PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression
Abstract
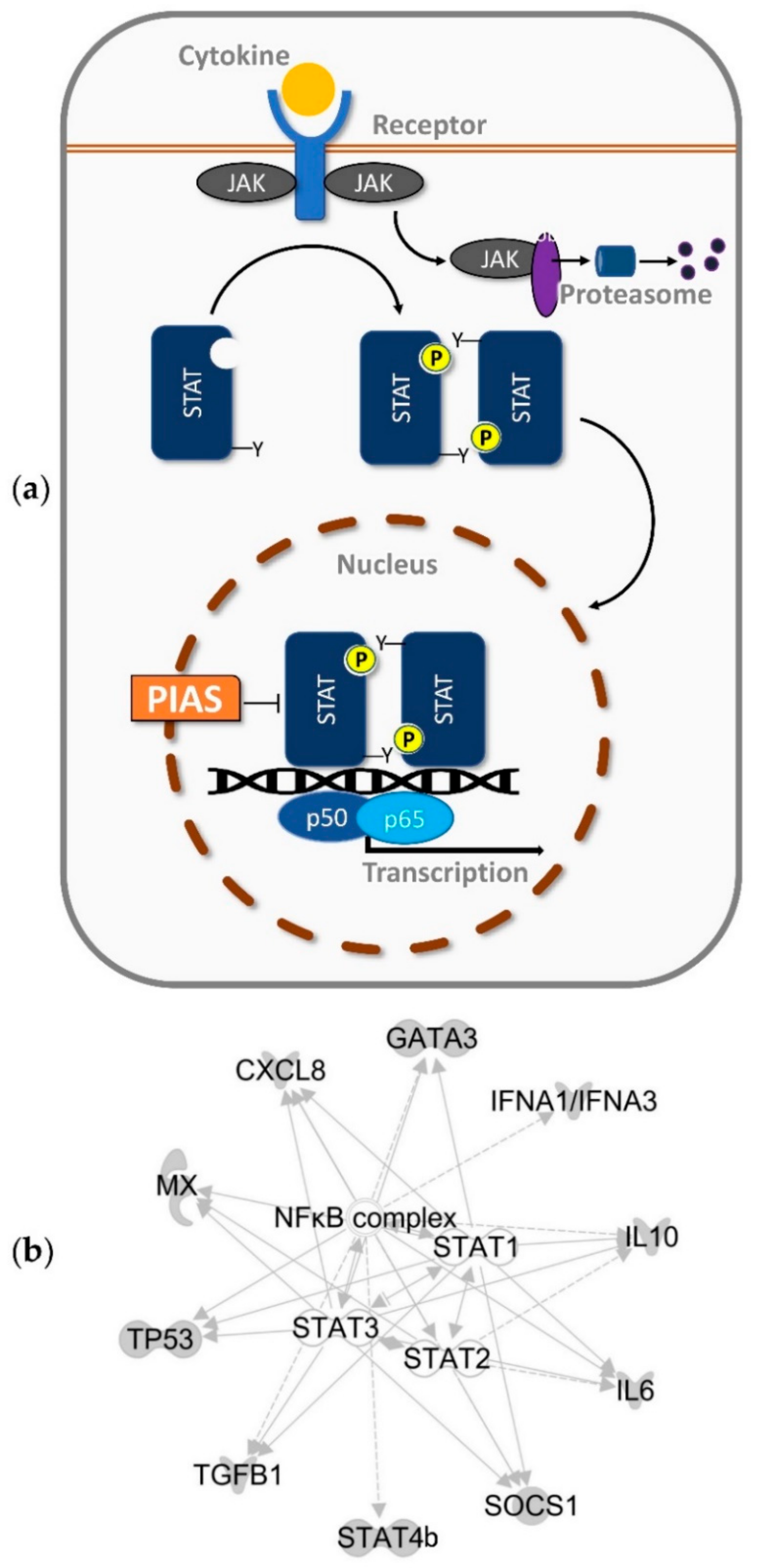
1. Introduction
2. Results
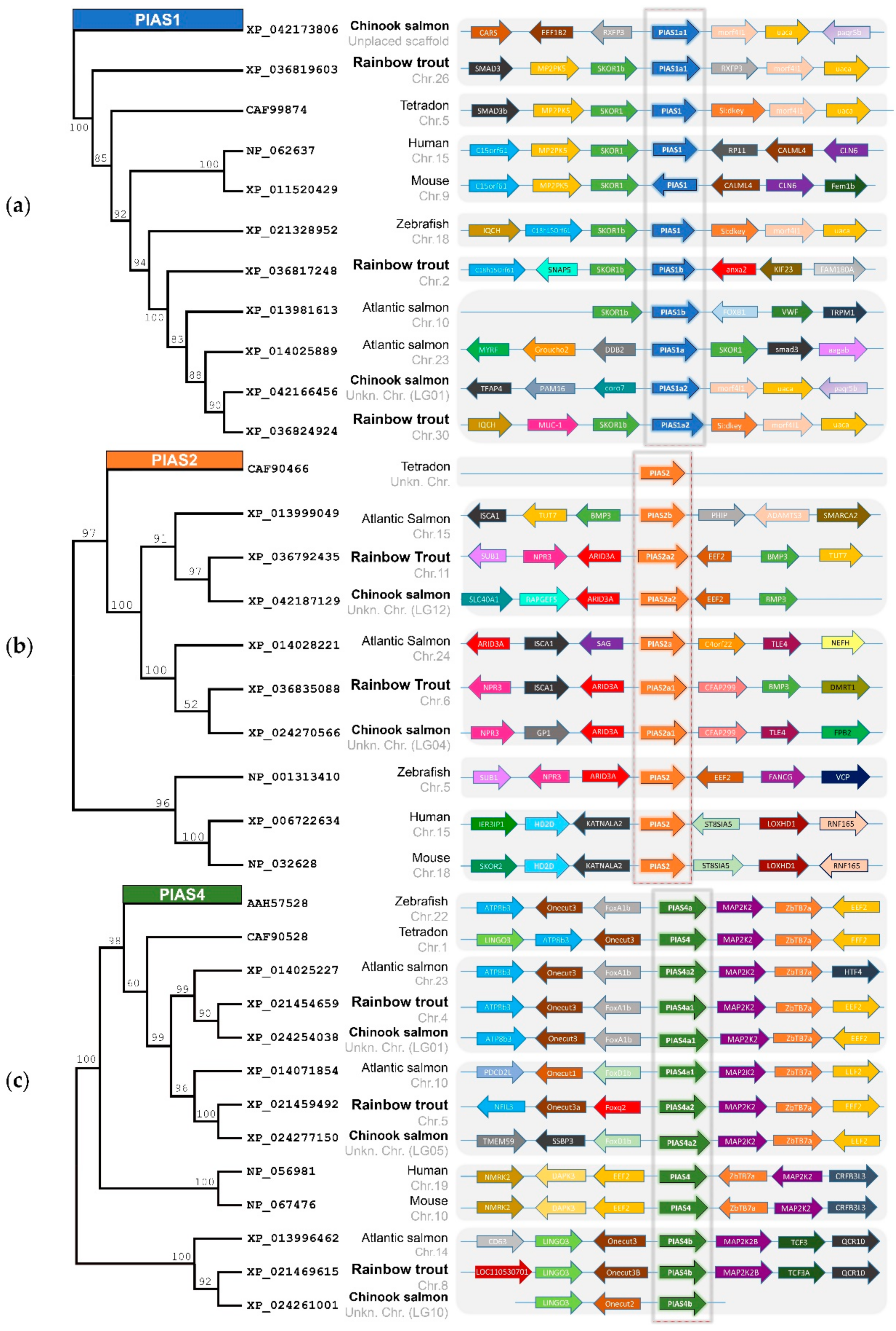
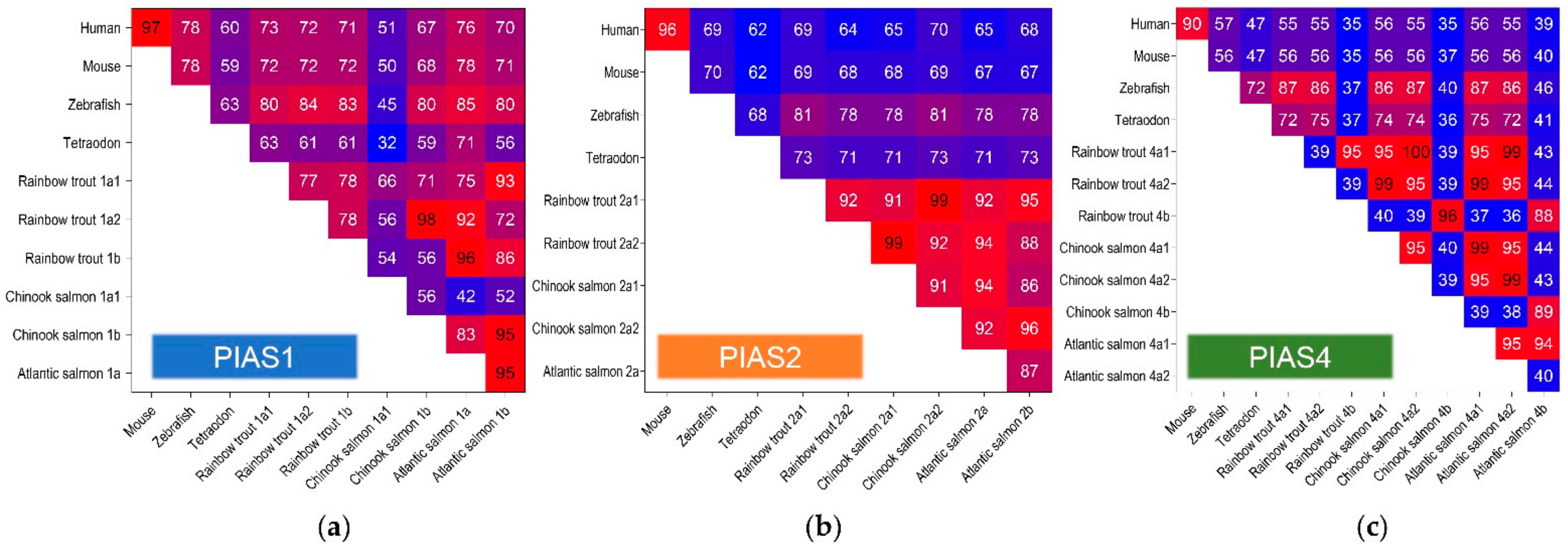
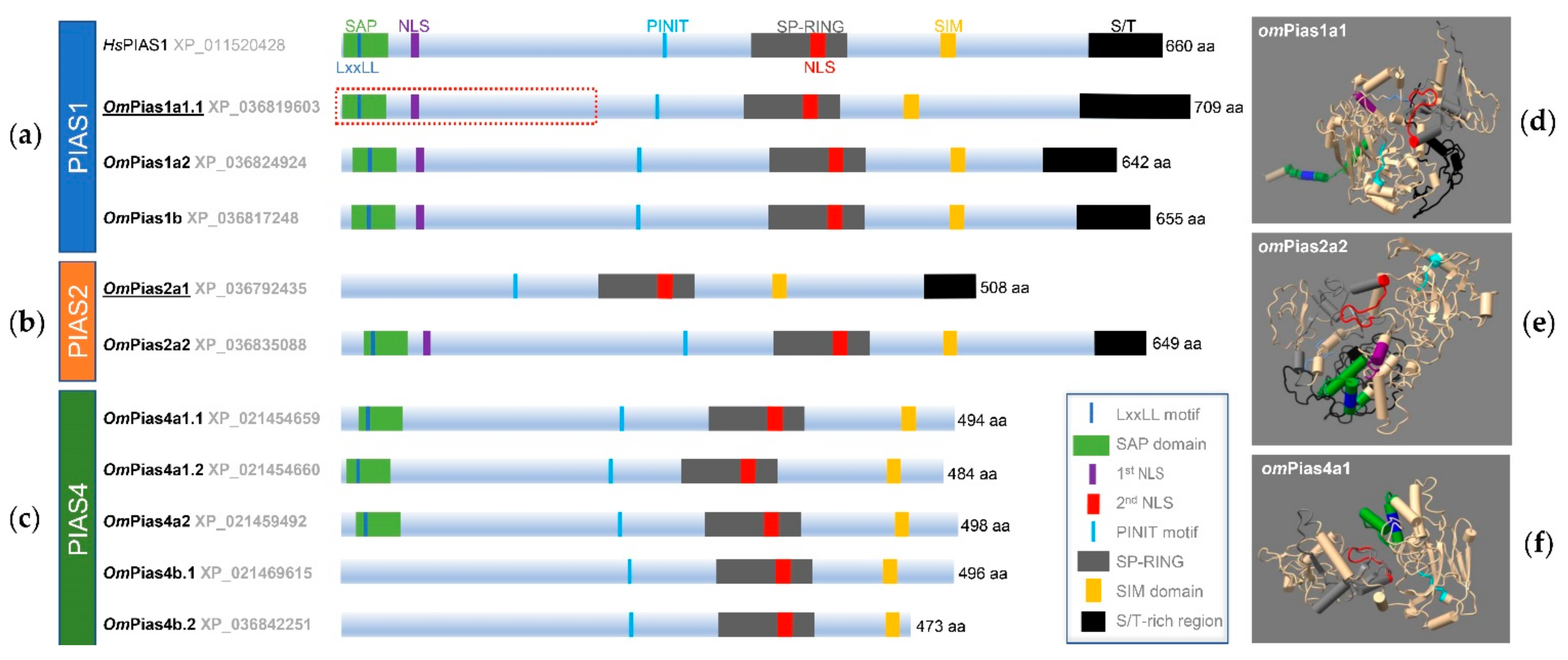
2.1. PIAS Genes Are Present as Multiple Gene Copies in Rainbow Trout and Chinook Salmon
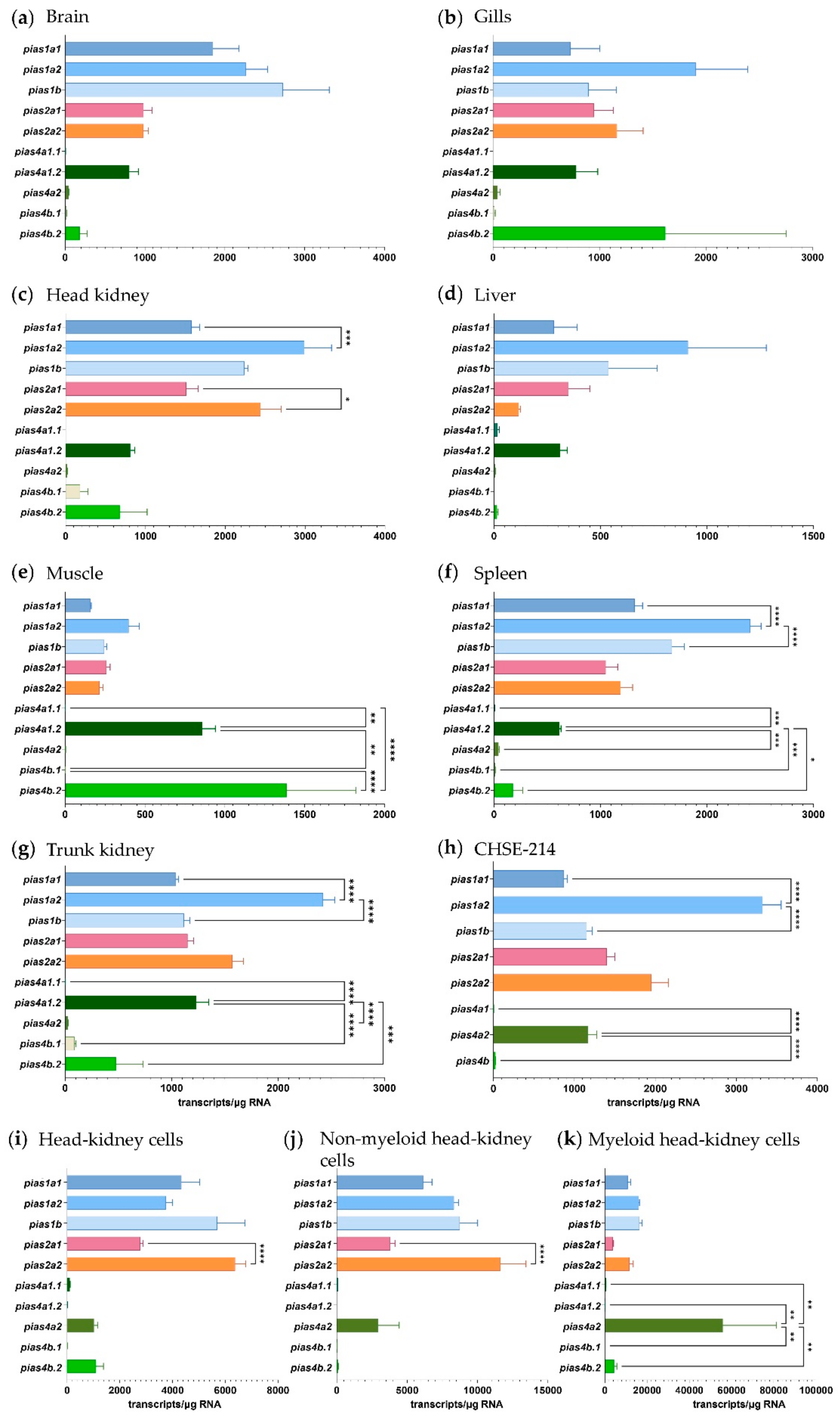
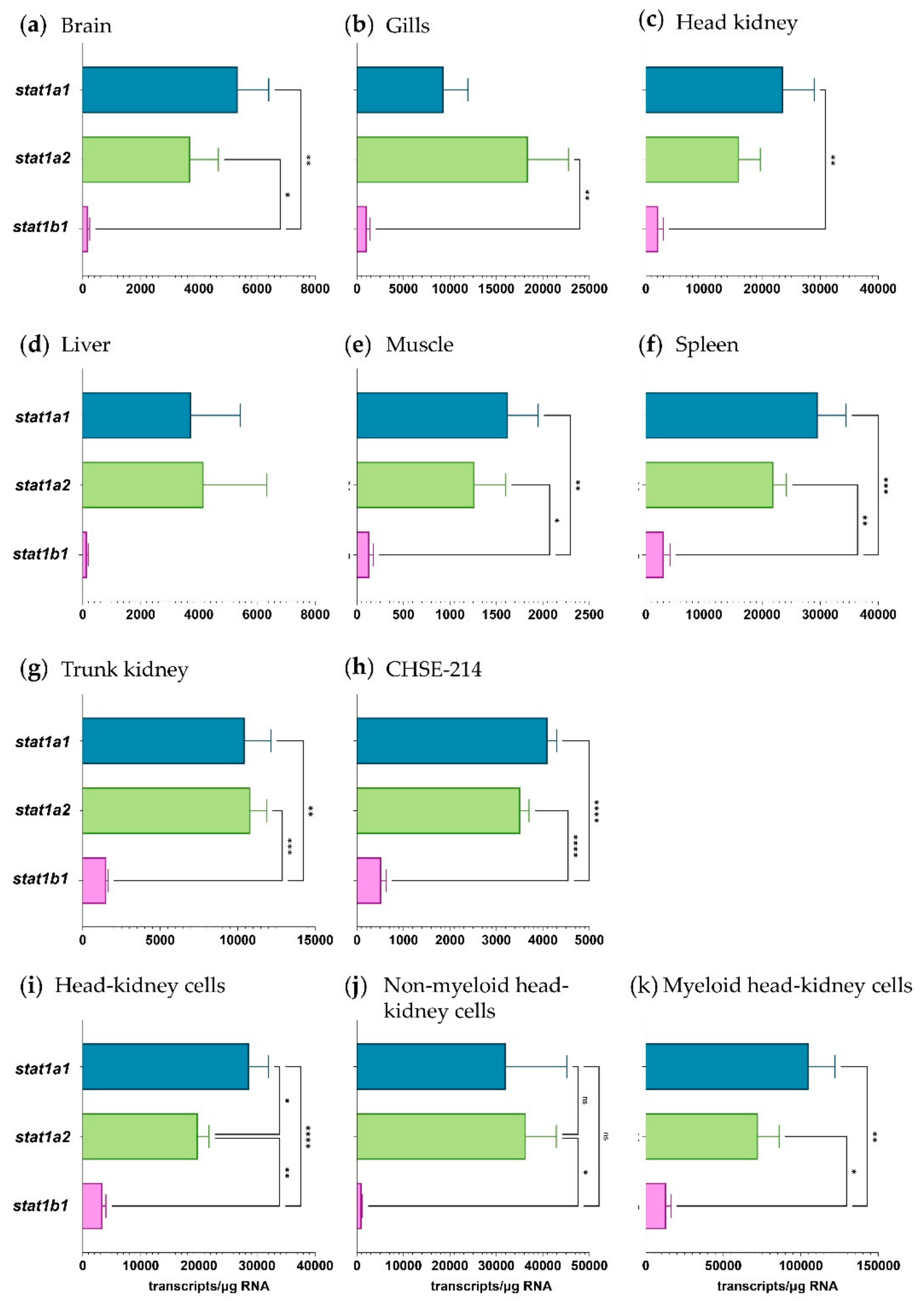
2.2. The Expression of Pias Genes Is Tissue and Cell-Type Specific
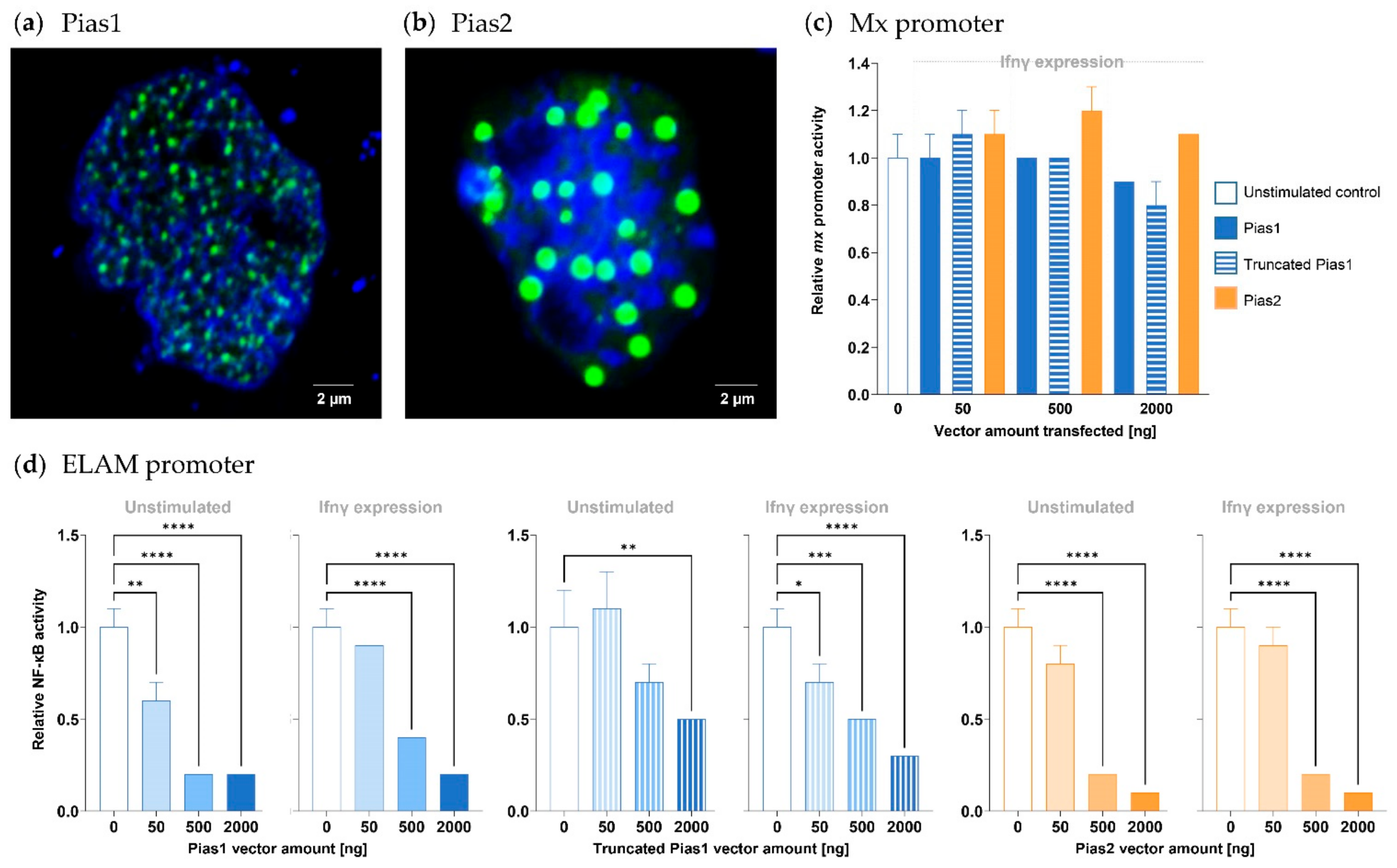
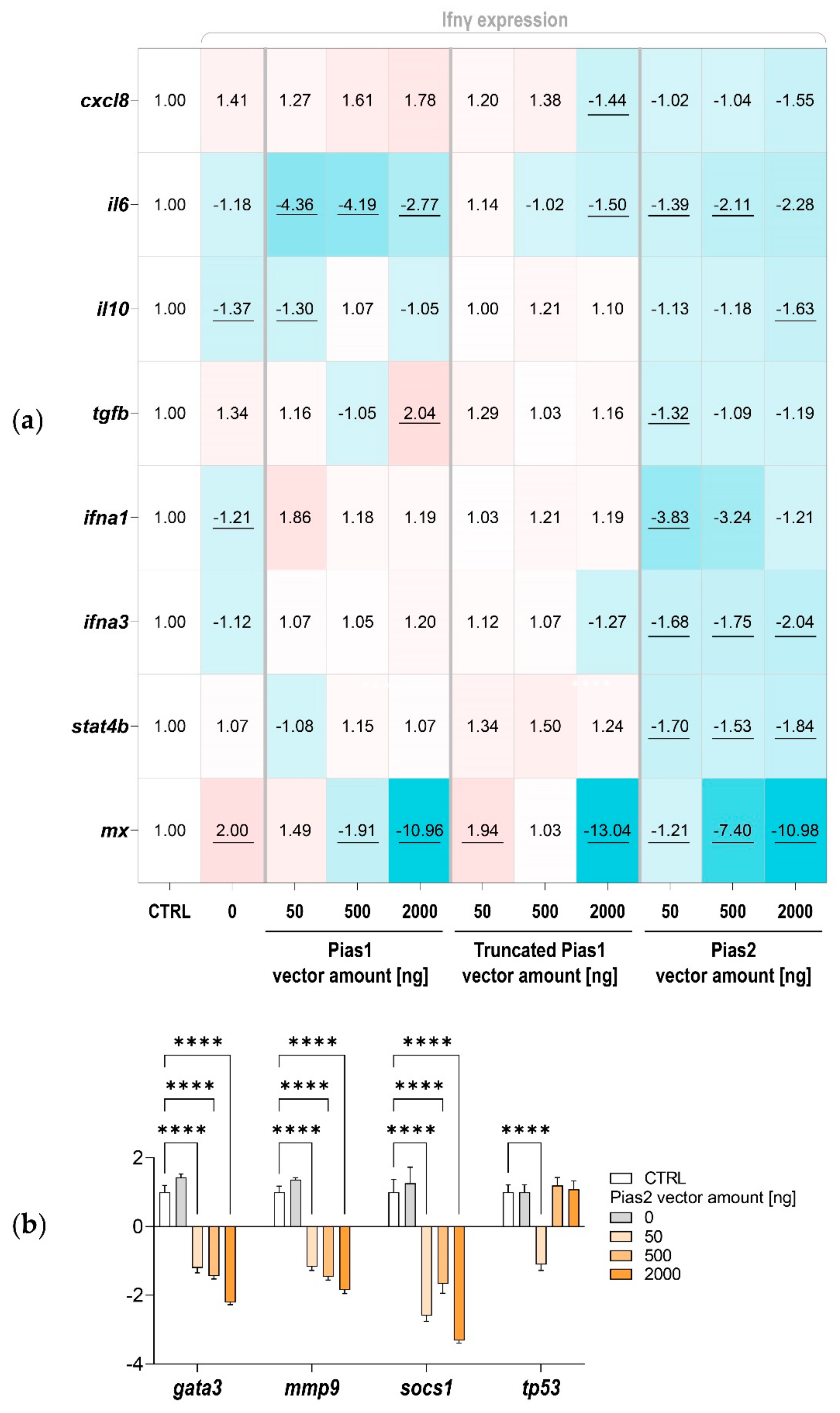
2.3. Pias1 and Pias2 Are Located in the Nucleus of Model Cells and Interact with NF-κB to Alter Transcriptional Responses
3. Discussion
4. Materials and Methods
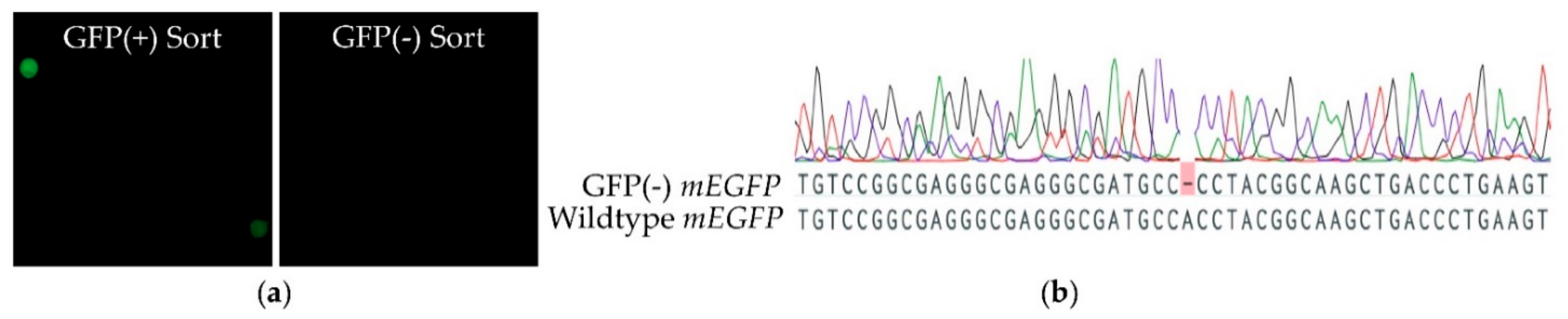
4.1. Sampling and Cell Sorting
4.2. RNA Isolation, cDNA Synthesis, and Quantitative PCR (qPCR) Analysis
4.3. Construction of Pias Expression Constructs
4.4. Transfection, Luciferase Assay, and Confocal Microscopy
4.5. Strategy for the Generation of a Pias-Knock-Out Cell Line
4.6. Data Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbrevations
References
- Wilks, A.F. Two putative protein-tyrosine kinases identified by application of the polymerase chain reaction. Proc. Natl. Acad. Sci. USA 1989, 86, 1603–1607. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.G.; Mangan, J.K.; Amanullah, A.; Wong, B.C.; Vora, R.K.; Liebermann, D.A.; Hoffman, B.; Graña, X.; Reddy, E.P. Activation of the Jak3 pathway is associated with granulocytic differentiation of myeloid precursor cells. Blood 2002, 100, 2753–2762. [Google Scholar] [CrossRef] [PubMed]
- Rawlings, J.S.; Rosler, K.M.; Harrison, D.A. The JAK/STAT signaling pathway. J. Cell Sci. 2004, 117, 1281–1283. [Google Scholar] [CrossRef] [PubMed]
- Bousoik, E.; Aliabadi, H.M. “Do We Know Jack” About JAK? A Closer Look at JAK/STAT Signaling Pathway. Front. Oncol. 2018, 8, 287. [Google Scholar] [CrossRef] [PubMed]
- Laurence, A. JAK Kinases in Health and Disease: An Update. Open Rheumatol. J. 2012, 6, 232–244. [Google Scholar] [CrossRef]
- Gilmore, T.D.; Wolenski, F.S. NF-κB: Where did it come from and why? Immunol. Rev. 2012, 246, 14–35. [Google Scholar] [CrossRef]
- Hiroi, M.; Ohmori, Y. Transcriptional Synergism between NF-κB and STAT1. J. Oral Biosci. 2005, 47, 230–242. [Google Scholar] [CrossRef]
- Platanitis, E.; Decker, T. Regulatory Networks Involving STATs, IRFs, and NFκB in Inflammation. Front. Immunol. 2018, 9, 2542. [Google Scholar] [CrossRef]
- Murphy, J.M.; Tannahill, G.M.; Hilton, D.J.; Greenhalgh, C.J. The Negative Regulation of Jak/stat Signaling, 2nd ed.; Elsevier Inc.: Amsterdam, The Netherlands, 2010. [Google Scholar] [CrossRef]
- Landsman, T.; Waxman, D.J. Role of the Cytokine-induced SH2 Domain-containing Protein CIS in Growth Hormone Receptor Internalization. J. Biol. Chem. 2005, 45, 37471–37480. [Google Scholar] [CrossRef]
- Feng, G.S.; Hui, C.C.; Pawson, T. Sh2-containing phosphotyrosine phosphatase as a target of protein-tyrosine kinases. Science 1993, 259, 1607–1611. [Google Scholar] [CrossRef]
- Kiu, H.; Nicholson, S.E. Biology and significance of the JAK/STAT signalling pathways. Growth Factors 2012, 30, 88–106. [Google Scholar] [CrossRef]
- Shuai, K.; Liu, B. Regulation of JAK-STAT signalling in the immune system. Nat. Rev. Immunol. 2003, 3, 900–911. [Google Scholar] [CrossRef]
- Rebl, A.; Goldammer, T. Under control: The innate immunity of fish from the inhibitors’ perspective. Fish Shellfish. Immunol. 2018, 77, 328–349. [Google Scholar] [CrossRef]
- Wang, R.; Huang, S.; Fu, X.; Huang, G.; Yan, X.; Yue, Z.; Chen, S.; Li, Y.; Xu, A. The conserved ancient role of chordate PIAS as a multilevel repressor of the NF-κB pathway. Sci. Rep. 2017, 7, 17063. [Google Scholar] [CrossRef]
- Liongue, C.; O’Sullivan, L.A.; Trengove, M.C.; Ward, A.C. Evolution of JAK-STAT pathway components: Mechanisms and role in immune system development. PLoS ONE 2012, 7, e32777. [Google Scholar] [CrossRef]
- Palvimo, J.J. PIAS proteins as regulators of small ubiquitin-related modifier (SUMO) modifications and transcription. Biochem. Soc. Trans. 2007, 35, 1405–1408. [Google Scholar] [CrossRef]
- Kipp, M.; Göhring, F.; Ostendorp, T.; van Drunen, C.M.; van Driel, R.; Przybylski, M.; Fackelmayer, F.O. SAF-Box, a conserved protein domain that specifically recognizes scaffold attachment region DNA. Mol. Cell. Biol. 2000, 20, 7480–7489. [Google Scholar] [CrossRef]
- Duval, D.; Duval, G.; Kedinger, C.; Poch, O.; Boeuf, H. The ‘PINIT’ motif, of a newly identified conserved domain of the PIAS protein family, is essential for nuclear retention of PIAS3L. FEBS Lett. 2003, 554, 111–118. [Google Scholar] [CrossRef]
- Joazeiro, C.A.; Weissman, A.M. RING finger proteins: Mediators of ubiquitin ligase activity. Cell 2000, 102, 549–552. [Google Scholar] [CrossRef]
- Lussier-Price, M.; Mascle, X.H.; Cappadocia, L.; Kamada, R.; Sakaguchi, K.; Wahba, H.M.; Omichinski, J.G. Characterization of a C-Terminal SUMO-Interacting Motif Present in Select PIAS-Family Proteins. Structure 2020, 28, 573–585. [Google Scholar] [CrossRef]
- Shuai, K.; Liu, B. Regulation of gene-activation pathways by pias proteins in the immune system. Nat. Rev. Immunol. 2005, 5, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Yang, R.; Wong, K.A.; Getman, C.; Stein, N.; Teitell, M.A.; Cheng, G.; Wu, H.; Shuai, K. Negative Regulation of NF- B Signaling by PIAS1. Mol. Cell. Biol. 2005, 25, 1113–1123. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Liao, J.; Rao, X.; Kushner, S.A.; Chung, C.D.; Chang, D.D.; Shuai, K.E. Inhibition of Stat1-mediated gene activation by PIAS1. Proc. Natl. Acad. Sci. USA 1998, 95, 10626–10631. [Google Scholar] [CrossRef] [PubMed]
- Chung, C.D.; Liao, J.; Liu, B.; Rao, X.; Jay, P.; Berta, P.; Shuai, K. Specific inhibition of Stat3 signal transduction by PIAS3. Science 1997, 278, 1803–1805. [Google Scholar] [CrossRef]
- Tussié-Luna, M.I.; Bayarsaihan, D.; Seto, E.; Ruddle, F.H.; Roy, A.L. Physical and functional interactions of histone deacetylase 3 with TFII-I family proteins and PIASx. Proc. Natl. Acad. Sci. USA 2002, 99, 12807–12812. [Google Scholar] [CrossRef]
- Sachdev, S.; Bruhn, L.; Sieber, H.; Pichler, A.; Melchior, F.; Grosschedl, R. PIASy, a nuclear matrix-associated SUMO E3 ligase, represses LEF1 activity by sequestration into nuclear bodies. Genes Dev. 2001, 15, 3088–3103. [Google Scholar] [CrossRef]
- Liu, B.; Gross, M.; Hoeve, J.T.; Shuai, K. A transcriptional corepressor of Stat1 with an essential LXXLL signature motif. Proc. Natl. Acad. Sci. USA 2001, 98, 3203–3207. [Google Scholar] [CrossRef]
- Liu, B.; Yang, Y.; Chernishof, V.; Loo, R.R.O.; Jang, H.; Tahk, S.; Yang, R.; Mink, S.; Shultz, D.; Bellone, C.J.; et al. Proinflammatory stimuli induce IKKalpha-mediated phosphorylation of PIAS1 to restrict inflammation and immunity. Cell 2007, 129, 903–914. [Google Scholar] [CrossRef]
- Liu, Y.; Bridges, R.; Wortham, A.; Kulesz-Martin, M. NF-κB repression by PIAS3 mediated RelA SUMOylation. PLoS ONE 2012, 7, e37636. [Google Scholar] [CrossRef]
- Jang, H.D.; Yoon, K.; Shin, Y.J.; Kim, J.; Lee, S.Y. PIAS3 suppresses NF-kappaB-mediated transcription by interacting with the p65/RelA subunit. J. Biol. Chem. 2004, 279, 24873–24880. [Google Scholar] [CrossRef]
- Gross, M.; Liu, B.; Tan, J.A.; French, F.S.; Carey, M.; Shuai, K. Distinct effects of PIAS proteins on androgen-mediated gene activation in prostate cancer cells. Oncogene 2001, 20, 3880–3887. [Google Scholar] [CrossRef]
- Mabb, A.M.; Wuerzberger-Davis, S.M.; Miyamoto, S. PIASy mediates NEMO sumoylation and NF-κB activation in response to genotoxic stress. Nat. Cell Biol. 2006, 8, 986–993. [Google Scholar] [CrossRef]
- Xiong, R.; Nie, L.; Xiang, L.; Shao, J. Characterization of a PIAS4 homologue from zebrafish: Insights into its conserved negative regulatory mechanism in the TRIF, MAVS, and IFN signaling pathways during vertebrate evolution. J. Immunol. 2012, 188, 2653–2668. [Google Scholar] [CrossRef]
- Boudinot, P.; Bird, S.; Pasquier, L.D.; Collet, B. The repertoire of vertebrate STAT transcription factors: Origin and variations in fish. Dev. Comp. Immunol. 2021, 116, 103929. [Google Scholar] [CrossRef]
- Levraud, J.-P.; Jouneau, L.; Briolat, V.; Laghi, V.; Boudinot, P. IFN-stimulated genes in zebrafish and humans define an ancient arsenal of antiviral immunity. J. Immunol. 2019, 203, 3361–3373. [Google Scholar] [CrossRef]
- Dehler, C.E.; Lester, K.; della Pelle, G.; Jouneau, L.; Houel, A.; Collins, C.; Dovgan, T.; Machat, R.; Zou, J.; Boudinot, P.; et al. Viral resistance and IFN signaling in STAT2 knockout fish cells. J. Immunol. 2019, 203, 465–475. [Google Scholar] [CrossRef]
- Valdez, B.C.; Henning, D.; Perlaky, L.; Busch, R.K.; Busch, H. Cloning and characterization of Gu/RH-II binding protein. Biochem. Biophys. Res. Commun. 1997, 234, 335–340. [Google Scholar] [CrossRef]
- Dehler, C.E.; Boudinot, P.; Martin, S.A.M.; Collet, B. Development of an efficient genome editing method by CRISPR/Cas9 in a fish cell line. Mar. Biotechnol. 2016, 18, 449–452. [Google Scholar] [CrossRef]
- Rytinki, M.M.; Kaikkonen, S.; Pehkonen, P.; Jääskeläinen, T.; Palvimo, J.J. PIAS proteins: Pleiotropic interactors associated with SUMO. Cell. Mol. Life Sci. 2009, 66, 3029–3041. [Google Scholar] [CrossRef]
- Collet, B.; Secombes, C.J. The rainbow trout (Oncorhynchus mykiss) Mx1 promoter: Structural and functional characterization. Eur. J. Biochem. 2001, 268, 1577–1584. [Google Scholar] [CrossRef]
- Johansen, A.; Collet, B.; Sandaker, E.; Secombes, C.J.; Jørgensen, J.B. Quantification of Atlantic salmon type-I interferon using an Mx1 promoter reporter gene assay. Fish Shellfish. Immunol. 2004, 16, 173–184. [Google Scholar] [CrossRef]
- Schindler, U.; Baichwal, V.R. Three NF-kappa B binding sites in the human E-selectin gene required for maximal tumor necrosis factor alpha-induced expression. Mol. Cell. Biol. 1994, 14, 5820–5831. [Google Scholar] [PubMed]
- Glasauer, S.M.K.; Neuhauss, S.C.F. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef]
- Macqueen, D.J.; Johnston, I.A. A well-constrained estimate for the timing of the salmonid whole genome duplication reveals major decoupling from species diversification. Proc. R. Soc. B Biol. Sci. 2014, 281, 20132881. [Google Scholar] [CrossRef]
- Rebl, A.; Rebl, H.; Verleih, M.; Haupt, S.; Köbis, J.M.; Goldammer, T.; Seyfert, H.M. At least two genes encode many variants of Irak3 in rainbow trout, but neither the full-length factor nor its variants interfere directly with the TLR-mediated stimulation of inflammation. Front. Immunol. 2019, 10, 2246. [Google Scholar] [CrossRef]
- Sarais, F.; Rebl, H.; Verleih, M.; Ostermann, S.; Krasnov, A.; Köllner, B.; Goldammer, T.; Rebl, A. Characterisation of the teleostean κB-Ras family: The two members NKIRAS1 and NKIRAS2 from rainbow trout influence the activity of NF-κB in opposite ways. Fish Shellfish. Immunol. 2020, 106, 1004–1013. [Google Scholar] [CrossRef]
- Munarriz, E.; Barcaroli, D.; Stephanou, A.; Townsend, P.A.; Maisse, C.; Terrinoni, A.; Neale, M.H.; Martin, S.J.; Latchman, D.S.; Knight, R.A.; et al. PIAS-1 is a checkpoint regulator which affects exit from G1 and G2 by sumoylation of p73. Mol. Cell. Biol. 2004, 24, 10593–10610. [Google Scholar] [CrossRef][Green Version]
- Heery, D.M.; Kalkhoven, E.; Hoare, S.; Parker, M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 1997, 387, 733–736. [Google Scholar] [CrossRef]
- Liu, B.; Yee, K.M.; Tahk, S.; Mackie, R.; Hsu, C.; Shuai, K. PIAS1 SUMO ligase regulates the self-renewal and differentiation of hematopoietic stem cells. EMBO J. 2014, 33, 101–113. [Google Scholar] [CrossRef]
- Prelich, G. Gene overexpression: Uses, mechanisms, and interpretation. Genetics 2012, 190, 841–854. [Google Scholar] [CrossRef]
- Rebl, A.; Rebl, H.; Köbis, J.M.; Goldammer, T.; Seyfert, H.-M. ST2 from rainbow trout quenches TLR signalling, localises at the nuclear membrane and allows the nuclear translocation of MYD88. Dev. Comp. Immunol. 2017, 67, 139–152. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Haller, O.; Arnheiter, H.; Pavlovic, J.; Staeheli, P. The discovery of the antiviral resistance gene Mx: A story of great ideas, great failures, and some success. Annu. Rev. Virol. 2018, 5, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Hirano, T.; Matsuda, T.; Nakajima, K. Signal transduction through gp130 that is shared among the receptors for the interleukin 6 related cytokine subfamily. Stem Cells 1994, 12, 262–277. [Google Scholar] [CrossRef]
- Kotaja, N.; Aittomäki, S.; Silvennoinen, O.; Palvimo, J.J.; Jänne, O.A. ARIP3 (androgen receptor-interacting protein 3) and other PIAS (protein inhibitor of activated STAT) proteins differ in their ability to modulate steroid receptor-dependent transcriptional activation. Mol. Endocrinol. 2000, 14, 1986–2000. [Google Scholar] [CrossRef]
- Bautista-Hernández, L.A.; Gómez-Olivares, J.L.; Buentello-Volante, B.; Bautista-de Lucio, V.M. Fibroblasts: The unknown sentinels eliciting immune responses against microorganisms. Eur. J. Microbiol. Immunol. 2017, 7, 151–157. [Google Scholar] [CrossRef]
- Estévez, R.A.; Mostazo, M.G.C.; Rodriguez, E.; Espinoza, J.C.; Kuznar, J.; Jónsson, Z.O.; Guðmundsson, G.H.; Maier, V.H. Inducers of salmon innate immunity: An in vitro and in vivo approach. Fish Shellfish. Immunol. 2018, 72, 247–258. [Google Scholar] [CrossRef]
- Köllner, B.; Blohm, U.; Kotterba, G.; Fischer, U. A monoclonal antibody recognising a surface marker on rainbow trout (Oncorhynchus mykiss) monocytes. Fish Shellfish Immunol. 2001, 11, 127–142. [Google Scholar] [CrossRef]
- Korytář, T.; Thi, H.D.; Takizawa, F.; Köllner, B. A multicolour flow cytometry identifying defined leukocyte subsets ofrainbow trout (Oncorhynchus mykiss). Fish Shellfish Immunol. 2013, 35, 2017–2019. [Google Scholar] [CrossRef]
- Bowers, R.M.; Lapatra, S.E.; Dhar, A.K. Detection and quantitation of infectious pancreatic necrosis virus by real-time reverse transcriptase-polymerase chain reaction using lethal and non-lethal tissue sampling. J. Virol. Methods. 2008, 147, 226–234. [Google Scholar] [CrossRef]
- Köbis, J.M.; Rebl, H.; Goldammer, T.; Rebl, A. Multiple gene and transcript variants encoding trout C-polysaccharide binding proteins are differentially but strongly induced after infection with Aeromonas salmonicida. Fish Shellfish Immunol. 2017, 60, 509–519. [Google Scholar] [CrossRef]
- Yang, W.; Zerbe, H.; Petzl, W.; Brunner, R.M.; Günther, J.; Draing, C.; von Aulock, S.; Schuberth, H.J.; Seyfert, H.M. Bovine TLR2 and TLR4 properly transduce signals from Staphylococcus aureus and E. coli, but S. aureus fails to both activate NF-kappaB in mammary epithelial cells and to quickly induce TNFalpha and interleukin-8 (CXCL8) expression in the udder. Mol. Immunol. 2008, 45, 1385–1397. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.; Ganne, G.; Collet, B. Isolation and activity of the promoters for STAT1 and 2 in Atlantic salmon Salmo salar. Fish Shellfish Immunol. 2014, 40, 644–647. [Google Scholar] [CrossRef] [PubMed]
- Huerta-Cepas, J.; Serra, F.; Bork, P. ETE 3: Reconstruction, analysis, and visualization of phylogenomic data. Mol. Biol. Evol. 2016, 33, 1635–1638. [Google Scholar] [CrossRef] [PubMed]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. Fasttree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol. Biol. Evol. 2009, 26, 1641–1650. [Google Scholar] [CrossRef]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developerS. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Ishida, T.; Kinoshita, K. PrDOS: Prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007, 35, W460–W464. [Google Scholar] [CrossRef]
| Nucleotide NCBI Accession Number | Chromosome | Gene | Transcript Isoform | CDS Length [nt] | UTR Length [bp] 5′ 3′ | Instability Motifs | Protein Length [aa] | Protein NCBI Accession Number | |
|---|---|---|---|---|---|---|---|---|---|
| Pias1 | |||||||||
| XM_036963708 | 26 (6) * | LOC110527003 | pias1a1.1 (X1) | 2130 | 41 | 1113 | 1 (3′ UTR) | 709 | XP_036819603 |
| XM_036963709 | 26 (6) | LOC110527003 | pias1a1.2 (X2) | 2112 | 41 | 1113 | 1 (3′ UTR) | 703 | XP_036819604 |
| XM_036961353 | 2 | pias1b | pias1b | 1968 | 78 | 3326 | 20 (3′ UTR) | 655 | XP_036817248 |
| XM_036969029 | 30 (4) | LOC110521158 | pias1a2 | 1929 | 140 | 1718 | 6 (3′ UTR) | 642 | XP_036824924 |
| Pias2 | |||||||||
| XM_036936540 | 11 | pias2 | pias2a2.1 (X1) | 1965 | 289 | 3152 | 14 (3′ UTR) | 654 | XP_036792435 |
| XM_036936542 | 11 | pias2 | pias2a2.2 (X2) | 1950 | 286 | 3134 | 14 (3′ UTR) | 649 | XP_036792437 |
| XM_036936543 | 11 | pias2 | pias2a2.3 (X3) | 1920 | 411 | 3134 | 14 (3′ UTR) | 639 | XP_036792438 |
| XM_036979193 | 6 | LOC110525143 | pias2a1.1 (X1) | 1527 | 572 | 2502 | 2 (5′ UTR), 3 (3′ UTR) | 508 | XP_036835088 |
| XM_036979194 | 6 | LOC110525143 | pias2a1.2 (X2) | 1512 | 572 | 2502 | 2 (5′ UTR), 3 (3′ UTR) | 503 | XP_036835089 |
| Pias4 | |||||||||
| XM_021598984 | 4 | LOC110521438 | pias4a1.1 (X1) | 1485 | 125 | 2933 | 24 (3′ UTR) | 494 | XP_021454659 |
| XM_021598985 | 4 | LOC110521438 | pias4a1.2 (X2) | 1455 | 60 | 2931 | 24 (3′ UTR) | 484 | XP_021454660 |
| XM_021603817 | 5 | LOC11052429 | pias4a2 | 1497 | 132 | 3617 | 26 (3′ UTR) | 498 | XP_021459492 |
| XM_021613940 | 8 | pias4b | pias4b.1 (X1) | 1491 | 410 | 696 | 1 (5′ UTR), 5 (3′ UTR) | 496 | XP_021469615 |
| XM_036986356 | 8 | pias4b | pias4b.2 (X2) | 1422 | 498 | 696 | 1 (5′ UTR), 5 (3′ UTR) | 473 | XP_036842251 |
| Gene Symbol | Primer Sequence 5′→3′ (Sense, Antisense) | Nucleotide NCBI Accession Number Used for Primer Design | Fragment Length [bp] | |
|---|---|---|---|---|
| O. mykiss | O. tshawytscha | |||
| Quantitative PCR Analysis (Oncorhynchus mykiss, O. tshawytscha) | ||||
| Pias1a1.1 | GTTGGAAGGCACCTTCTGTGTT, CTACGGTCCAAAGGCATCAGG | XM_036963708 | XM_031812568 | 108 |
| Pias1b | GGAGCTACTCTATGGCGGTGT, ATCAGGAACCCAGACCATTCCA | XM_036961353 | XM_042317872 | 99 |
| Pias1a2 | TAGGCAGGAATTTCTCCATGGC, AGAGAAGTTAACAGCTGACCCG | XM_036969029 | XM_042310862 | 140 |
| Pias2a1.1 | GTGTGCATCTCCAGGGACTTTT, CTAAGAATGGAGTGGAACAGAAG | XM_036979193 | XM_024414775 | 195 |
| Pias2a2.1 | GAGCTACGGAGCATGGTGTCA, AACTTTATCGACGCCGCTATCC | XM_036936540 | XM_042331195 | 185 |
| Pias4a1.1 | ATTGGAAGCAGAGAACCGTCGA, ATTTTCGGGTGTCTGACCTGCA | XM_021598984 | XM_024398270 | 158 |
| Pias4a1.2 | GCCTGCTAGGCTGGGAAACTA, CGCAGTAAAAGTGGTCTGAAGC | XM_021598985 | ― | 99 |
| Pias4a2 | AGGAGGAGGGGGGAGGAGG, CGGACTGACCCCACAAACTGA | XM_021603817 | XM_024421382 | 144 |
| Pias4b.1 | ACATAGCAGAAGCAATTAGGTTGT, AATCTGCTGGTGAGGGCAGTG | XM_021613940 | XM_024421383 | 146 |
| Pias4b.2 | ACAAAGGCCCCGGAGTGAACA, GGGAGGGGAGTCAAGCTACAT | XM_021613941 | ― | 129 |
| Stat1a1 (stat1-1) | GAGAGCATCGACTGGGAAAATGT, AAACAACTTCCTGCTACAACACAA | NM_001124707 | XM_024426102 | 131 |
| Stat1a2 (stat1-2) | CCCCGTTCACATGGCCATGAT, CATAGAGACCGACAGAGAAAACA | XM_021608237 | XM_042324083 | 95 |
| Stat1b1 (stat1ab) | GGCCATGATAATCTGTAACTGTC, ACGTTAAAGACCTGAGGAACCG | XM_021579196 | XM_042306329 | 150 |
| Cxcl8 | ATATAACACTTGTTACCAGCGAGA, ATTACTGAGGAGATGAGTCTGAG | HG917307 | XM_024415648 | 106 |
| Il6 | GTGTTAGTTAAGGGGAATCCAGT, CCTTGCGGAACCAACAGTTTGT | NM_001124657 | XM_024404411 | 128 |
| Il10 | TGCCCAGTGCAGACGTGTACC, TACACCACTTGAAGAGCCCCG | NM_001245099 | XM_042324963 | 137 |
| Tgfb | ATCAGGGATGAACAAGCTGAGG, CGGAGAGTTGCTGTGTGCGAA | XM_021591332 | XM_024397891 | 161 |
| Ifna1 | TTGAAGAGAGCAAATGTATGATGG, TCCTGTACAGCCTACAGTTCATT | XM_024434105 (representative for LOC110538045, LOC110538046, LOC110538047,LOC110538053, LOC110538937, LOC118937709) | XM_024434105 (representative for LOC112259401, LOC112259404, LOC121847201, LOC121847202, LOC112258510) | 173 |
| Ifna3 | CCAACATCACTTTACAGACACATA, GGGACAAGAAAAACCTGGACGA | XM_024432928 (representative for LOC110511235, LOC110517168, LOC110538043, LOC110538058) | XM_024389910 (representative for LOC121838839, LOC112225816, LOC112258507, LOC112258508) | 140 |
| Stat4b | ACCTCATCAAAAGCTCCTTTGTG, TTCACCACCAAAGTCAGATTGCT | XM_024388828 | XM_024388828 | 112 |
| Mx | GTAGCGGTATTGTAACACGATGC, TCGTGAAGCCCAGGATGAAATG | XM_036958922 | XM_024415949 | 158 |
| Gata3 | CCACCTCCTCCACATAGTAGTC, GACCTGCCGGGGAACCGTG | XM_036957437 | XM_042311479 | 160 |
| Mmp9 | TGCCAAGATAGAGGCTACAGTC, TGTCTTGGACCCATAGAGATAGT | XM_036986917 | XM_024376362 | 181 |
| Socs1 | ACGGATTCTGCGTCGGAAAATAT, ACACAGTTCCCTGGCATCCGT | XM_036973400 | XM_042313671 | 91 |
| Tp53 | GAATTTGAACCTGGTGGCAGTTC, CACCTCAAACAGACTCGGATCA | NM_001124692 | XM_024394883 | 115 |
| Construction of PIAS-expression constructs | ||||
| Pias1a1 | ATGCAAGCTTATGGCGGAGAGTGCGGAACT, CATACCAGACGTGATCTCGTTAGACGAATTCGCAT | XM_036963708 | 1968 | |
| Pias2a1 | ATGCAAGCTTATGATCCTGACAAGAAAAATGGCGG, ACATCATCTCAGACATCATCTCATTGGACGAATTCGCAT | XM_036936540 | 1965 | |
| SgRNA target sequence | ||||
| Pias1a2 | AGTAACACTTGTAGCTCTGA GCGTAGCCTAGTAACACTTG TGTTTGTTGCGTCCTGCGTA | XM_024407180 | 20 | |
| Pias1b | TCTACAATAACACAAAAAGA ACACGACTCTGCAAGAGGGT TCTGTCAATCCATCTACAAT | XM_024410569 | 20 | |
| Pias2a1.1 | ACACGTCGTAGAACGGGAGA GAGGGATGAGAGGGGCGGGC CCAGCAGCCCGCCCCTCTCA | XM_024414775 | 20 | |
| Pias2a2 | CCATTTTTCTTGTCAGGATC GCGAAGCCCAGTAACACTTG TAGCTCCTCAAATTCCGCCA | XM_024407486 | 20 | |
| mEGFP | GGCGAGGGCGATGCCACCTA | [39] | 20 | |
| Sequencing primers for GFP(-) cells | ||||
| Pias1a2 | TTAGTTGTTCTATTCGTGTGTCCTA, TTACACACACTGTGTGTACAAAACA | XM_024407180 | 500 | |
| Pias1b | GACCCCACTGCCTTTGTTTCAAACC, CATTCCTCCAAGGAGACAACCACCAG | XM_024410569 | 500 | |
| Pias2a2 | AGTCTAAGCTTGACATCCATGAAAG, GTGTAGGCATTGGCTTAGCAATGC | XM_024407486 | 360 | |
| Pias2a1.1 | CCCAAGGCGGTAGACAGTAGTCT, ACTGGGCTTTATGTTTCTGGTGACG | XM_024414775 | 500 | |
| mEGFP | ATGGTGAGCAAGGGCGAGCTG, GTCCTCCTTGAAGTCGATGCCCT | [39] | 500 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sarais, F.; Kummerow, S.; Montero, R.; Rebl, H.; Köllner, B.; Goldammer, T.; Collet, B.; Rebl, A. PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression. Int. J. Mol. Sci. 2021, 22, 12815. https://doi.org/10.3390/ijms222312815
Sarais F, Kummerow S, Montero R, Rebl H, Köllner B, Goldammer T, Collet B, Rebl A. PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression. International Journal of Molecular Sciences. 2021; 22(23):12815. https://doi.org/10.3390/ijms222312815
Chicago/Turabian StyleSarais, Fabio, Sophia Kummerow, Ruth Montero, Henrike Rebl, Bernd Köllner, Tom Goldammer, Bertrand Collet, and Alexander Rebl. 2021. "PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression" International Journal of Molecular Sciences 22, no. 23: 12815. https://doi.org/10.3390/ijms222312815
APA StyleSarais, F., Kummerow, S., Montero, R., Rebl, H., Köllner, B., Goldammer, T., Collet, B., & Rebl, A. (2021). PIAS Factors from Rainbow Trout Control NF-κB- and STAT-Dependent Gene Expression. International Journal of Molecular Sciences, 22(23), 12815. https://doi.org/10.3390/ijms222312815







