrullin, R. Sequence Does Not Matter: The Biomedical Applications of DNA-Based Coatings and Cores. Int. J. Mol. Sci. 2021, 22, 12884. https://doi.org/10.3390/ijms222312884
Batasheva S, Fakhrullin R. Sequence Does Not Matter: The Biomedical Applications of DNA-Based Coatings and Cores. International Journal of Molecular Sciences. 2021; 22(23):12884. https://doi.org/10.3390/ijms222312884
Chicago/Turabian StyleBatasheva, Svetlana, and Rawil Fakhrullin. 2021. "Sequence Does Not Matter: The Biomedical Applications of DNA-Based Coatings and Cores" International Journal of Molecular Sciences 22, no. 23: 12884. https://doi.org/10.3390/ijms222312884
APA StyleBatasheva, S., & Fakhrullin, R. (2021). Sequence Does Not Matter: The Biomedical Applications of DNA-Based Coatings and Cores. International Journal of Molecular Sciences, 22(23), 12884. https://doi.org/10.3390/ijms222312884






