Small Molecule Inhibitors Targeting Nuclear Factor κB Activation Markedly Reduce Expression of Interleukin-2, but Not Interferon-γ, Induced by Phorbol Esters and Calcium Ionophores
Abstract
:1. Introduction
2. Results
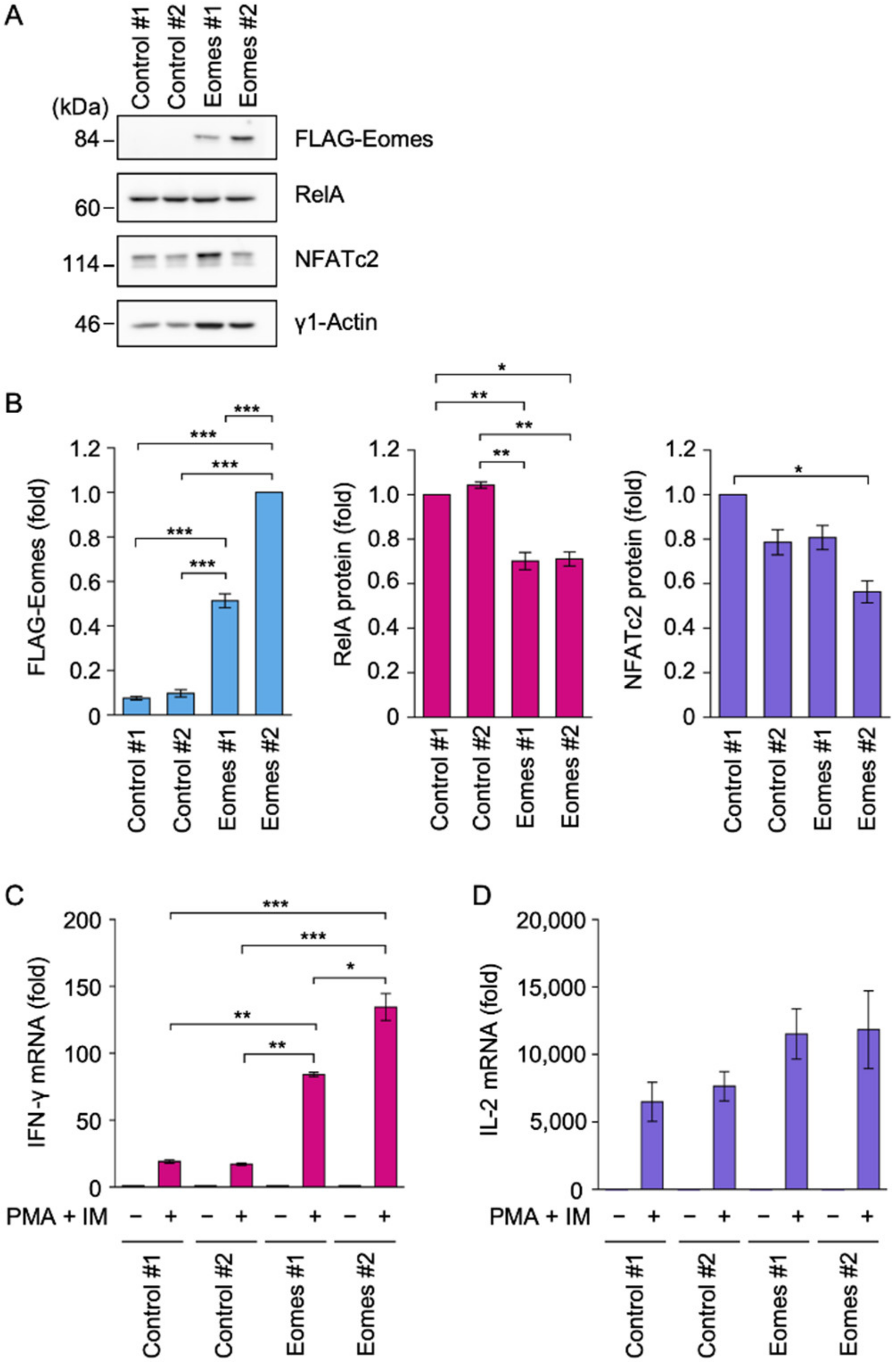
2.1. Eomes Promoted IFN-γ mRNA Expression in EL4 Cells
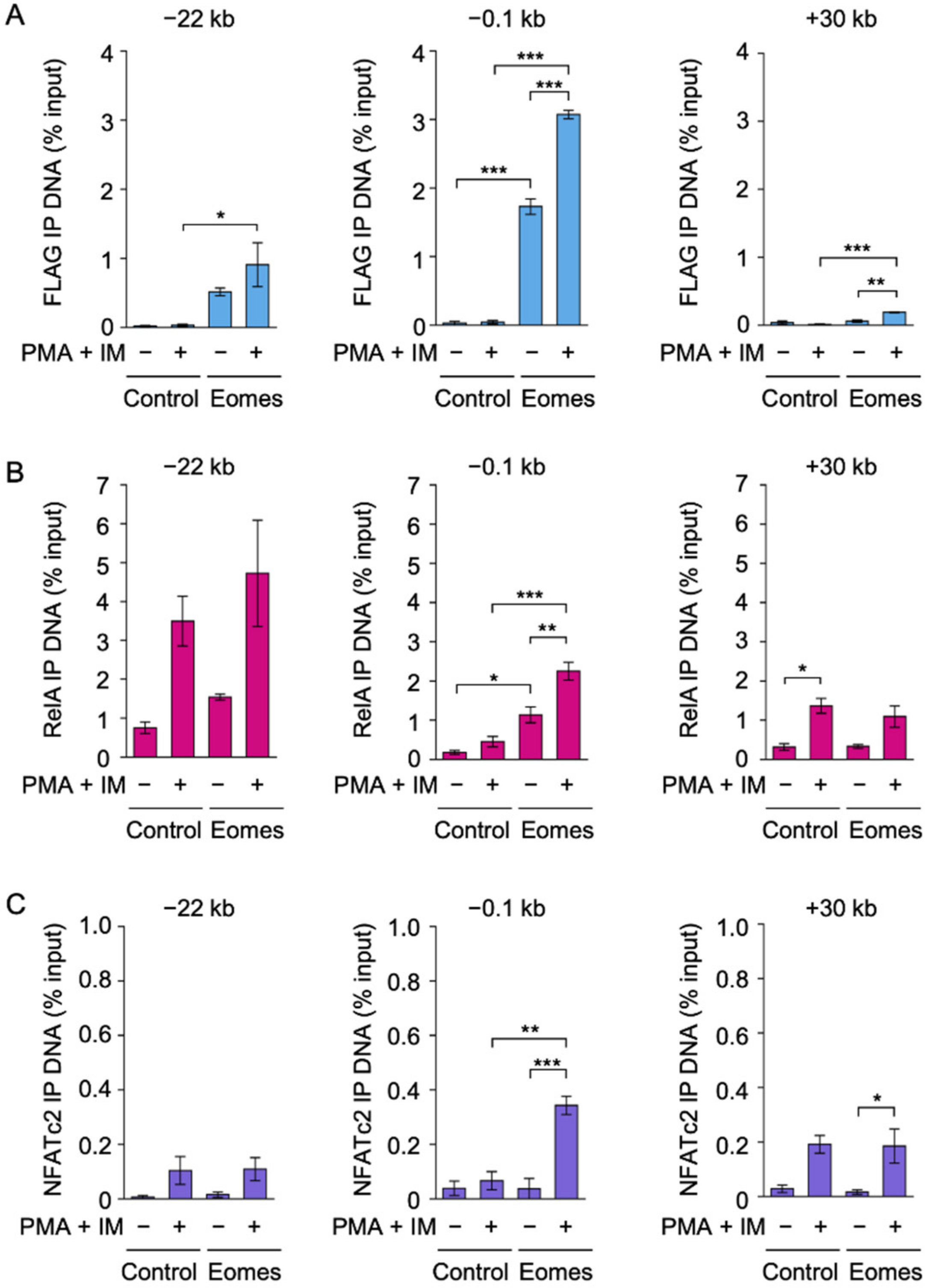
2.2. Eomes Augmented the Binding of RelA and NFATc2 to the IFN-γ Promoter in EL4 Cells
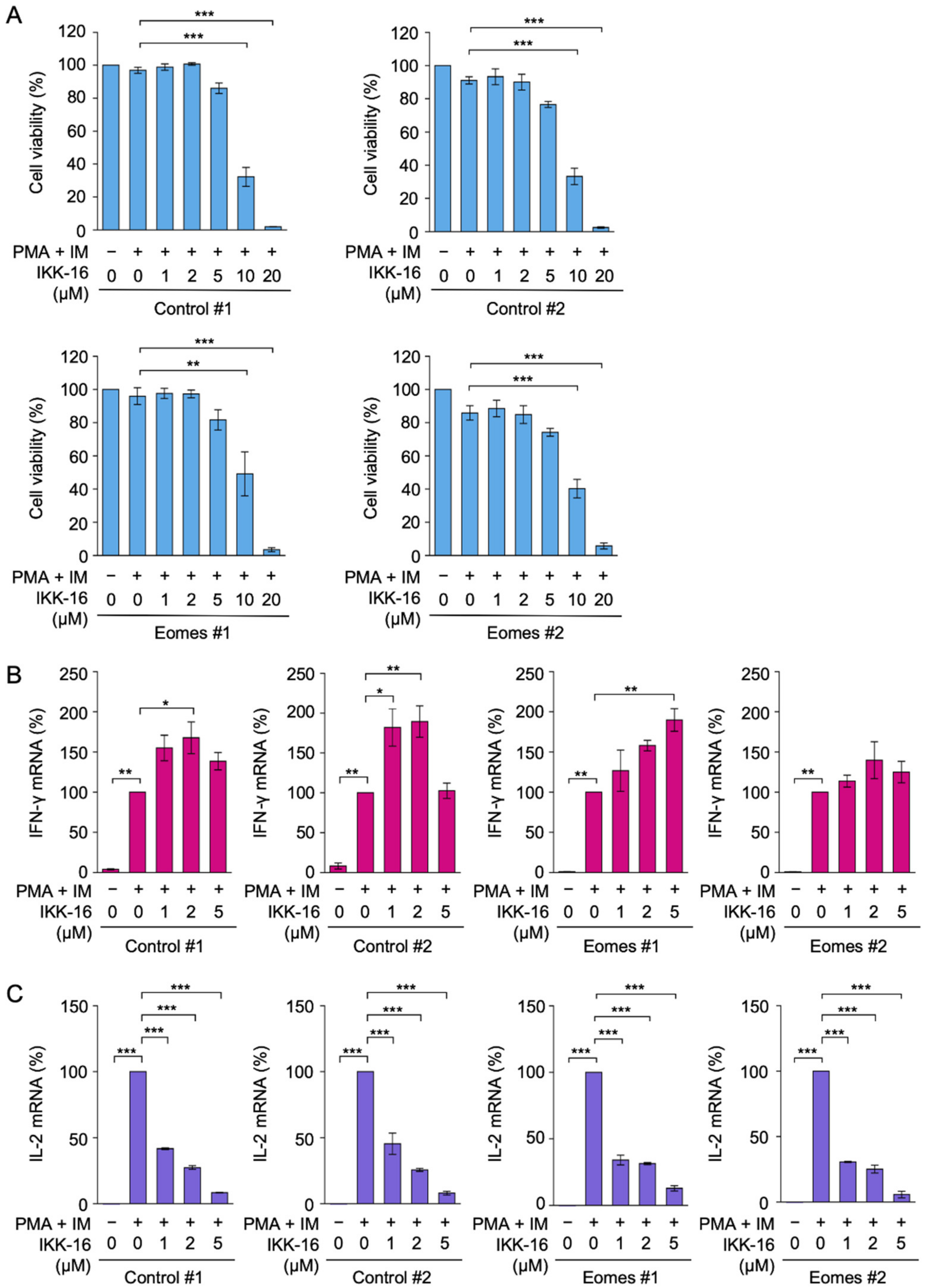
2.3. TPCA-1 Inhibited PMA- and IM-Induced IL-2 mRNA Expression, but Not IFN-γ mRNA Expression in EL4 Cells
2.4. IKK-16 Inhibited PMA- and IM-Induced IL-2 mRNA Expression, but Not IFN-γ mRNA Expression in EL4 Cells
2.5. TPCA-1 Inhibited the Binding of RelA, but Not NFATc2 or Eomes, to the IFN-γ Promoter in EL4 Cells
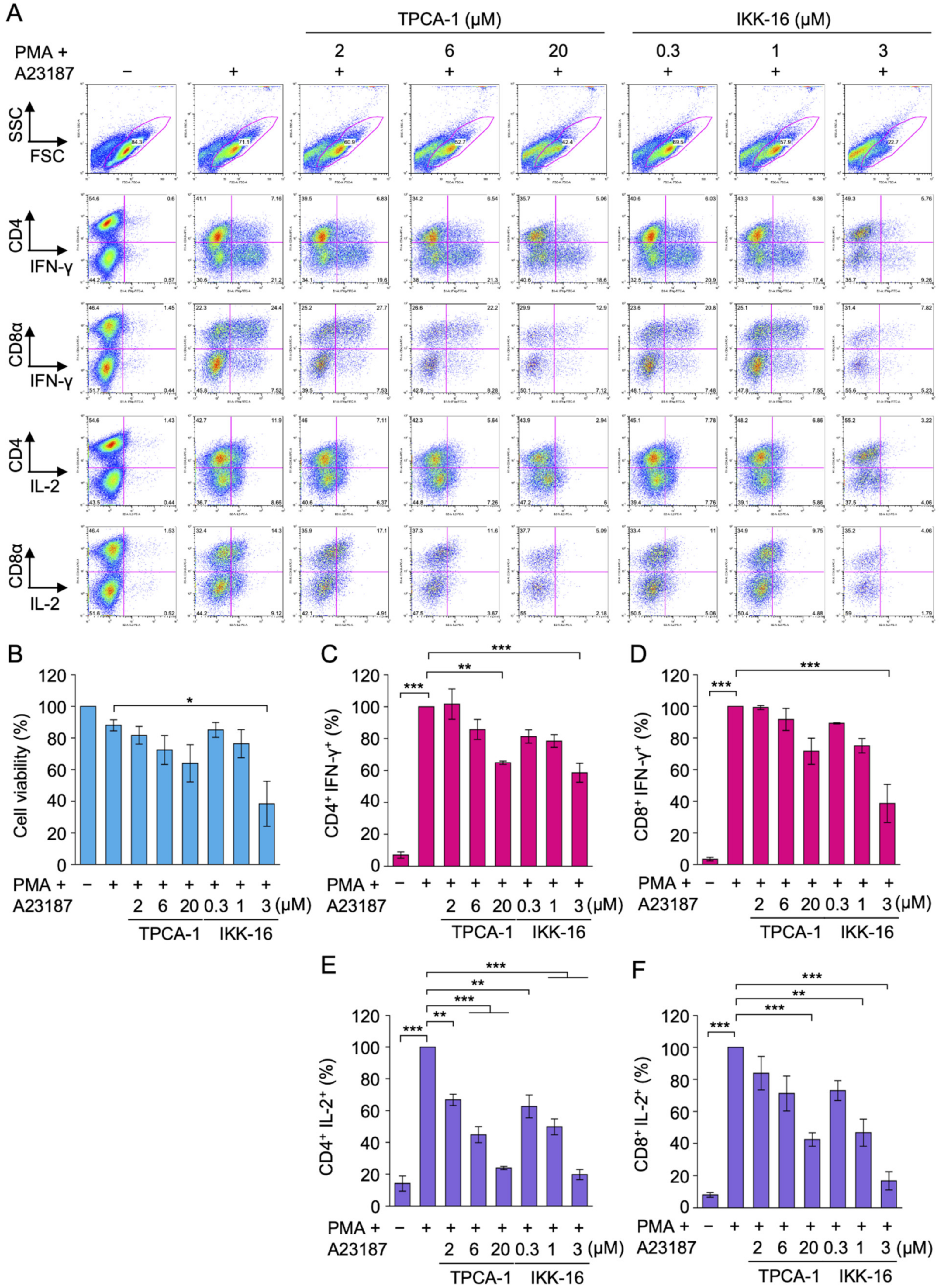
2.6. TPCA-1 and IKK-16 Inhibited IL-2 Expression, but Not IFN-γ Expression in Primary Effector CD4+ and CD8+ T Cells
3. Discussion
4. Materials and Methods
4.1. Mice
4.2. Cells
4.3. Reagents
4.4. Antibodies
4.5. Transfection
4.6. Western Blotting
4.7. Quantitative PCR
4.8. ChIP Assay
4.9. Cell Viability Assay
4.10. Purification and Culture of Primary T Cells
4.11. Intracellular Staining
4.12. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rožman, P.; Švajger, U. The tolerogenic role of IFN-γ. Cytokine Growth Factor Rev. 2018, 41, 40–53. [Google Scholar] [CrossRef]
- Burke, J.D.; Young, H.A. IFN-γ: A cytokine at the right time, is in the right place. Semin. Immunol. 2019, 43, 101280. [Google Scholar] [CrossRef]
- Mojic, M.; Takeda, K.; Hayakawa, Y. The dark side of IFN-γ: Its role in promoting cancer immunoevasion. Int. J. Mol. Sci. 2018, 19, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kursunel, M.A.; Esendagli, G. The untold story of IFN-γ in cancer biology. Cytokine Growth Factor Rev. 2016, 31, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Badovinac, V.P.; Harty, J.T. Programming, demarcating, and manipulating CD8+ T-cell memory. Immunol. Rev. 2006, 211, 67–80. [Google Scholar] [CrossRef]
- Hadjur, S.; Williams, L.M.; Ryan, N.K.; Cobb, B.S.; Sexton, T.; Fraser, P.; Fisher, A.G.; Merkenschlager, M. Cohesions form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 2009, 460, 410–413. [Google Scholar] [CrossRef] [PubMed]
- Sekimata, M.; Pérez-Melgosa, M.; Miller, S.A.; Weinmann, A.S.; Sabo, P.J.; Sandstrom, R.; Dorschner, M.O.; Stamatoyannopoulos, J.A.; Wilson, C.B. CCCTC-binding factor and the transcription factor T-bet orchestrate T helper 1 cell-specific structure and function at the interferon-γ locus. Immunity 2009, 31, 551–564. [Google Scholar] [CrossRef] [Green Version]
- Wilson, C.B.; Rowell, E.; Sekimata, M. Epigenetic control of T-helper-cell differentiation. Nat. Rev. Immunol. 2009, 9, 91–105. [Google Scholar] [CrossRef] [PubMed]
- Balasubramani, A.; Mukasa, R.; Hatton, R.D.; Weaver, C.T. Regulation of the Ifng locus in the context of T-lineage specification and plasticity. Immunol. Rev. 2010, 238, 216–232. [Google Scholar] [CrossRef]
- Aune, T.M.; Collins, P.L.; Collier, S.P.; Henderson, M.A.; Chang, S. Epigenetic activation and silencing of the gene that encodes IFN-γ. Front. Immunol. 2013, 4, 112. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Marotel, M.; Fauteux-Daniel, S.; Mathieu, A.L.; Viel, S.; Marçais, A.; Walzer, T. T-bet and Eomes govern differentiation and function of mouse and human NK cells and ILC1. Eur. J. Immunol. 2018, 48, 738–750. [Google Scholar] [CrossRef] [Green Version]
- Szabo, S.J.; Kim, S.T.; Costa, G.L.; Zhang, X.; Fathman, C.G.; Glimcher, L.H. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell 2000, 100, 655–669. [Google Scholar] [CrossRef] [Green Version]
- Pearce, E.L.; Mullen, A.C.; Martins, G.A.; Krawczyk, C.M.; Hutchins, A.S.; Zediak, V.P.; Banica, M.; DiCioccio, C.B.; Gross, D.A.; Mao, C.A.; et al. Control of effector CD8+ T cell function by the transcription factor Eomesodermin. Science 2003, 302, 1041–1043. [Google Scholar] [CrossRef]
- Pritchard, G.H.; Kedl, R.M.; Hunter, C.A. The evolving role of T-bet in resistance to infection. Nat. Rev. Immunol. 2019, 19, 398–410. [Google Scholar] [CrossRef]
- Takemoto, N.; Intlekofer, A.M.; Northrup, J.T.; Wherry, E.J.; Reiner, S.L. IL-12 inversely regulates T-bet and eomesodermin expression during pathogen-induced CD8+ T cell differentiation. J. Immunol. 2006, 177, 7515–7519. [Google Scholar] [CrossRef] [Green Version]
- Joshi, N.S.; Cui, W.; Chandele, A.; Lee, H.K.; Urso, D.R.; Hagman, J.; Gapin, L.; Kaech, S.M. Inflammation directs memory precursor and short-lived effector CD8+ T cell fates via the graded expression of T-bet transcription factor. Immunity 2007, 27, 281–295. [Google Scholar] [CrossRef] [Green Version]
- Cruz-Guilloty, F.; Pipkin, M.E.; Djuretic, I.M.; Levanon, D.; Lotem, J.; Lichtenheld, M.G.; Groner, Y.; Rao, A. Runx3 and T-box proteins cooperate to establish the transcriptional program of effector CTLs. J. Exp. Med. 2009, 206, 51–59. [Google Scholar] [CrossRef]
- Pipkin, M.E.; Sacks, J.A.; Cruz-Guilloty, F.; Lichtenheld, M.G.; Bevan, M.J.; Rao, A. Interleukin-2 and inflammation induce distinct transcriptional programs that promote the differentiation of effector cytolytic T cells. Immunity 2010, 32, 79–90. [Google Scholar] [CrossRef] [Green Version]
- Hatton, R.D.; Harrington, L.E.; Luther, R.J.; Wakefield, T.; Janowski, K.M.; Oliver, J.R.; Lallone, R.L.; Murphy, K.M.; Weaver, C.T. A distal conserved sequence element controls Ifng gene expression by T cells and NK cells. Immunity 2006, 25, 717–729. [Google Scholar] [CrossRef] [Green Version]
- Yagi, R.; Junttila, I.S.; Wei, G.; Urban, J.F.; Zhao, K.; Paul, W.E.; Zhu, J. The transcription factor GATA3 actively represses RUNX3 protein-regulated production of interferon-γ. Immunity 2010, 32, 507–517. [Google Scholar] [CrossRef] [Green Version]
- Schoenborn, J.R.; Dorschner, M.; Sekimata, M.; Santer, D.; Shnyreva, M.; Fitzpatrick, D.R.; Stamatoyannopoulos, J.A.; Wilson, C.B. Comprehensive epigenetic profiling identifies multiple distal regulatory elements directing Ifng transcription. Nat. Immunol. 2007, 8, 732–742. [Google Scholar] [CrossRef] [Green Version]
- Mukasa, R.; Balasubramani, A.; Lee, Y.K.; Whitley, S.K.; Weaver, B.T.; Shibata, Y.; Crawford, G.E.; Hatton, R.D.; Weaver, C.T. Epigenetic instability of cytokine and transcription factor gene loci underlies plasticity of the T helper 17 cell lineage. Immunity 2010, 32, 616–627. [Google Scholar] [CrossRef] [Green Version]
- Balasubramani, A.; Winstead, C.J.; Turner, H.; Janowski, K.M.; Harbour, S.N.; Shibata, Y.; Crawford, G.E.; Hatton, R.D.; Weaver, C.T. Deletion of a conserved cis-element in the Ifng locus highlights the role of acute histone acetylation in modulating inducible gene transcription. PLoS Genet. 2014, 10, e1003969. [Google Scholar] [CrossRef]
- Gruarin, P.; Maglie, S.; De Simone, M.; Häringer, B.; Vasco, C.; Ranzani, V.; Bosotti, R.; Noddings, J.S.; Larghi, P.; Facciotti, F.; et al. Eomesodermin controls a unique differentiation program in human IL-10 and IFN-γ coproducing regulatory T cells. Eur. J. Immunol. 2019, 49, 96–111. [Google Scholar] [CrossRef] [Green Version]
- Fukuoka, N.; Harada, M.; Nishida, A.; Ito, Y.; Shiota, H.; Kataoka, T. Eomesodermin promotes interferon-γ expression and binds to multiple conserved noncoding sequences across the Ifng locus in mouse thymoma cell lines. Genes Cells 2016, 21, 146–162. [Google Scholar] [CrossRef]
- Brownlie, R.J.; Zamoyska, R. T cell receptor signalling networks: Branched, diversified and bounded. Nat. Rev. Immunol. 2013, 13, 257–269. [Google Scholar] [CrossRef]
- Fric, J.; Zelante, T.; Wong, A.Y.W.; Mertes, A.; Yu, H.B.; Ricciardi-Castagnoli, P. NFAT control of innate immunity. Blood 2012, 120, 1380–1389. [Google Scholar] [CrossRef]
- Kiani, A.; García-Cózar, F.J.; Habermann, I.; Laforsch, S.; Aebischer, T.; Ehninger, G.; Rao, A. Regulation of interferon-γ gene expression by nuclear factor of activated T cells. Blood 2001, 98, 1480–1488. [Google Scholar] [CrossRef] [Green Version]
- Peng, S.L.; Gerth, A.J.; Ranger, A.M.; Glimcher, L.H. NFATc1 and NFATc2 together control both T and B cell activation and differentiation. Immunity 2001, 14, 13–20. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, L.K.; Fonseca, B.P.F.; Vieira-de-Abreu, A.; Barboza, B.A.; Robbs, B.K.; Bozza, P.T.; Viola, J.P.B. IFN-γ production by CD8+ T cells depends on NFAT1 transcription factor and regulates Th differentiation. J. Immunol. 2005, 175, 5931–5939. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.U.; Avni, O.; Chen, L.; Rao, A. A distal enhancer in the interferon-γ (IFN-γ) locus revealed by genome sequence comparison. J. Biol. Chem. 2004, 279, 4802–4810. [Google Scholar] [CrossRef] [Green Version]
- Hayden, M.S.; Ghosh, S. Regulation of NF-κB by TNF family cytokines. Semin. Immunol. 2014, 26, 253–266. [Google Scholar] [CrossRef] [Green Version]
- Balasubramani, A.; Shibata, Y.; Crawford, G.E.; Baldwin, A.S.; Hatton, R.D.; Weaver, C.T. Modular utilization of distal cis-regulatory elements controls Ifng gene expression in T cells activated by distinct stimuli. Immunity 2010, 33, 35–47. [Google Scholar] [CrossRef] [Green Version]
- Corn, R.A.; Aronica, M.A.; Zhang, F.; Tong, Y.; Stanley, S.A.; Kim, S.R.A.; Stephenson, L.; Enerson, B.; McCarthy, S.; Mora, A.; et al. T cell-intrinsic requirement for NF-κB induction in postdifferentiation IFN-γ production and clonal expansion in a Th1 response. J. Immunol. 2003, 171, 1816–1824. [Google Scholar] [CrossRef] [Green Version]
- Waelchli, R.; Bollbuck, B.; Bruns, C.; Buhl, T.; Eder, J.; Feifel, R.; Hersperger, R.; Janser, P.; Revesz, L.; Zerwes, H.G.; et al. Design and preparation of 2-benzamido-pyrimidines as inhibitors of IKK. Bioorg. Med. Chem. Lett. 2006, 16, 108–112. [Google Scholar] [CrossRef]
- Shashkova, E.V.; Trivedi, J.; Cline-Smith, A.B.; Ferris, C.; Buchwald, Z.S.; Gibbs, J.; Novack, D.; Aurora, R. Osteoclast-primed Foxp3+ CD8 T cells induce T-bet, Eomesodermin, and IFN-γ to regulate bone resorption. J. Immunol. 2016, 197, 726–735. [Google Scholar] [CrossRef] [Green Version]
- Shimizu, K.; Sato, Y.; Kawamura, M.; Nakazato, H.; Watanabe, T.; Ohara, O.; Fujii, S.I. Eomes transcription factor is required for the development and differentiation of invariant NKT cells. Commun. Biol. 2019, 2, 150. [Google Scholar] [CrossRef] [Green Version]
- Harada, M.; Vo, N.T.; Nakao, A.; Tanigaki, R.; Fukuoka, N.; Nishida, A.; Kataoka, T. Eomesodermin promotes interaction of RelA and NFATc2 with the Ifng promoter and multiple conserved noncoding sequences across the Ifng locus in mouse lymphoma BW5147 cells. Immunol. Lett. 2020, 225, 33–43. [Google Scholar] [CrossRef]
- Podolin, P.L.; Callahan, J.F.; Bolognese, B.J.; Li, Y.H.; Carlson, K.; Davis, T.G.; Mellor, G.W.; Evans, C.; Roshak, A.K. Attenuation of murine collagen-induced arthritis by a novel, potent, selective small molecule inhibitor of IκB kinase 2, TPCA-1 (2-[(aminocarbonyl)amino]-5-(4-fluorophenyl)-3-thiophenecarboxamide), occurs via reduction of proinflammatory cytokines and antigen-induced T cell proliferation. J. Pharmacol. Exp. Ther. 2005, 312, 373–381. [Google Scholar]
- Nan, J.; Du, Y.; Chen, X.; Bai, Q.; Wang, Y.; Zhang, X.; Zhu, N.; Zhang, J.; Hou, J.; Wang, Q.; et al. TPCA-1 is a direct dual inhibitor of STAT3 and NF-κB and regresses mutant EGFR-associated human non-small cell lung cancers. Mol. Cancer Ther. 2014, 13, 617–629. [Google Scholar] [CrossRef] [Green Version]
- Lazarevic, V.; Glimcher, L.H.; Lord, G.M. T-bet: A bridge between innate and adaptive immunity. Nat. Rev. Immunol. 2013, 13, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Suto, A.; Wurster, A.L.; Reiner, S.L.; Grusby, M.J. IL-21 inhibits IFN-γ production in developing Th1 cells through the repression of Eomesodermin expression. J. Immunol. 2006, 177, 3721–3727. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Mohn, S.E.; Weinmann, A.S. Jmjd3 and UTX play a demethylase-independent role in chromatin remodeling to regulate T-box family member-dependent gene expression. Mol. Cell 2010, 40, 594–605. [Google Scholar] [CrossRef] [Green Version]
- Eshima, K.; Chiba, S.; Suzuki, H.; Kokubo, K.; Kobayashi, H.; Iizuka, M.; Iwabuchi, K.; Shinohara, N. Ectopic expression of a T-box transcription factor, eomesodermin, renders CD4+ Th cells cytotoxic by activation both perforin- and FasL-pathways. Immunol. Lett. 2012, 144, 7–15. [Google Scholar] [CrossRef]
- Mazzoni, A.; Maggi, L.; Siracusa, F.; Ramazzotti, M.; Rossi, M.C.; Santarlasci, V.; Montaini, G.; Capone, M.; Rossettini, B.; De Palma, R.; et al. Eomes controls the development of Th17-derived (non-classic) Th1 cells during chronic inflammation. Eur. J. Immunol. 2019, 49, 79–95. [Google Scholar] [CrossRef] [Green Version]
- Cho, J.Y.; Grigura, V.; Murphy, T.L.; Murphy, K. Identification of cooperative monomeric Brachyury sites conferring T-bet responsiveness to the proximal IFN-γ promoter. Int. Immunol. 2003, 15, 1149–1160. [Google Scholar] [CrossRef] [Green Version]
- Sica, A.; Dorman, L.; Viggiano, V.; Cippitelli, M.; Ghosh, P.; Rice, N.; Young, H.A. Interaction of NF-κB and NFAT with the interferon-γ promoter. J. Biol. Chem. 1997, 272, 30412–30420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaplanski, G. Interleukin-18: Biological properties and role in disease pathogenesis. Immunol. Rev. 2018, 281, 138–153. [Google Scholar] [CrossRef] [Green Version]
- Yasuda, K.; Nakanishi, K.; Tsutsui, H. Interleukin-18 in health and disease. Int. J. Mol. Sci. 2019, 20, 649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, O.; Akira, S. Pattern recognition receptors and inflammation. Cell 2010, 140, 805–820. [Google Scholar] [CrossRef] [Green Version]
- Imanishi, T.; Unno, M.; Kobayashi, W.; Yoneda, N.; Akira, S.; Saito, T. mTORC1 signaling controls TLR2-mediated T-cell activation by inducing TIRAP expression. Cell Rep. 2020, 32, 107911. [Google Scholar] [CrossRef] [PubMed]
- Thome, M.; Charton, J.E.; Pelzer, C.; Hailfinger, S. Antigen receptor signaling to NF-κB via CARMA1, BCL10, and MALT1. Cold Spring Harb. Perspect. Biol. 2010, 2, a003004. [Google Scholar] [CrossRef]
- Huang, D.C.S.; Cory, S.; Strasser, A. Bcl-2, Bcl-xL and adenovirus protein E1B19kD are functionally equivalent in their ability to inhibit cell death. Oncogene 1997, 14, 405–414. [Google Scholar] [CrossRef] [Green Version]
- Kondo, T.; Takeda, K.; Muko, R.; Ito, A.; Chang, Y.C.; Magae, J.; Kataoka, T. 4-O-Methylascochlorin inhibits the prolyl hydroxylation of hypoxia-inducible factor-1α, which is attenuated by ascorbate. J. Antibiot. 2019, 72, 271–281. [Google Scholar] [CrossRef] [PubMed]
- Harada, M.; Morimoto, K.; Kondo, T.; Hiramatsu, R.; Okina, Y.; Muko, R.; Matsuda, I.; Kataoka, T. Quinacrine inhibits ICAM-1 transcription by blocking DNA binding of the NF-κB subunit p65 and sensitizes human lung adenocarcinoma A549 cells to TNF-α and the Fas ligand. Int. J. Mol. Sci. 2017, 18, 2603. [Google Scholar] [CrossRef] [Green Version]
- Casteels, K.M.; Mathieu, C.; Waer, M.; Valckx, D.; Overbergh, L.; Laureys, J.M.; Bouillon, R. Prevention of type I diabetes in nonobese diabetic mice by late intervention with nonhypercalcemic analogs of 1,25-dihydroxyvitamin D3 in combination with a short induction course of cyclosporin A. Endocrinology 1998, 139, 95–102. [Google Scholar] [CrossRef] [PubMed]
- Overbergh, L.; Valckx, D.; Waer, M.; Mathieu, C. Quantification of murine cytokine mRNAs using real time quantitative reverse transcriptase PCR. Cytokine 1999, 11, 305–312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamada, Y.; Taketani, S.; Osada, H.; Kataoka, T. Cytotorienin A, a translation inhibitor that induces ectodomain shedding of TNF receptor 1 via activation of ERK and p38 MAP kinase. Eur. J. Pharmacol. 2011, 667, 113–119. [Google Scholar] [CrossRef]
- Mitsuda, S.; Yokomichi, T.; Yokoigawa, J.; Kataoka, T. Ursolic acid, a natural pentacyclic triterpenoid, inhibits intercellular trafficking of proteins and induces accumulation of intercellular adhesion molecule-1 linked to high-mannose-type glycans in the endoplasmic reticulum. FEBS Open Bio 2014, 4, 229–239. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tanaka, Y.; Nakao, A.; Miyake, Y.; Higashi, Y.; Tanigaki, R.; Kataoka, T. Small Molecule Inhibitors Targeting Nuclear Factor κB Activation Markedly Reduce Expression of Interleukin-2, but Not Interferon-γ, Induced by Phorbol Esters and Calcium Ionophores. Int. J. Mol. Sci. 2021, 22, 13098. https://doi.org/10.3390/ijms222313098
Tanaka Y, Nakao A, Miyake Y, Higashi Y, Tanigaki R, Kataoka T. Small Molecule Inhibitors Targeting Nuclear Factor κB Activation Markedly Reduce Expression of Interleukin-2, but Not Interferon-γ, Induced by Phorbol Esters and Calcium Ionophores. International Journal of Molecular Sciences. 2021; 22(23):13098. https://doi.org/10.3390/ijms222313098
Chicago/Turabian StyleTanaka, Yumiko, Ayaka Nakao, Yasunobu Miyake, Yukina Higashi, Riho Tanigaki, and Takao Kataoka. 2021. "Small Molecule Inhibitors Targeting Nuclear Factor κB Activation Markedly Reduce Expression of Interleukin-2, but Not Interferon-γ, Induced by Phorbol Esters and Calcium Ionophores" International Journal of Molecular Sciences 22, no. 23: 13098. https://doi.org/10.3390/ijms222313098
APA StyleTanaka, Y., Nakao, A., Miyake, Y., Higashi, Y., Tanigaki, R., & Kataoka, T. (2021). Small Molecule Inhibitors Targeting Nuclear Factor κB Activation Markedly Reduce Expression of Interleukin-2, but Not Interferon-γ, Induced by Phorbol Esters and Calcium Ionophores. International Journal of Molecular Sciences, 22(23), 13098. https://doi.org/10.3390/ijms222313098





