Profiling the Proteome of Cyst Nematode-Induced Syncytia on Tomato Roots
Abstract
:1. Introduction
2. Results
2.1. Optimization of the Fixation Method, Tissue Embedding, and Sectioning Procedure
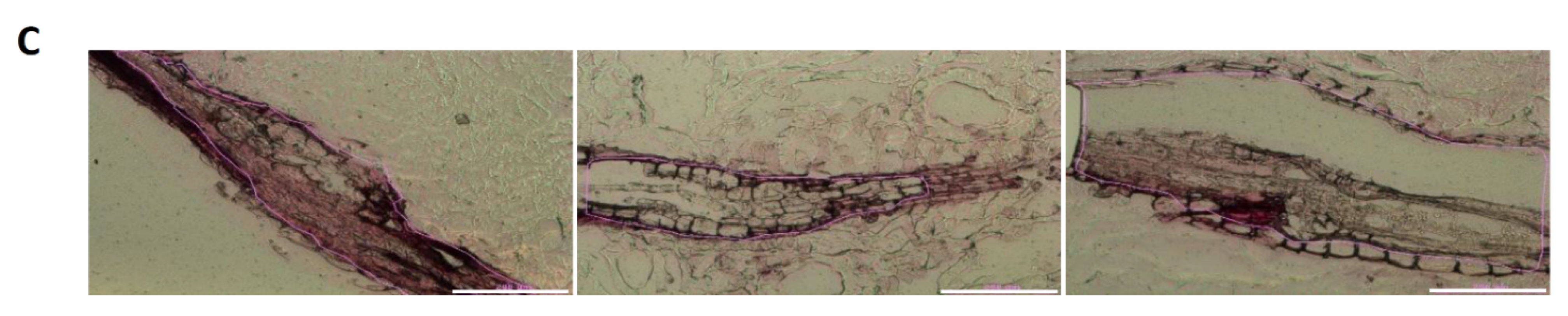
2.2. Tissue Analysis and LCM
2.3. Protein Isolation
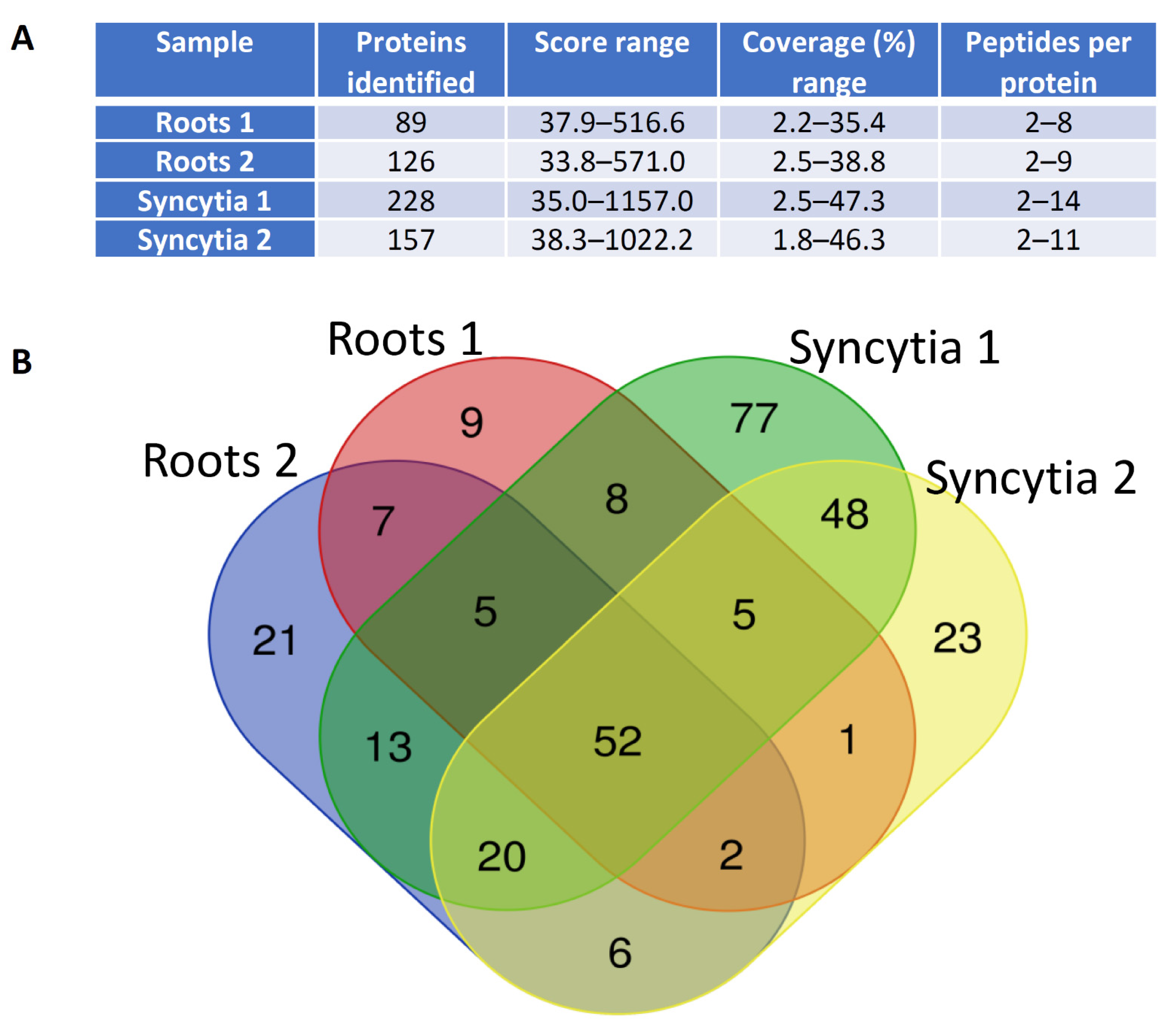
2.4. Mass Spectrometry Results
3. Discussion
4. Materials and Methods
4.1. Plant Material and Specimen Preparation
4.2. Laser-Capture Microdissection and Protein Sample Preparation
4.3. Mass Spectrometry
4.4. RNA Isolation, cDNA Synthesis, and qPCR Reaction
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Grundler, F.M.W.; Sobczak, M.; Golinowski, W. Formation of wall openings in root cells of Arabidopsis thaliana following infection by the plant-parasitic nematode Heterodera schachtii. Eur. J. Plant Pathol. 1998, 104, 545–551. [Google Scholar] [CrossRef]
- Grundler, F.M.W.; Sobczak, M.; Lange, S. Defence responses of Arabidopsis thaliana during invasion and feeding site induction by the plant-parasitic nematode Heterodera glycines. Physiol. Mol. Plant Pathol. 1997, 50, 419–429. [Google Scholar] [CrossRef]
- Dubreuil, G.; Magliano, M.; Deleury, E.; Abad, P.; Rosso, M.N. Transcriptome analysis of root-knot nematode functions induced in the early stage of parasitism. New Phytol. 2007, 176, 426–436. [Google Scholar] [CrossRef]
- Święcicka, M.; Filipecki, M.; Lont, D.; van Vliet, J.; Qin, L.; Goverse, A.; Bakker, J.; Helder, J. Dynamics in the tomato root transcriptome on infection with the potato cyst nematode Globodera rostochiensis. Mol. Plant Pathol. 2009, 10, 487–500. [Google Scholar] [CrossRef] [PubMed]
- Szakasits, D.; Heinen, P.; Wieczorek, K.; Hofmann, J.; Wagner, F.; Kreil, D.P.; Sykacek, P.; Grundler, F.M.W.; Bohlmann, H. The transcriptome of syncytia induced by the cyst nematode Heterodera schachtii in Arabidopsis roots. Plant J. 2009, 57, 771–784. [Google Scholar] [CrossRef] [Green Version]
- Kooliyottil, R.; Dandurand, L.-M.; Kuhl, J.C.; Caplan, A.; Xiao, F.; Mimee, B.; Lafond-Lapalme, J. Transcriptome analysis of Globodera pallida from the susceptible host Solanum tuberosum or the resistant plant Solanum sisymbriifolium. Sci. Rep. 2019, 9, 13256. [Google Scholar] [CrossRef] [PubMed]
- Labudda, M.; Różańska, E.; Czarnocka, W.; Sobczak, M.; Dzik, J.M. Systemic changes in photosynthesis and reactive oxygen species homeostasis in shoots of Arabidopsis thaliana infected with the beet cyst nematode Heterodera schachtii. Mol. Plant Pathol. 2018, 19, 1690–1704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gleason, C.A.; Liu, Q.L.; Williamson, V.M. Silencing a candidate nematode effector gene corresponding to the tomato resistance gene Mi-1 leads to acquisition of virulence. Mol. Plant Microbe Interact. 2008, 21, 576–585. [Google Scholar] [CrossRef] [Green Version]
- Jones, J.T.; Kumar, A.; Pylypenko, L.A.; Thirugnanasambandam, A.; Castelli, L.; Chapman, S.; Cock, P.J.A.; Grenier, E.; Lilley, C.J.; Phillips, M.S.; et al. Identification and functional characterization of effectors in expressed sequence tags from various life cycle stages of the potato cyst nematode Globodera pallida. Mol. Plant Pathol. 2009, 10, 815–828. [Google Scholar] [CrossRef]
- Postma, W.J.; Slootweg, E.J.; Rehman, S.; Finkers-Tomczak, A.; Tytgat, T.O.G.; van Gelderen, K.; Lozano-Torres, J.L.; Roosien, J.; Pomp, R.; van Schaik, C.; et al. The effector SPRYSEC-19 of Globodera rostochiensis suppresses CC-NB-LRR-mediated disease resistance in plants. Plant Physiol. 2012, 160, 944–954. [Google Scholar] [CrossRef] [Green Version]
- Quentin, M.; Abad, P.; Favery, B. Plant parasitic nematode effectors target host defense and nuclear functions to establish feeding cells. Front. Plant Sci. 2013, 4, 53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eves-van den Akker, S.; Lilley, C.J.; Jones, J.T.; Urwin, P.E. Identification and characterisation of a hyper-variable apoplastic effector gene family of the potato cyst nematodes. PLoS Pathog. 2014, 10, e1004391. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mejias, J.; Truong, N.M.; Abad, P.; Favery, B.; Quentin, M. Plant proteins and processes targeted by parasitic nematode effectors. Front. Plant Sci. 2019, 10, 970. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.; Khan, A.U. Plant parasitic nematodes effectors and their crosstalk with defense response of host plants: A battle underground. Rhizosphere 2021, 17, 100288. [Google Scholar] [CrossRef]
- Matuszkiewicz, M.; Sobczak, M.; Cabrera, J.; Escobar, C.; Karpiński, S.; Filipecki, M. The role of programmed cell death regulator LSD1 in nematode-induced syncytium formation. Front. Plant Sci. 2018, 9, 314. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, J.; Barcala, M.; García, A.; Rio-Machín, A.; Medina, C.; Jaubert-Possamai, S.; Favery, B.; Maizel, A.; Ruiz-Ferrer, V.; Fenoll, C.; et al. Differentially expressed small RNAs in Arabidopsis galls formed by Meloidogyne javanica: A functional role for miR390 and its TAS3-derived tasiRNAs. New Phytol. 2016, 209, 1625–1640. [Google Scholar] [CrossRef] [Green Version]
- Święcicka, M.; Skowron, W.; Cieszyński, P.; Dąbrowska-Bronk, J.; Matuszkiewicz, M.; Filipecki, M.; Koter, M.D. The suppression of tomato defence response genes upon potato cyst nematode infection indicates a key regulatory role of miRNAs. Plant Physiol. Biochem. 2017, 113, 51–55. [Google Scholar] [CrossRef]
- Koter, M.D.; Święcicka, M.; Matuszkiewicz, M.; Pacak, A.; Derebecka, N.; Filipecki, M. The miRNAome dynamics during developmental and metabolic reprogramming of tomato root infected with potato cyst nematode. Plant Sci. 2018, 268, 18–29. [Google Scholar] [CrossRef]
- Hammond-Kosack, K.E.; Atkinson, H.J.; Bowles, D.J. Changes in abundance of translatable mRNA species in potato roots and leaves following root invasion by cyst-nematode G. rostochiensis pathotypes. Physiol. Mol. Plant Pathol. 1990, 37, 339–354. [Google Scholar] [CrossRef]
- Callahan, F.E.; Jenkins, J.N.; Creech, R.G.; Lawrence, G.W. Changes in cotton root proteins correlated with resistance to root knot nematode development. J. Cotton Sci. 1997, 1, 38–47. [Google Scholar]
- Hütten, M.; Geukes, M.; Misas-Villamil, J.C.; van der Hoorn, R.A.L.; Grundler, F.M.W.; Siddique, S. Activity profiling reveals changes in the diversity and activity of proteins in Arabidopsis roots in response to nematode infection. Plant Physiol. Biochem. 2015, 97, 36–43. [Google Scholar] [CrossRef]
- Jain, A.; Singh, H.B.; Das, S. Deciphering plant-microbe crosstalk through proteomics studies. Microbiol. Res. 2021, 242, 126590. [Google Scholar] [CrossRef]
- Valot, B.; Negroni, L.; Zivy, M.; Gianinazzi, S.; Dumas-Gaudot, E. A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 2006, 6, S145–S155. [Google Scholar] [CrossRef] [PubMed]
- Rep, M.; Dekker, H.L.; Vossen, J.H.; de Boer, A.D.; Houterman, P.M.; Speijer, D.; Back, J.W.; de Koster, C.G.; Cornelissen, B.J.C. Mass spectrometric identification of isoforms of PR proteins in xylem sap of fungus-infected tomato. Plant Physiol. 2002, 130, 904–917. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Yang, L.; Xu, H.; Li, Q.; Ma, Z.; Chu, C. Differential proteomic analysis of proteins in wheat spikes induced by Fusarium graminearum. Proteomics 2005, 5, 4496–4503. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Eudes, F.; Laroche, A. Identification of differentially regulated proteins in response to a compatible interaction between the pathogen Fusarium graminearum and its host, Triticum aestivum. Proteomics 2006, 6, 4599–4609. [Google Scholar] [CrossRef] [PubMed]
- Escobar, C.; Brown, S.; Mitchum, M.G. Transcriptomic and proteomic analysis of the plant response to nematode infection. In Genomics and Molecular Genetics of Plant-Nematode Interactions; Jones, J., Gheysen, G., Fenoll, C., Eds.; Springer: Dordrecht, The Netherlands, 2011; pp. 157–173. [Google Scholar] [CrossRef]
- Peck, S.C.; Nühse, T.S.; Hess, D.; Iglesias, A.; Meins, F.; Boller, T. Directed proteomics identifies a plant-specific protein rapidly phosphorylated in response to bacterial and fungal elicitors. Plant Cell 2001, 13, 1467–1475. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klink, V.P.; Alkharouf, N.; MacDonald, M.; Matthews, B. Laser capture microdissection (LCM) and expression analyses of Glycine max (soybean) syncytium containing root regions formed by the plant pathogen Heterodera glycines (soybean cyst nematode). Plant Mol. Biol. 2005, 59, 965–979. [Google Scholar] [CrossRef]
- Klink, V.P.; Overall, C.C.; Alkharouf, N.W.; MacDonald, M.H.; Matthews, B.F. Laser capture microdissection (LCM) and comparative microarray expression analysis of syncytial cells isolated from incompatible and compatible soybean (Glycine max) roots infected by the soybean cyst nematode (Heterodera glycines). Planta 2007, 226, 1389–1409. [Google Scholar] [CrossRef]
- Klink, V.P.; Thibaudeau, G. Laser microdissection of semi-thin sections from plastic-embedded tissue for studying plant-organism developmental processes at single-cell resolution. J. Plant Interact. 2014, 9, 610–617. [Google Scholar] [CrossRef]
- Anjam, M.S.; Ludwig, Y.; Hochholdinger, F.; Miyaura, C.; Inada, M.; Siddique, S.; Grundler, F.M.W. An improved procedure for isolation of high-quality RNA from nematode-infected Arabidopsis roots through laser capture microdissection. Plant Methods 2016, 12, 25. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, Y.; Hochholdinger, F. Laser microdissection of plant cells. In Plant Cell Morphogenesis; Methods in Molecular Biology (Methods and Protocols); Žárský, V., Cvrčková, F., Eds.; Humana Press: Totowa, NJ, USA, 2014; Volume 1080, pp. 249–258. [Google Scholar] [CrossRef]
- Gautam, V.; Sarkar, A.K. Laser assisted microdissection, an efficient technique to understand tissue specific gene expression patterns and functional genomics in plants. Mol. Biotechnol. 2015, 57, 299–308. [Google Scholar] [CrossRef]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Portillo, M.; Lindsey, K.; Casson, S.; García-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Isolation of RNA from laser-capture-microdissected giant cells at early differentiation stages suitable for differential transcriptome analysis. Mol. Plant Pathol. 2009, 10, 523–535. [Google Scholar] [CrossRef]
- Barcala, M.; García, A.; Cabrera, J.; Casson, S.; Lindsey, K.; Favery, B.; García-Casado, G.; Solano, R.; Fenoll, C.; Escobar, C. Early transcriptomic events in microdissected Arabidopsis nematode-induced giant cells. Plant J. 2010, 61, 698–712. [Google Scholar] [CrossRef] [PubMed]
- Ji, H.; Gheysen, G.; Denil, S.; Lindsey, K.; Topping, J.F.; Nahar, K.; Haegeman, A.; de Vos, W.H.; Trooskens, G.; van Criekinge, W.; et al. Transcriptional analysis through RNA sequencing of giant cells induced by Meloidogyne graminicola in rice roots. J. Exp. Bot. 2013, 64, 3885–3898. [Google Scholar] [CrossRef] [Green Version]
- Srinivasan, M.; Sedmak, D.; Jewell, S. Effect of fixatives and tissue processing on the content and integrity of nucleic acids. Am. J. Pathol. 2002, 161, 1961–1971. [Google Scholar] [CrossRef] [Green Version]
- Böckenhoff, A.; Grundler, F.M.W. Studies on the nutrient uptake by the beet cyst nematode Heterodera schachtii by in situ microinjection of fluorescent probes into the feeding structures in Arabidopsis thaliana. Parasitology 1994, 109, 249–255. [Google Scholar] [CrossRef]
- Cabrera, J.; Bustos, R.; Favery, B.; Fenoll, C.; Escobar, C. NEMATIC: A simple and versatile tool for the in silico analysis of plant-nematode interactions. Mol. Plant Pathol. 2014, 15, 627–636. [Google Scholar] [CrossRef] [PubMed]
- Nakazono, M.; Qiu, F.; Borsuk, L.A.; Schnable, P.S. Laser-capture microdissection, a tool for the global analysis of gene expression in specific plant cell types: Identification of genes expressed differentially in epidermal cells or vascular tissues of maize. Plant Cell 2003, 15, 583–596. [Google Scholar] [CrossRef] [Green Version]
- Kerk, N.M.; Ceserani, T.; Tausta, S.L.; Sussex, I.M.; Nelson, T.M. Laser capture microdissection of cells from plant tissues. Plant Physiol. 2003, 132, 27–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsay, K.; Wang, Z.; Jones, M.G.K. Using laser capture microdissection to study gene expression in early stages of giant cells induced by root-knot nematodes. Mol. Plant Pathol. 2004, 5, 587–592. [Google Scholar] [CrossRef] [Green Version]
- Baranowski, Ł.; Różańska, E.; Sańko-Sawczenko, I.; Matuszkiewicz, M.; Znojek, E.; Filipecki, M.; Grundler, F.M.W.; Sobczak, M. Arabidopsis tonoplast intrinsic protein and vacuolar H+-adenosinetriphosphatase reflect vacuole dynamics during development of syncytia induced by the beet cyst nematode Heterodera schachtii. Protoplasma 2019, 256, 419–429. [Google Scholar] [CrossRef]
- Sobczak, M.; Avrova, A.; Jupowicz, J.; Phillips, M.S.; Ernst, K.; Kumar, A. Characterization of susceptibility and resistance responses to potato cyst nematode (Globodera spp.) infection of tomato lines in the absence and presence of the broad-spectrum nematode resistance Hero gene. Mol. Plant Microbe Interact. 2005, 18, 158–168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W.; Horgan, G.W.; Dempfle, L. Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res. 2002, 30, e36. [Google Scholar] [CrossRef] [PubMed]


| Collection Buffer | Tissue Homogenization | Protein Sample Preparation | Polymer Purification | Sample Size |
|---|---|---|---|---|
| 8 M urea, 1× protease inhibitors cocktail, 1× phosphatase inhibitors cocktail | Homogenizer and glass beads | PAGE-separated gel sections | Not included | 4 mm2 |
| 8M urea, 1× protease inhibitors cocktail, 1× phosphatase inhibitors cocktail, 100 mM Tris, 150 mM NaCl, 5 mM EDTA, 10 mM DTT, 0.5% (v/v) Triton X-100 | Pestle and Eppendorf tube | Methanol–chloroform precipitate | Included | 15 mm2 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filipecki, M.; Żurczak, M.; Matuszkiewicz, M.; Święcicka, M.; Kurek, W.; Olszewski, J.; Koter, M.D.; Lamont, D.; Sobczak, M. Profiling the Proteome of Cyst Nematode-Induced Syncytia on Tomato Roots. Int. J. Mol. Sci. 2021, 22, 12147. https://doi.org/10.3390/ijms222212147
Filipecki M, Żurczak M, Matuszkiewicz M, Święcicka M, Kurek W, Olszewski J, Koter MD, Lamont D, Sobczak M. Profiling the Proteome of Cyst Nematode-Induced Syncytia on Tomato Roots. International Journal of Molecular Sciences. 2021; 22(22):12147. https://doi.org/10.3390/ijms222212147
Chicago/Turabian StyleFilipecki, Marcin, Marek Żurczak, Mateusz Matuszkiewicz, Magdalena Święcicka, Wojciech Kurek, Jarosław Olszewski, Marek Daniel Koter, Douglas Lamont, and Mirosław Sobczak. 2021. "Profiling the Proteome of Cyst Nematode-Induced Syncytia on Tomato Roots" International Journal of Molecular Sciences 22, no. 22: 12147. https://doi.org/10.3390/ijms222212147
APA StyleFilipecki, M., Żurczak, M., Matuszkiewicz, M., Święcicka, M., Kurek, W., Olszewski, J., Koter, M. D., Lamont, D., & Sobczak, M. (2021). Profiling the Proteome of Cyst Nematode-Induced Syncytia on Tomato Roots. International Journal of Molecular Sciences, 22(22), 12147. https://doi.org/10.3390/ijms222212147






