Physiological and Dual Transcriptional Analysis of Microalga Graesiella emersonii–Amoeboaphelidium protococcarum Pathosystem Uncovers Conserved Defense Response and Robust Pathogenicity
Abstract
:1. Introduction
2. Results
2.1. Analysis of the Symptom and Features Associated with Infection by A. protococcarum
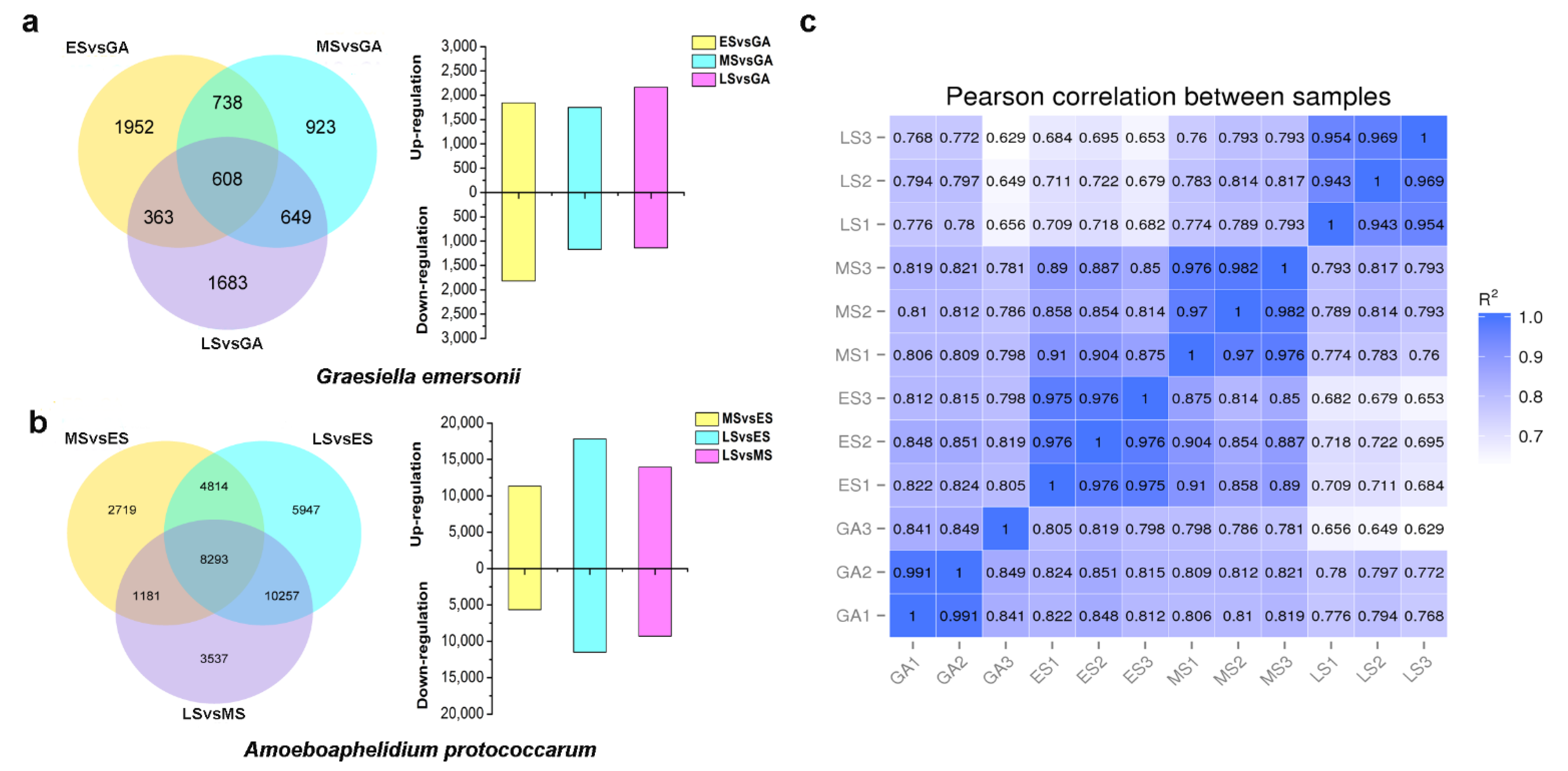
2.2. A Global View of Transcriptomic Analysis of Microalga and Endoparasite during Their Interaction
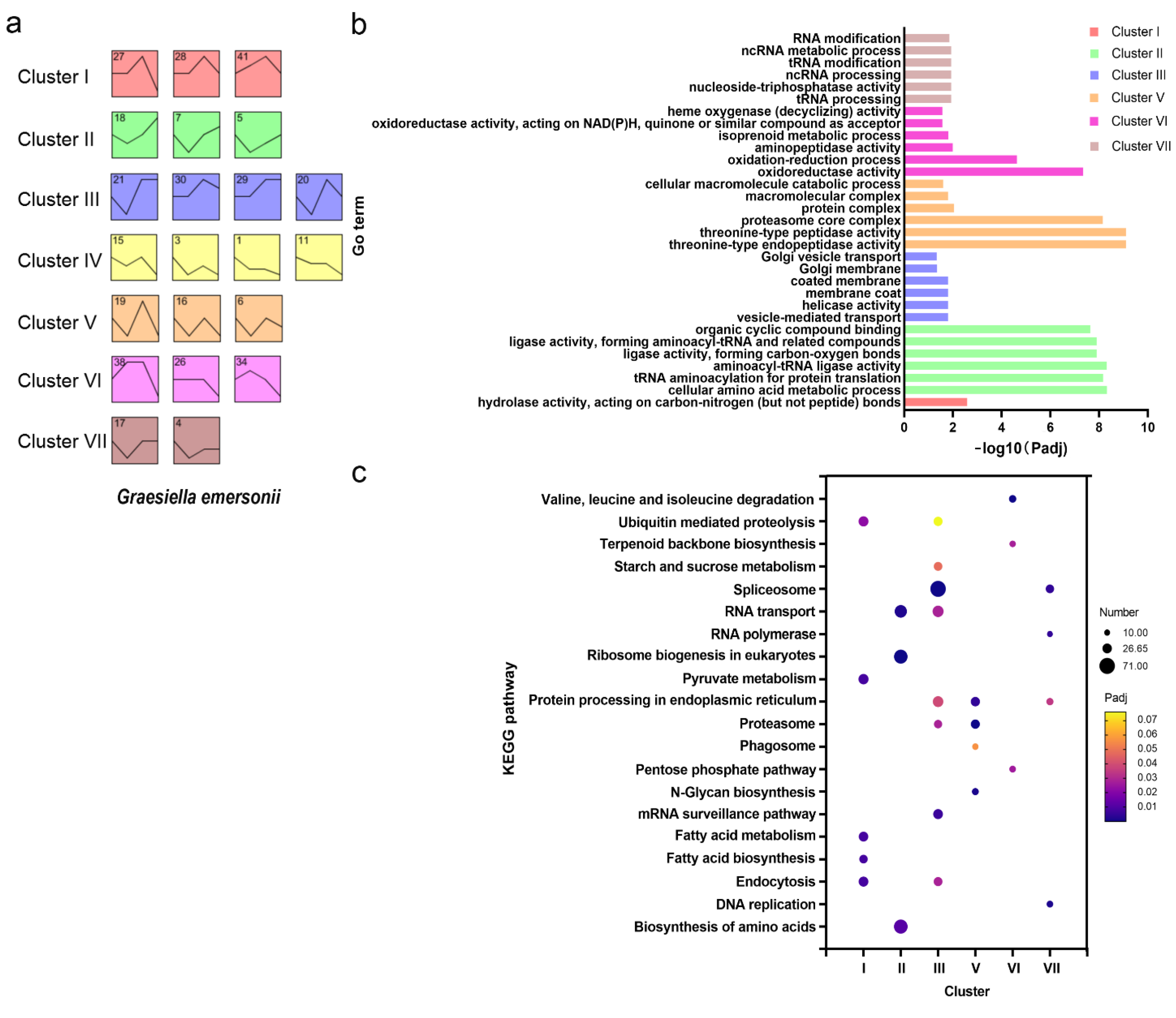
2.3. Gene Expression Profile of G. emersonii during Infection
2.4. Ubiquitin Mediated Proteolysis and Endocytosis in Response to Infection
2.5. Potential Pathogen Receptors and Putative R Proteins in G. emersonii
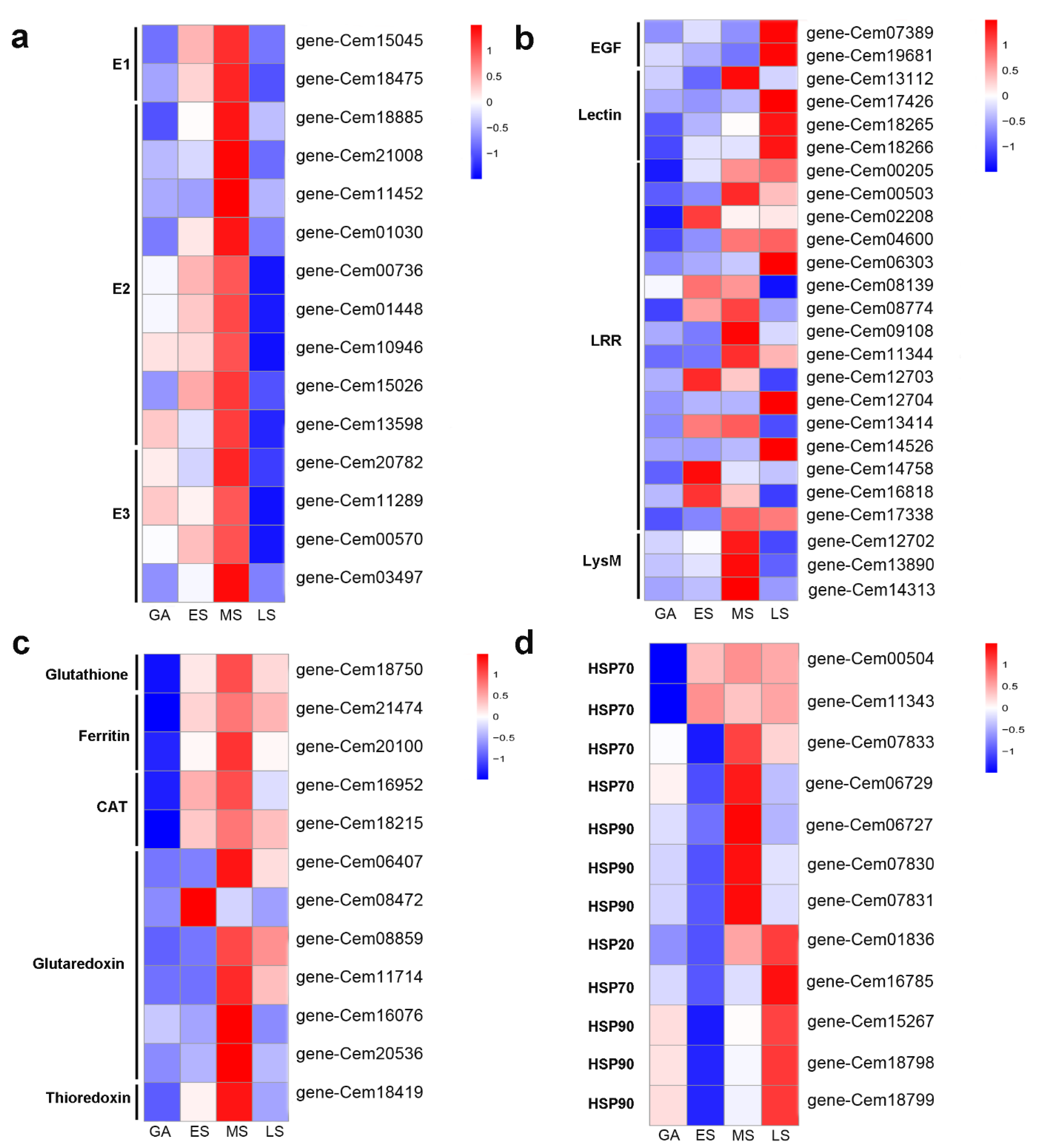
2.6. ROS-Related Genes in Response to Infection
2.7. Some Heat Shock Proteins (HSPs) and Transcription Factors (TFs) Involved during Infection
2.8. Gene Expression Profile of A. protococcarum during Infection
2.9. Expression of CAZymes and Pathogen–Host Interaction Genes in A. protococcarum during Infection
3. Discussion
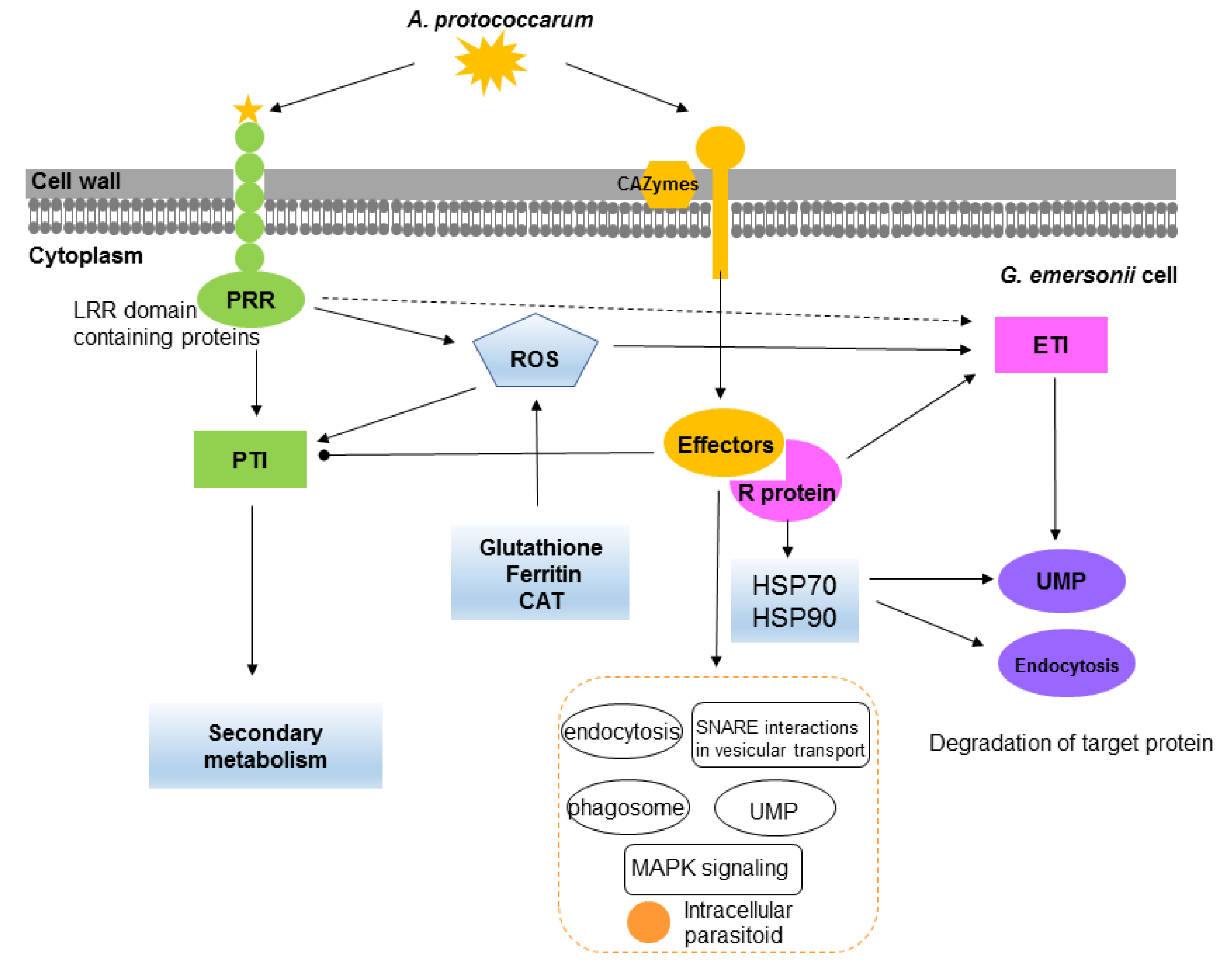
3.1. Defense Strategies in G. emersonii
3.2. Pathogenicity of A. protococcarum
4. Materials and Methods
4.1. Microalga and Parasite Laboratory Culturing
4.2. Infection Test and Sampling
4.3. Chlorophyll Fluorescence Parameter and ROS Measurement
4.4. RNA Isolation, cDNA Library Construction, and Sequencing
4.5. Transcriptome Data Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| CAZymes | carbohydrate-active enzymes |
| DEGs | differentially expressed genes |
| EGF | epidermal growth factor |
| ETI | effector-triggered immunity |
| ES | early stage |
| Fv/Fm | maximum photochemical yield |
| GA | control |
| HSPs | large heat shock proteins |
| LRR | leucine-rich repeat |
| LysM | lysine motif |
| LS | late stage |
| MS | medium stage |
| MAPK | mitogen-activated protein kinase |
| NBS | nucleotide binding site |
| PTI | PAMP-triggered immunity |
| PRRs | pattern recognition receptors |
| R proteins | resistance proteins |
| ROS | reactive oxygen species |
| STEM | short time-series expression miner |
| UPS | ubiquitin–proteasome system |
References
- Chen, J.; Li, J.; Dong, W.; Zhang, X.; Tyagi, R.D.; Drogui, P.; Surampalli, R.Y. The potential of microalgae in biodiesel production. Renew. Sustain. Energy Rev. 2018, 90, 336–346. [Google Scholar] [CrossRef]
- Kondaveeti, S.; Abu-Reesh, I.M.; Mohanakrishna, G.; Bulut, M.; Pant, D. Advanced routes of biological and bio-electrocatalytic carbon dioxide (CO2) mitigation toward carbon neutrality. Front. Energy Res. 2020, 8, 94. [Google Scholar] [CrossRef]
- Kumsiri, B.; Pekkoh, J.; Pathom-aree, W.; Lumyong, S.; Phinyo, K.; Pumas, C.; Srinuanpan, S. Enhanced production of microalgal biomass and lipid as an environmentally friendly biodiesel feedstock through actinomycete co-culture in biogas digestate effluent. Bioresour. Technol. 2021, 337, 125446. [Google Scholar] [CrossRef]
- Ding, Y.; Wen, X.; Peng, X.; Zhang, A.; Wang, Z.; Geng, Y.; Li, Y. Surfactants as fungal parasite control agents in oleaginous microalga, Graesiella sp. WBG-1, mass culture. Algal Res. 2019, 41, 101539. [Google Scholar] [CrossRef]
- Sime-Ngando, T. Phytoplankton chytridiomycosis: Fungal parasites of phytoplankton and their imprints on the food web dynamics. Front. Microbiol. 2012, 3, 361. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Peng, X.; Wang, Z.; Wen, X.; Geng, Y.; Zhang, D.; Li, Y. Occurrence and characterization of an epibiotic parasite in cultures of oleaginous microalga Graesiella sp. WBG-1. J. Appl. Phycol. 2018, 30, 819–830. [Google Scholar] [CrossRef]
- Letcher, P.M.; Lopez, S.; Schmieder, R.; Lee, P.A.; Behnke, C.; Powell, M.J.; Mcbride, R.C. Characterization of Amoeboaphelidium protococcarum, an algal parasite new to the cryptomycota isolated from an outdoor algal pond used for the production of biofuel. PLoS ONE 2013, 8, e56232. [Google Scholar] [CrossRef]
- Frenken, T.; Alacid, E.; Berger, S.A.; Bourne, E.C.; Gerphagnon, M.; Grossart, H.P.; Gsell, A.S.; Ibelings, B.W.; Kagami, M.; Kupper, F.C.; et al. Integrating chytrid fungal parasites into plankton ecology: Research gaps and needs. Environ. Microbiol. 2017, 9, 3802–3822. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, J.; Yan, H.; Zhao, L.; Li, Y.; Nahidian, B.; Zhu, M.; Hu, Q.; Han, D. Interaction between the cell walls of microalgal host and fungal carbohydrate-activate enzymes is essential for the pathogenic parasitism process. Environ. Microbiol. 2021, 23, 5114–5130. [Google Scholar] [CrossRef]
- Wen, X.; Tao, H.; Peng, X.; Wang, Z.; Ding, Y.; Xu, Y.; Liang, L.; Du, K.; Zhang, A.; Liu, C.; et al. Sequential phototrophic–mixotrophic cultivation of oleaginous microalga Graesiella sp. WBG-1 in a 1000 m2 open raceway pond. Biotechnol. Biofuels 2019, 12, 27. [Google Scholar] [CrossRef] [PubMed]
- Ding, Y.; Peng, X.; Wang, Z.; Wen, X.; Geng, Y.; Li, Y. Isolation and characterization of an endoparasite from the culture of oleaginous microalga Graesiella sp. WBG-1. Algal Res. 2017, 26, 371–379. [Google Scholar] [CrossRef]
- Strittmatter, M.; Grenville-Briggs, L.J.; Breithut, L.; Van West, P.; Gachon, C.M.M.; Küpper, F.C. Infection of the brown alga Ectocarpus siliculosus by the oomycete Eurychasma dicksoniiinduces oxidative stress and halogen metabolism. Plant Cell Environ. 2016, 39, 259–271. [Google Scholar] [CrossRef] [Green Version]
- Im, S.H.; Klochkova, T.A.; Lee, D.J.; Gachon, C.M.M.; Kim, G.H. Genetic toolkits of the red alga Pyropia tenera against the three most common diseases in Pyropia farms. J. Phycol. 2019, 55, 801–815. [Google Scholar] [CrossRef]
- Tang, L.; Qiu, L.P.; Liu, C.; Du, G.Y.; Mo, Z.L.; Tang, X.H.; Mao, Y.X. Transcriptomic insights into innate immunity responding to red rot disease in red alga Pyropia yezoensis. Int. J. Mol. Sci. 2019, 20, 5970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, V.H.; McBride, R.C.; Shurin, J.B.; Bever, J.D.; Crews, T.E.; Tilman, G.D. Crop diversification can contribute to disease risk control in sustainable biofuels production. Front. Ecol. Environ. 2015, 13, 561–567. [Google Scholar] [CrossRef] [Green Version]
- Dangl, J.L.; Horvath, D.M.; Staskawicz, B.J. Pivoting the plant immune system from dissection to deployment. Science 2013, 341, 746–751. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laflamme, B.; Dillon, M.M.; Martel, A.; Almeida, R.N.D.; Desveaux, D.; Guttman, D.S. The pan-genome effector-triggered immunity landscape of a host-pathogen interaction. Science 2020, 367, 763–768. [Google Scholar] [CrossRef]
- Presti, L.L.; Lanver, D.; Schweizer, G.; Tanaka, S.; Liang, L.; Tollot, M.; Zuccaro, A.; Reissmann, S.; Kahmann, R. Fungal Effectors and Plant Susceptibility. Annu. Rev. Plant Biol. 2015, 66, 513–545. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Xue, J.Y.; Wang, Q.; Wang, B.; Chen, J.Q. Revisiting the origin of plant NBS-LRR genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef]
- Thao, N.P.; Chen, L.; Nakashima, A.; Hara, S.I.; Umemura, K.; Takahashi, A.; Shirasu, K.; Kawasaki, T.; Shimamoto, K. RAR1 and HSP90 form a complex with Rac/Rop GTPase and function in innate-immune responses in rice. Plant Cell. 2007, 19, 4035–4045. [Google Scholar] [CrossRef] [Green Version]
- Yoshioka, H.; Adachi, H.; Nakano, T.; Miyagawa, N.; Asai, S.; Ishihama, N.; Yoshioka, M. Hierarchical regulation of NADPH oxidase by protein kinases in plant immunity. Physiol. Mol. Plant Pathol. 2016, 95, 20–26. [Google Scholar] [CrossRef]
- Yuan, M.; Jiang, Z.; Bi, G.; Nomura, K.; Liu, M.; Wang, Y.; Cai, B.; Zhou, J.M.; He, S.Y.; Xin, X.F. Pattern-recognition receptors are required for NLR-mediated plant immunity. Nature 2021, 592, 105–109. [Google Scholar] [CrossRef]
- Hu, C.; Cui, D.; Sun, X.; Shi, J.; Song, L.; Li, Y.; Xu, N. Transcriptomic analysis unveils survival strategies of autotrophic Haematococcus pluvialis against high light stress. Aquaculture 2019, 513, 734430. [Google Scholar] [CrossRef]
- Dubiella, U.; Serrano, I. The Ubiquitin proteasome system as a double agent in plant-virus interactions. Plants 2021, 10, 928. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Yang, M.Y.; Wang, P.P.; Zhao, Y.X.; Ma, C.L. Interplay between the ubiquitin proteasome system and ubiquitin-mediated autophagy in plants. Cells 2020, 9, 2219. [Google Scholar] [CrossRef]
- Gupta, I.; Singh, K.; Varshney, N.K.; Khan, S. Delineating crosstalk mechanisms of the ubiquitin proteasome system that regulate apoptosis. Front. Cell Dev. Biol. 2018, 6, 11. [Google Scholar] [CrossRef]
- Kud, J.; Wang, W.J.; Gross, R.; Fan, Y.H.; Huang, L.; Yuan, Y.L.; Gray, A.; Duarte, A.; Kuhl, J.C.; Caplan, A.; et al. The potato cyst nematode effector RHA1B is a ubiquitin ligase and uses two distinct mechanisms to suppress plant immune signaling. PLoS Pathog. 2019, 15, e1007720. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Huang, L.; Luo, W.; Jin, Y.; Gong, F.; He, J.; Liu, D.; Zheng, Y.; Wu, B. Transcriptome analysis provides insights into the mechanisms underlying wheat cultivar Shumai 126 responding to stripe rust. Gene 2021, 768, 145290. [Google Scholar] [CrossRef]
- Du, F.; Hu, C.; Sun, X.; Xu, N. Transcriptome analysis reveals pathways responsible for the promoting effect of sucrose on astaxanthin accumulation in Haematococcus pluvialis under high light condition. Aquaculture 2021, 530, 735757. [Google Scholar] [CrossRef]
- De Vries, S.; de Vries, J. A global survey of carbohydrate esterase families 1 and 10 in oomycetes. Front. Genet. 2020, 11, 756. [Google Scholar] [CrossRef] [PubMed]
- Sophie de Vries, J.d.V.; Archibald, J.M.; Slamovits, C.H. Comparative analyses of saprotrophy in Salisapilia sapeloensis and diverse plant pathogenic oomycetes reveal lifestyle-specific gene expression. FEMS Microbiol. Ecol. 2020, 96, fiaa184. [Google Scholar] [CrossRef]
- Grams, N.; Komar, H.; Jainchill, D.; Ospina-Giraldo, M. Comparative expression analysis of Phytophthora sojae polysaccharide lyase family 3 (pectate lyase) genes during infection of the soybean Glycine max. Phytopathol. Res. 2019, 1, 15. [Google Scholar] [CrossRef] [Green Version]
- Baddal, B. Next-generation technologies for studying host-pathogen interactions: A focus on dual transcriptomics, CRISPR/Cas9 screening and organs-on-chips. Pathog. Dis. 2019, 77, ftz060. [Google Scholar] [CrossRef]
- Arasaki, K.; Roy, C.R. Legionella pneumophila promotes functional interactions between plasma membrane syntaxins and sec22b. Traffic 2010, 11, 587–600. [Google Scholar] [CrossRef] [Green Version]
- Cheng, B.; Yu, X.; Ma, Z.; Dong, S.; Dou, D.; Wang, Y.; Zheng, X. Phytophthora sojae effector Avh331 suppresses the plant defence response by disturbing the MAPK signalling pathway. Physiol. Mol. Plant Pathol. 2012, 77, 1–9. [Google Scholar] [CrossRef]
- Wen, X.; Zhang, A.; Zhu, X.; Liang, L.; Huo, Y.; Wang, K.; Yu, Y.; Geng, Y.; Ding, Y.; Li, Y. Controlling of two destructive zooplanktonic predators in Chlorella mass culture with surfactants. Biotechnol. Biofuels 2021, 14, 21. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [Green Version]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urban, M.; Cuzick, A.; Seager, J.; Wood, V.; Rutherford, K.; Venkatesh, S.Y.; De Silva, N.; Martinez, M.C.; Pedro, H.; Yates, A.D.; et al. PHI-base: The pathogen-host interactions database. Nucleic Acids Res. 2020, 48, D613–D620. [Google Scholar] [CrossRef]


| No. | Gene ID | Types | Log2FC(ESvsGA) | Log2FC(MSvsGA) | Log2FC(LSvsGA) |
|---|---|---|---|---|---|
| 1 | gene-Cem00854 | PLATZ | 1.65 | 3.61 | 3.06 |
| 2 | gene-Cem04263 | SNF2 | 1.53 | 1.31 | 1.65 |
| 3 | gene-Cem15036 | FHA | 2.91 | 2.45 | 1.58 |
| 4 | gene-Cem16321 | SNF2 | 2.08 | 3.57 | 2.51 |
| 5 | gene-Cem19050 | Orphans | 1.41 | 2.68 | 3.18 |
| No. | Gene ID | ProteinID | PHI ID | Gene Name | Phenotype | log2FC(MS vs. ES) | log2FC(LS vs. ES) | log2FC(LS vs. MS) |
|---|---|---|---|---|---|---|---|---|
| 1 | Cluster-8366.85734 | A0A0H3HVK0 | PHI:5335 | clpV-5 | Effector | 1.729 | 3.8033 | 1.8221 |
| 2 | Cluster-8366.8318 | Q8RP09 | PHI:981 | hopI1 | Effector | 2.0296 | 6.6465 | 4.3176 |
| 3 | Cluster-8366.82670 | Q79LY0 | PHI:992/PHI:7237/PHI:7265 | hopPtoD2/hopAO1/HopPtoD2 | Effector | 1.6504 | 4.9986 | 3.0935 |
| 4 | Cluster-8366.80929 | P17778 | PHI:6101/PHI:6824/PHI:6830 | YopM | Effector | 6.0254 | 11.636 | 3.4623 |
| 5 | Cluster-8366.7571 | C5BD30 | PHI:6294 | EseM | Effector | 2.5059 | 7.5318 | 4.6832 |
| 6 | Cluster-8366.72579 | Q8PC98 | PHI:7945 | pip | Effector | 4.4021 | 9.7346 | 2.9476 |
| 7 | Cluster-8366.6633 | Q8PC98 | PHI:7945 | pip | Effector | 3.5808 | 10.373 | 4.6194 |
| 8 | Cluster-8366.43382 | Q8XTK9 | PHI:5119 | RSp0099 | Effector | 1.7086 | 4.8665 | 2.9109 |
| 9 | Cluster-8366.14241 | P17778 | PHI:6101/PHI:6824/PHI:6830 | YopM | Effector | 1.6503 | 5.6023 | 3.6416 |
| 10 | Cluster-8366.11799 | Q8PI08 | PHI:2703 | xac3090 | Effector | 1.5342 | 5.0159 | 3.2262 |
| 11 | Cluster-8366.11711 | Q8XZN9 | PHI:5173 | RSc1356 | Effector | 2.971 | 6.0476 | 2.8247 |
| 12 | Cluster-8366.11710 | Q8XZN9 | PHI:5173 | RSc1356 | Effector | 2.3996 | 5.6692 | 3.0096 |
| 13 | Cluster-8366.11150 | Q8XZN9 | PHI:5173 | RSc1356 | Effector | 3.3433 | 7.5956 | 3.9299 |
| 14 | Cluster-8366.11149 | Q8XZN9 | PHI:5173 | RSc1356 | Effector | 3.8592 | 11.787 | 3.2499 |
| 15 | Cluster-8366.11148 | Q8XZN9 | PHI:5173 | RSc1356 | Effector | 3.1417 | 7.4909 | 4.0076 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, Y.; Wang, Z.; Wang, Y.; Geng, Y.; Wen, X.; Li, Y. Physiological and Dual Transcriptional Analysis of Microalga Graesiella emersonii–Amoeboaphelidium protococcarum Pathosystem Uncovers Conserved Defense Response and Robust Pathogenicity. Int. J. Mol. Sci. 2021, 22, 12847. https://doi.org/10.3390/ijms222312847
Ding Y, Wang Z, Wang Y, Geng Y, Wen X, Li Y. Physiological and Dual Transcriptional Analysis of Microalga Graesiella emersonii–Amoeboaphelidium protococcarum Pathosystem Uncovers Conserved Defense Response and Robust Pathogenicity. International Journal of Molecular Sciences. 2021; 22(23):12847. https://doi.org/10.3390/ijms222312847
Chicago/Turabian StyleDing, Yi, Zhongjie Wang, Yali Wang, Yahong Geng, Xiaobin Wen, and Yeguang Li. 2021. "Physiological and Dual Transcriptional Analysis of Microalga Graesiella emersonii–Amoeboaphelidium protococcarum Pathosystem Uncovers Conserved Defense Response and Robust Pathogenicity" International Journal of Molecular Sciences 22, no. 23: 12847. https://doi.org/10.3390/ijms222312847
APA StyleDing, Y., Wang, Z., Wang, Y., Geng, Y., Wen, X., & Li, Y. (2021). Physiological and Dual Transcriptional Analysis of Microalga Graesiella emersonii–Amoeboaphelidium protococcarum Pathosystem Uncovers Conserved Defense Response and Robust Pathogenicity. International Journal of Molecular Sciences, 22(23), 12847. https://doi.org/10.3390/ijms222312847






