1. Introduction
One of the most important problems of cancer patients is metastasis. It contributes to almost 66.7% of cancer mortality [
1]. It also applies to breast cancers tumors, 81% of which are invasive. Although in patients with lesions limited only to the primary site the 5-year survival is relatively high, in the case of metastases it drops to only 30%. Thus, the search for new therapy goals based on broadly understood cancer cell biology, with particular emphasis on molecular factors involved in the emergence of secondary tumor outbreaks is reasonable. Highly invasive cancer cells can change the phenotype from epithelial to mesenchymal. These changes are called the epithelial-mesenchymal transition (EMT), during which cells acquire motor skills. The formation of metastases is a cascade of closely consecutive molecular and morphological changes, including the reconstruction of the cytoskeleton and extracellular matrix (ECM) [
2,
3]. Metalloproteinases (MMPs) are zinc-dependent endopeptidases responsible for physiological and pathophysiological tissue remodeling [
4]. According to available literature reports, breast cancer cells of various invasiveness are characterized by variable expression of MMP, e.g., MMP-1, MMP-2, MMP-9, MMP-7, MMP-10, and MMP-19 [
5]. Individual MMP isoenzymes have different substrate specificity, e.g., MMP-1 shows hydrolytic activity against collagens (collagenases). During metastasis, MMPs degrade ECM proteins. It opens pathways for cancer cells that move using actin-rich protrusions, e.g., filopodia [
6,
7]. Changes in MMPs levels have been observed in many cancers, such as ovarian and colorectal cancer [
8]. MMPs enzymatic activity also correlates with the stage of cancer, increased invasiveness, and lower survival rate. These factors emphasize the value of MMPs in the prognosis and monitoring of the effectiveness of cancer treatment [
9,
10]. MMP-1 and MMP-9 are also very important in these processes, especially in cancers with different receptor profiles, e.g., progesterone positive (ER+) and triple-negative breast cancer (TNBC) [
11,
12]. MMP-9 is crucial in metastasis due to the high potential for ECM degradation (various substrates: gelatin types I and V, collagen types IV and V, fibronectin). Moreover, its high expression correlates with a worse prognosis among patients with breast, kidney, lung, and ovarian cancers [
9]. The importance of cytokeratins (CK) is also of great interest, both in the context of the migration process and their prognostic value. The relationship between MMPs and CKs raises many questions regarding the nature and function of this interaction. The common point was indicated in the research on sentinel lymph nodes specimens obtained from breast cancer patients. The study presents the prognostic value of MMP-9 but also its correlation with CK19 [
13].
Our study aimed to investigate how changes in the expression level and enzymatic activity of MMPs affect the proliferative and migratory capacity and thus metastatic potential of breast cancer cells. Additionally, we treated cells with Cyclophosphamide (CP) used in chemotherapy of various types of cancer, including breast cancer. Recent reports indicate high toxicity of CP and increased migration of the CP-treated breast cancer cells [
14,
15]. Therefore, we wanted to investigate how reduced MMP-9 expression affects the response of breast cancer cells (MDA-MB-231 and MCF-7) to CP treatment and whether there is a correlation between MMP-9 and CK19.
3. Discussion
Inhibition of tumor metastasis is a desirable goal of anti-cancer therapy. It is especially important in triple-negative breast cancer, which is characterized by very high aggressiveness and mortality [
19]. Invasive cells can move through the reorganization of the cytoskeleton, which allows the formation of migratory protrusions. MMPs facilitate this process by digesting the extracellular matrix, which paves the way for cancer cells. In vitro studies and analysis of available data on breast cancer patients presented in this paper indicate high levels of MMP-9 in breast tumors [
8,
9,
12]. MMP-9 is classified as a tumor biomarker and prognostic factor, and its regulation may become a therapeutic target [
20]. In 2016, Moirangthem et al. showed that knockdown of MMP-9 in the breast cancer cell line MDA-MB-231 may reduce progression by increasing cell-to-cell adhesion and regulating EMT markers, including urokinase receptor (uPAR), E-cadherin, vimentin, and Snail. In addition, an enhanced anti-metastatic effect after a simultaneous knockdown of the plasminogen activator urokinase (uPA) and MMP-9 was indicated [
21].
Under unfavorable conditions such as hypoxia or inflammation, intracellular signaling pathways trigger the expression of MMP-9 [
22]. These include the phosphoinositide 3 (PI3K)/protein kinase B (AKT), mitogen-activated protein kinases (MAPK)/extracellular signal-regulated kinases (ERK), and nuclear factor-kappa B (NF-kB) pathways [
22]. Numerous scientific reports indicate that different compounds target PI3K/AKT/NFκB, AKT/mTOR pathways reducing MMP-9 expression. Wu et al. noticed that natural herbal flavonoid—luteolin suppresses triple-negative breast cancer cell proliferation and metastasis by the downregulation of MMP-9 expression via the AKT/mTOR signaling pathway [
23]. On the other hand, Zeng et al. indicated the reduction of MMP-9 levels in MDA-MB-231 and MCF-7 cells after treatment with tectorigenin [
24]. Furthermore, piceatannol inhibits the invasion of breast cancer cells through the PI3K/AKT and NF-κB pathways and inhibition of MMP-9 [
25]. It is in line with our results as MMP-9 downregulation reduced the migration potential of breast cancer cells of different aggressiveness. Moreover, we decided to check whether CP, a cytostatic commonly used in breast cancer, may intensify the changes observed after MMP-9 silencing or abolish it. This aspect was of particular interest as there were some disturbing reports on the pro-cancerous effect of CP [
14]. Studies by Hung et al. indicate that CP can induce breast cancer metastasis. They assessed the results of CP treatment on the migration of cancer cells and its correlation with the chemokine receptor 4 (CXCR4), which is considered a biomarker of cancer metastasis [
14]. The obtained results indicate that in MDA-MB-231 cells, increasing concentrations of CP induced the expression of CXCR4 and thus facilitated cell migration. Moreover, the expression of MMP-9 in CP-treated cells was elevated. Zhang et al. also observed that CP increases the levels of MMP-9. By examining the effect of CP in the kidney tissue of rats with Diabetic nephropathy, they indicated decreased expression of TGF-β1 and increased level of MMP-9 [
26]. Our data also suggest that CP can indeed increase the expression of MMP-9, and its activity in monotherapy does not result in a statistically significant anti-tumor effect. Perhaps the use of combination therapy along with the agent targeting MMP-9 would be more effective.
On the other hand, Guo et al. showed that a newly described chemical synthetic peptide (E5) inhibits the CXCR4/CXCL12 axis in breast cancer both in vitro and in vivo. E5 was able to bind specifically to the 4T1 breast cancer line, inhibit migration by reducing CXCR4 expression. Its combination with paclitaxel or CP significantly inhibited tumor growth in a breast cancer model [
27]. Moreover, the preclinical studies indicate the therapeutic effect and low toxicity of the combined chemotherapy with CP and celecoxib in the case of breast tumors. The combination was anti-angiogenic and was well tolerated [
28]. CP was also used in combination with Doxorubicin, 5-fluorouracil, Methotrexate, Docetaxel, and Paclitaxel [
29,
30]. Currently, the application of such therapy is limited due to serious side effects, liver damage in particular. However, the possible combination with other compounds or factors altering the tumor environment makes CP an important cytostatic in anti-cancer therapy. Cancer cell apoptosis and suppression of tumor growth were observed after implantation of nude mice with MDA-MB-231 cells and treatment with a combination of CP with Chalone 19 peptide [
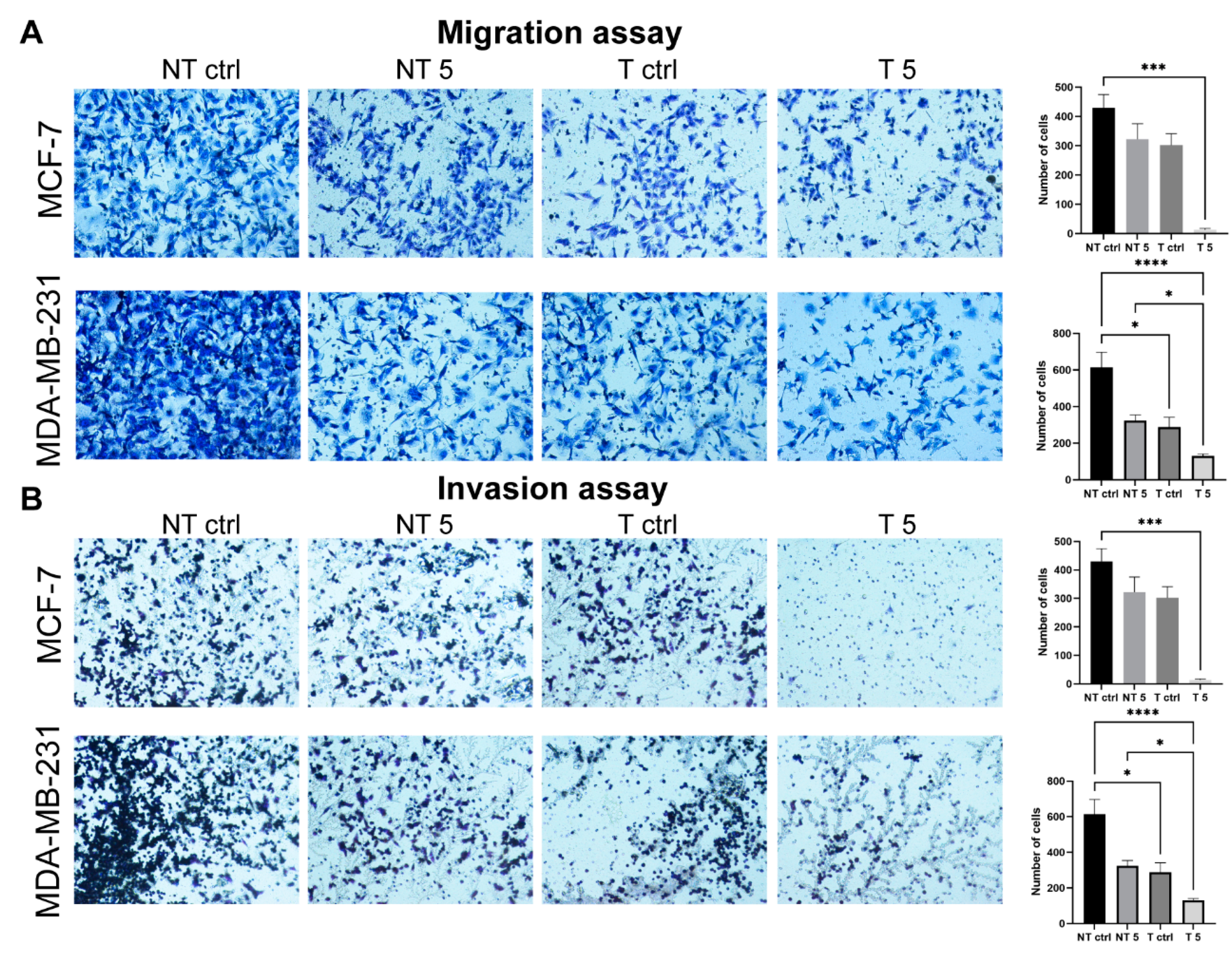
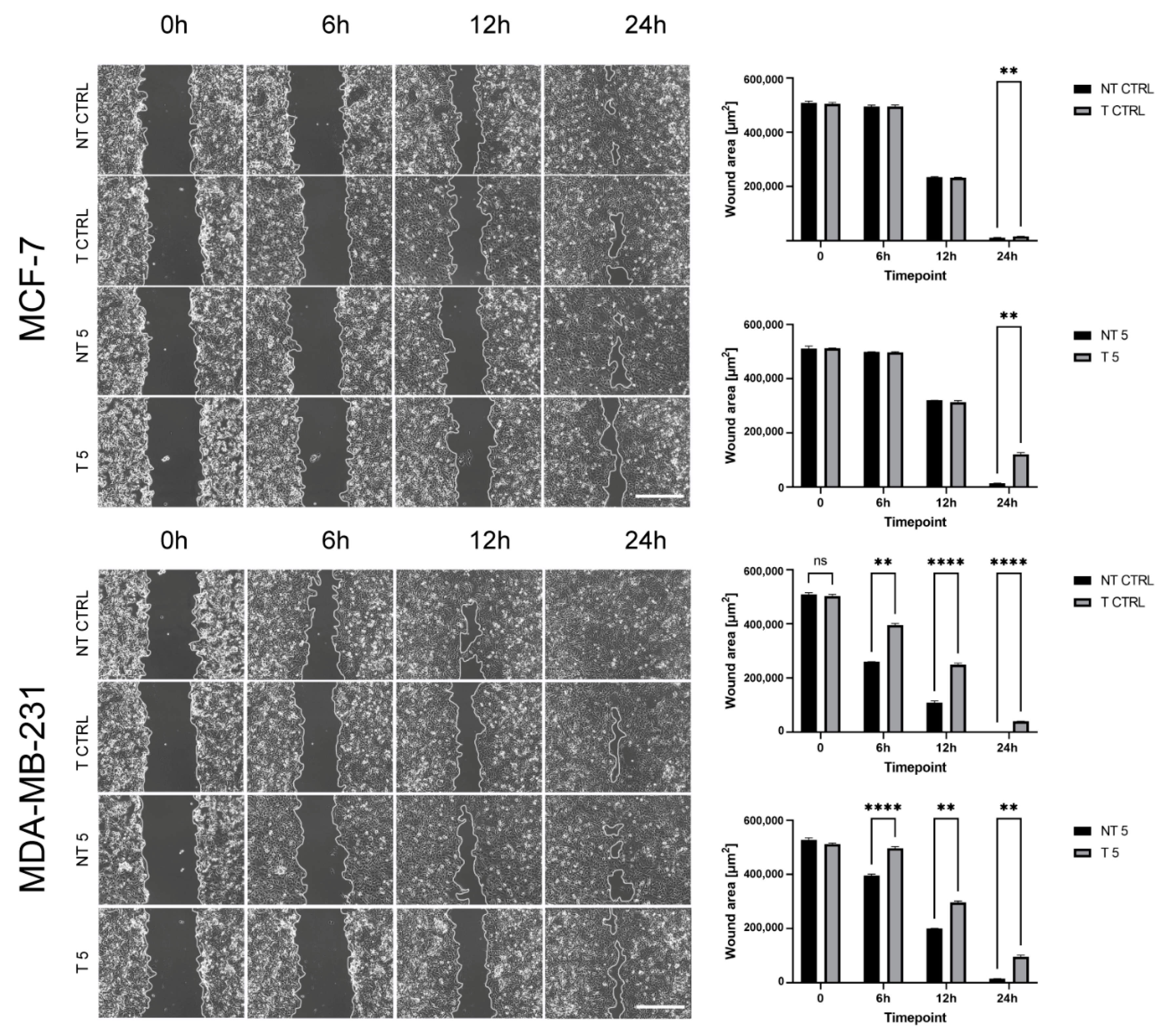
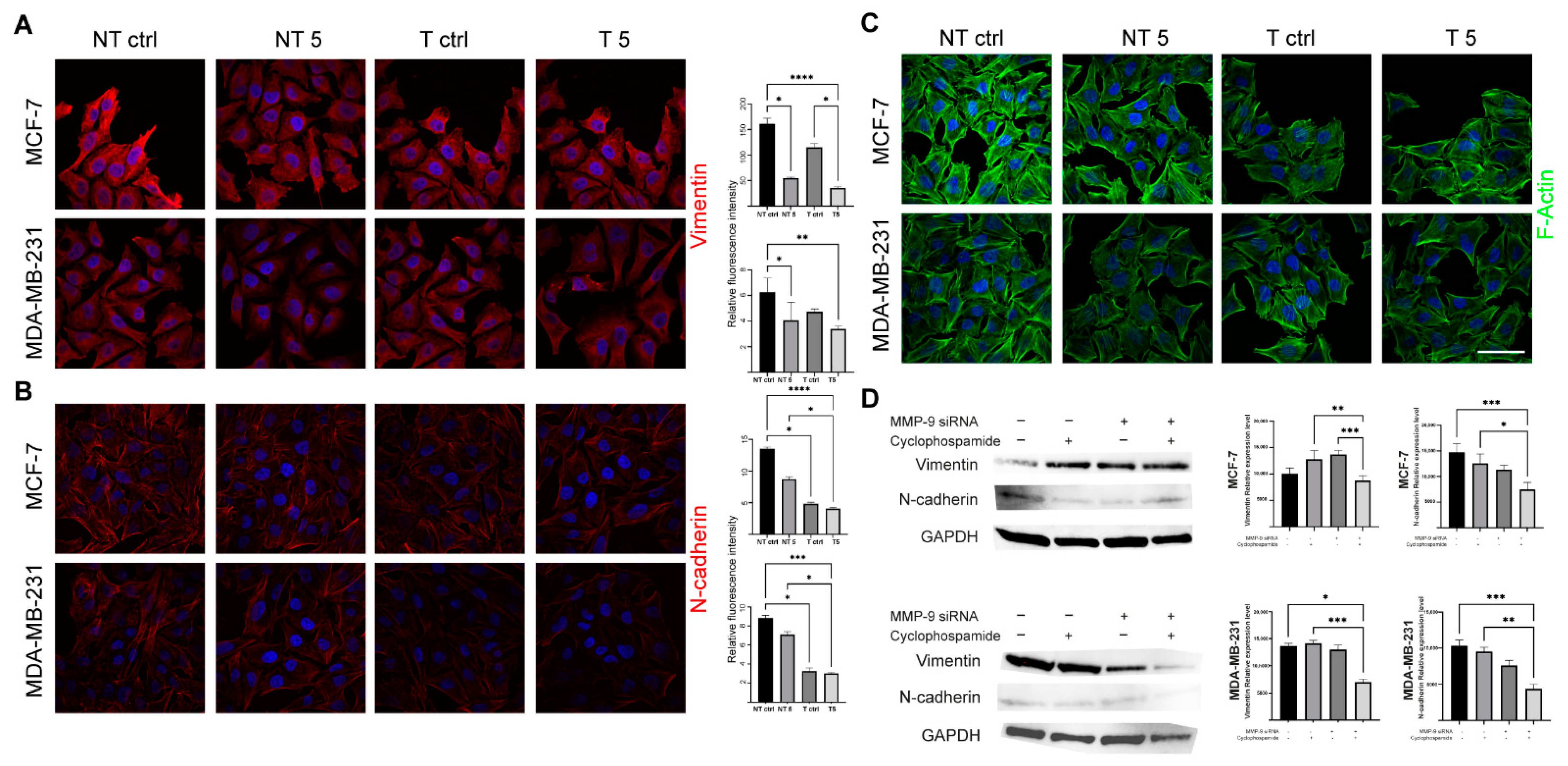
31]. In turn, Agrawal et al. found that insulin increases the cytotoxic effect of 5-fluorouracil and CP and enhances the apoptosis in the MCF-7 cell line. We also confirmed pro-apoptotic and antimetastatic effects of a 5 mM CP concentration in MCF-7 and MDA-MB-231 cells with reduced MMP-9 expression. Analysis of the percentage of apoptotic cells showed a statistically significant increase only in the group of cells transfected with MMP-9 siRNA treated with CP. Moreover, the downregulation of MMP-9 followed by the cytostatic treatment reduced migration and invasion (wound healing assay, chemotaxis test) and influenced the level of EMT markers in cells of both breast cancer cell lines. Vimentin is an intermediate filament protein and an established EMT marker. Similarly, the high level of N-cadherin indicates the cell’s pro-migratory phenotype [
32]. The observed decrease of the EMT markers in cells transfected with MMP-9 siRNA treated with CP proves the effectiveness of such approach.
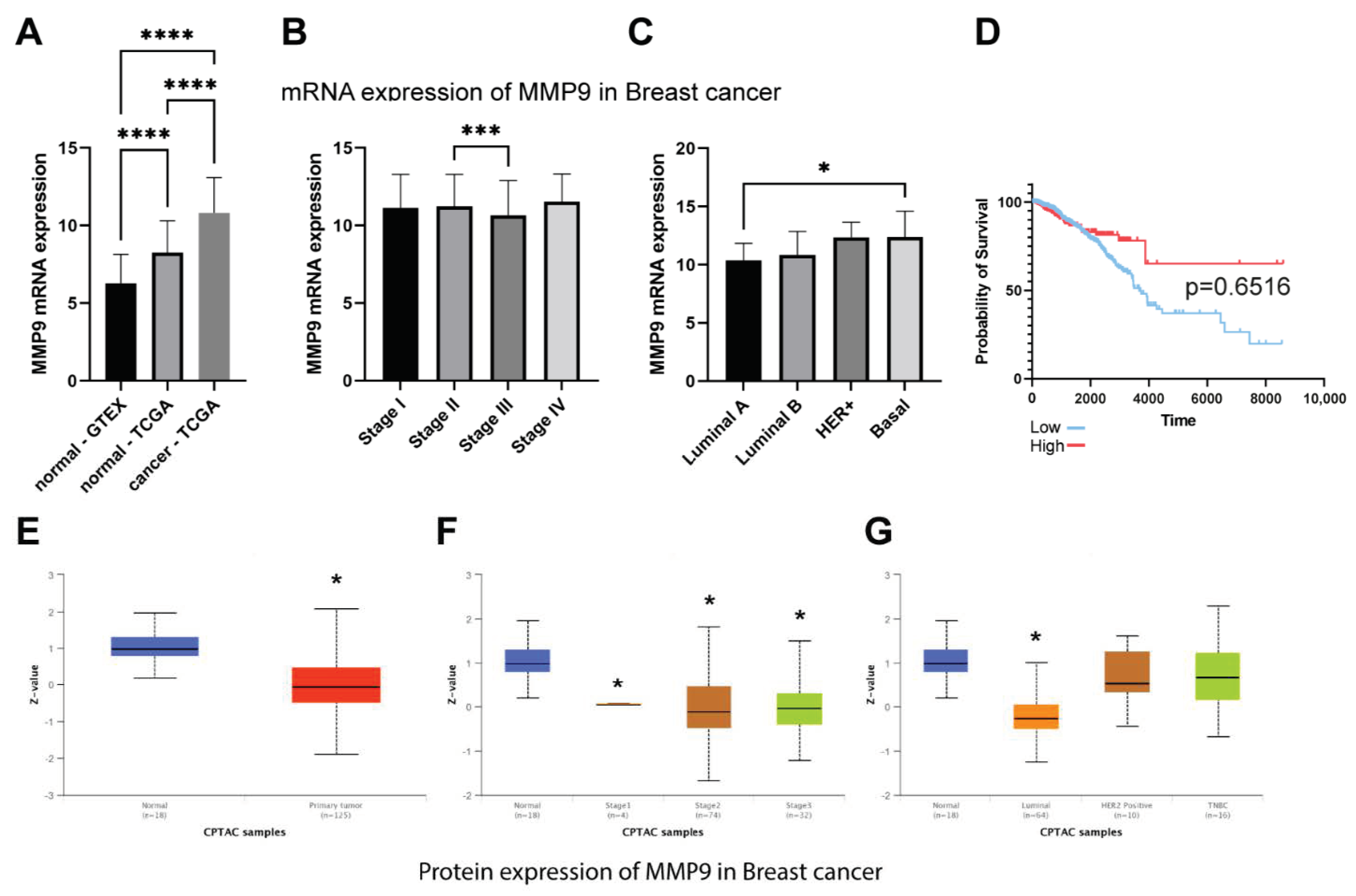
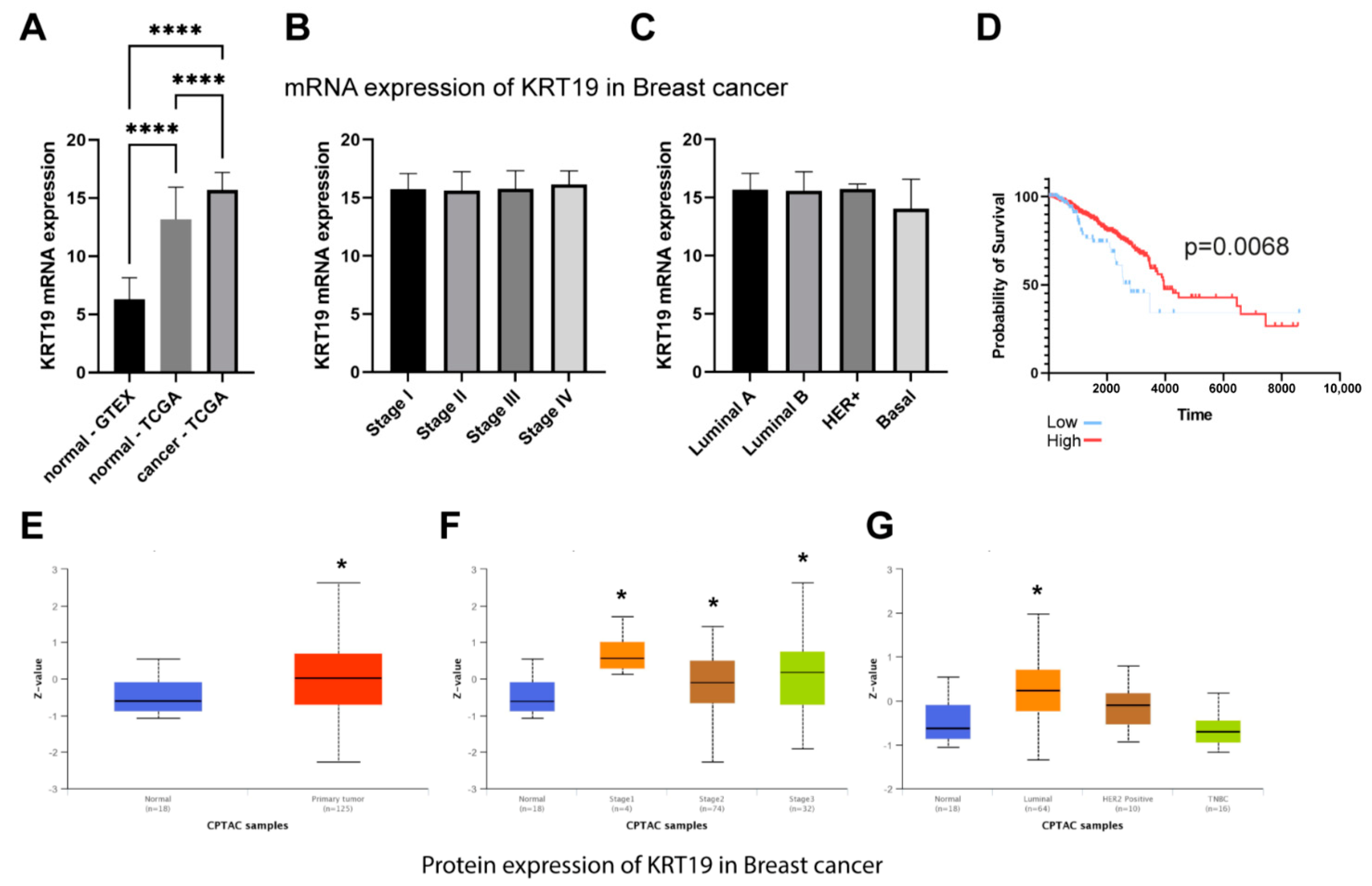
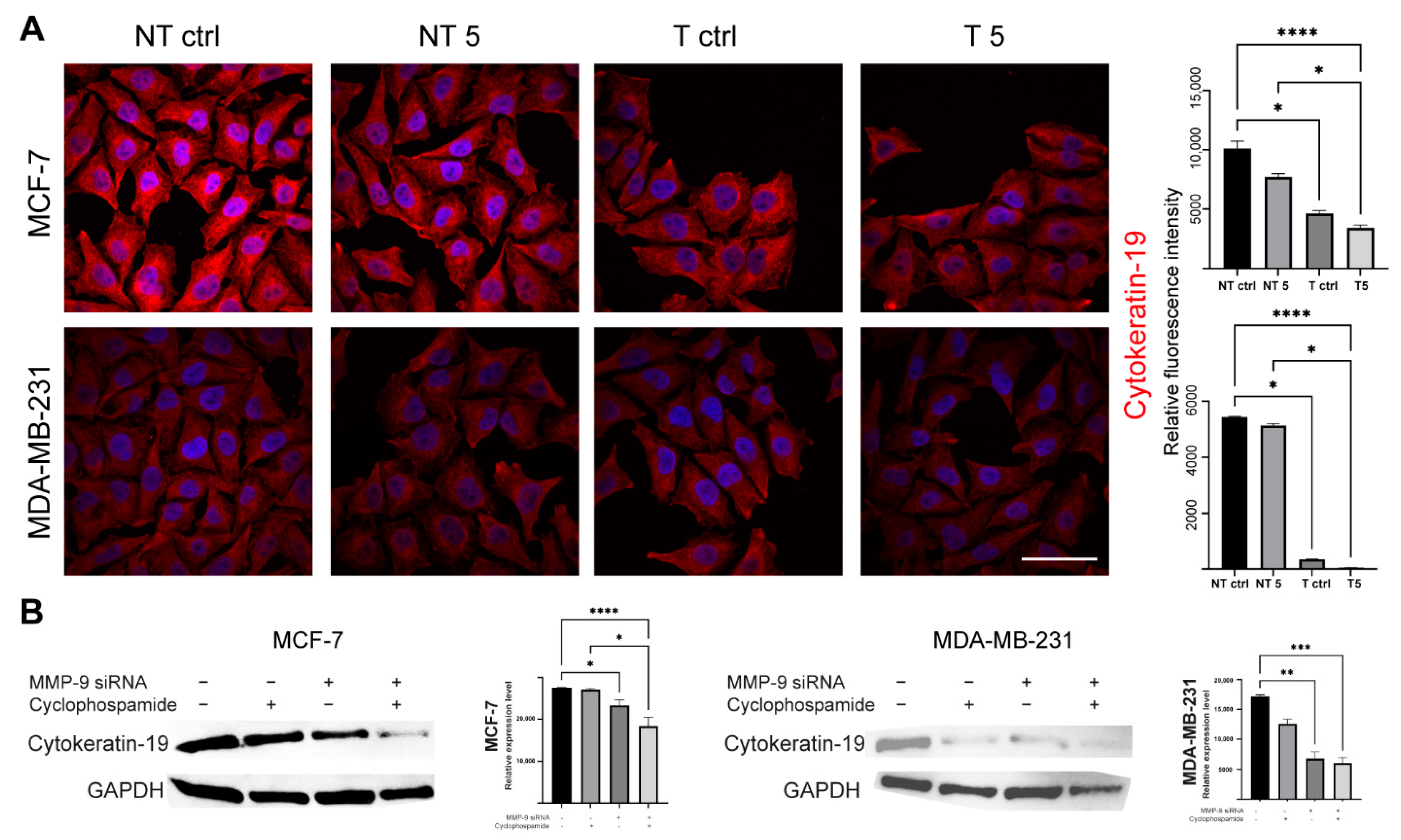
What we also found interesting was a decrease in the level of CK19 after the downregulation of MMP-9. TCGA dataset confirmed that breast cancer is characterized by the overexpression of MMP-9 and CK19 mRNA. However, it has no impact on breast patients’ survival. Although we observe the significant difference between low and high KRT19 (CK19) expression survival, the accuracy of these findings is limited due to the large disproportion between-group strength (929 vs. 118). In turn, CPTAC data suggest the downregulation of MMP-9 in breast cancer patients and upregulation of KRT19 (CK19). However, because of the size of the cohort and the lack of other clinical parameters, we cannot provide any conclusive remarks from this analysis. Both MMP-9 and CK19 are considered negative prognostic markers, but the effect of CK19 in breast cancers seems to be more complex and ambiguous. Some studies show the oncogenic functions of CK19 associated with the stabilization of the HER2/ERK signaling pathway [
33]. On the other hand, CK19 can bind β-catenin/RAC1 and control NUMB transcriptional activity, a protein that is an upstream Notch pathway inhibitor [
34]. Moreover, the silencing of CK19 in breast cancer cells results in Akt/PTEN pathway [
35].
Murawski et al. assessed the importance of MMP-9 expression as a supporting marker for CK19 mRNA in sentinel lymph nodes in breast cancer patients and the link between this expression and clinical-pathological data. The results indicate a strong correlation of MMP-9 with CK19 mRNA in breast carcinoma [
13]. In turn, Joseph et al. indicate an increased level of CK17, which was associated with the increased expression of MMP-9. Patients with breast tumors characterized by high expression of CK17 and CK5/6 show high mortality, which indicates the correlation between cytokeratins and metalloproteinases [
12].
4. Materials and Methods
4.1. Cell Culture
The human breast cancer MCF-7 (HTB-22) and MDA-MB-231 (HTB-26) cell lines were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). The control was a non-tumorigenic epithelial MCF-10A (EP-CL-0525) cell line purchased from Elabscience Biotechnology Inc. All cell lines were cultured according to the manufacturer’s instructions. MCF-7 cells were maintained in Eagle’s Minimum Essential Medium (EMEM, cat. No. 10-009-CV, Corning, New York, NY, USA) supplemented with 0.01 mg/mL Recombinant Human Insulin (rHu Insulin, cat. No. HY-P1156/CS-7906, MedChemExpress, Monmouth Junction, NJ, USA), 10% Fetal Bovine Serum (FBS, cat. No. 35-079-CV, Corning, New York, NY, USA) and 1% Penicillin-Streptomycin Solution (P/S, cat. No. 30-002-CI, Corning, New York, NY, USA). The MDA-MB-231 cell line was cultured in Dulbecco’s Modified Eagle Medium (DMEM, cat. No. 12-604F, Lonza, Basel, Switzerland) with 10% FBS and 1% P/S. MCF-10A cell line was grown in monolayer in DMEM/F12 (1:1 mixture of DMEM and Ham’s F12, cat. No. 10-090-CV, Corning, New York, NY, USA) with 5% Horse Serum (HS, cat. No. 16050-122, Thermo Fisher Scientific, Waltham, MA, USA), 20 ng/mL Human Epidermal Growth Factor (EGF, cat. No. AF-100-15-100UG, Peprotech Ec Ltd., London, UK), 0.5 µg/mL Hydrocortisone (cat. No. HY-N0583/CS-2226, MedChemExpress, Monmouth Junction, NJ, USA), 10 µg/mL rHu Insulin (cat. No. HY-P1156/CS-7906, MedChemExpress, Monmouth Junction, NJ, USA), 1% Non-Essential Amino Acid (NEAA, cat. No. M7145-100 mL, Merck KGaA, Darmstadt, Germany) and 1% P/S. The culture was carried out in a 95% humidified atmosphere, 5% CO2 at 37 °C. The cells were cultivated up to five passages and regularly tested for mycoplasma contamination using 4′,6-Diamidino-2-phenylindole dihydrochloride-staining solution (DAPI, D9542, Merck KGaA, Darmstadt, Germany). The cells were subcultured after reaching about 80% confluency using Trypsin-Ethylenediamine tetraacetic acid (EDTA, cat. No. 25-053-CI, Corning, New York, NY, USA) and on T-25 cm2 flasks (cat. No. 156367, Thermo Fisher Scientific, Waltham, MA, USA) or 24, 12- and 6-well plates (cat. No. SIAL0524, cat. No. SIAL0513, cat. No. SIAL0516, Merck KGaA, Darmstadt, Germany) depending on the assay and in cell type-dependent density. Before the experiments, optimal density was investigated by seeding cells at different concentrations and evaluated visually in light Axio Observer Z1 inverted motorized microscope (Zeiss, Oberkochen, Germany). Twenty-four hours after seeding, the cells were treated with different concentrations of CP (cat. No. T0707, TargetMol, Wellesley Hills, MA, USA) selected based on the available literature reports including declared CP concentrations in plasma and the Thiazol-2-yl-2,5-diphenyl tetrazolium bromide (MTT) assay and incubated for the next twenty-four hours. Control cell’s populations were cultured in the same conditions and time without the addition of CP.
4.2. Cell Transfection
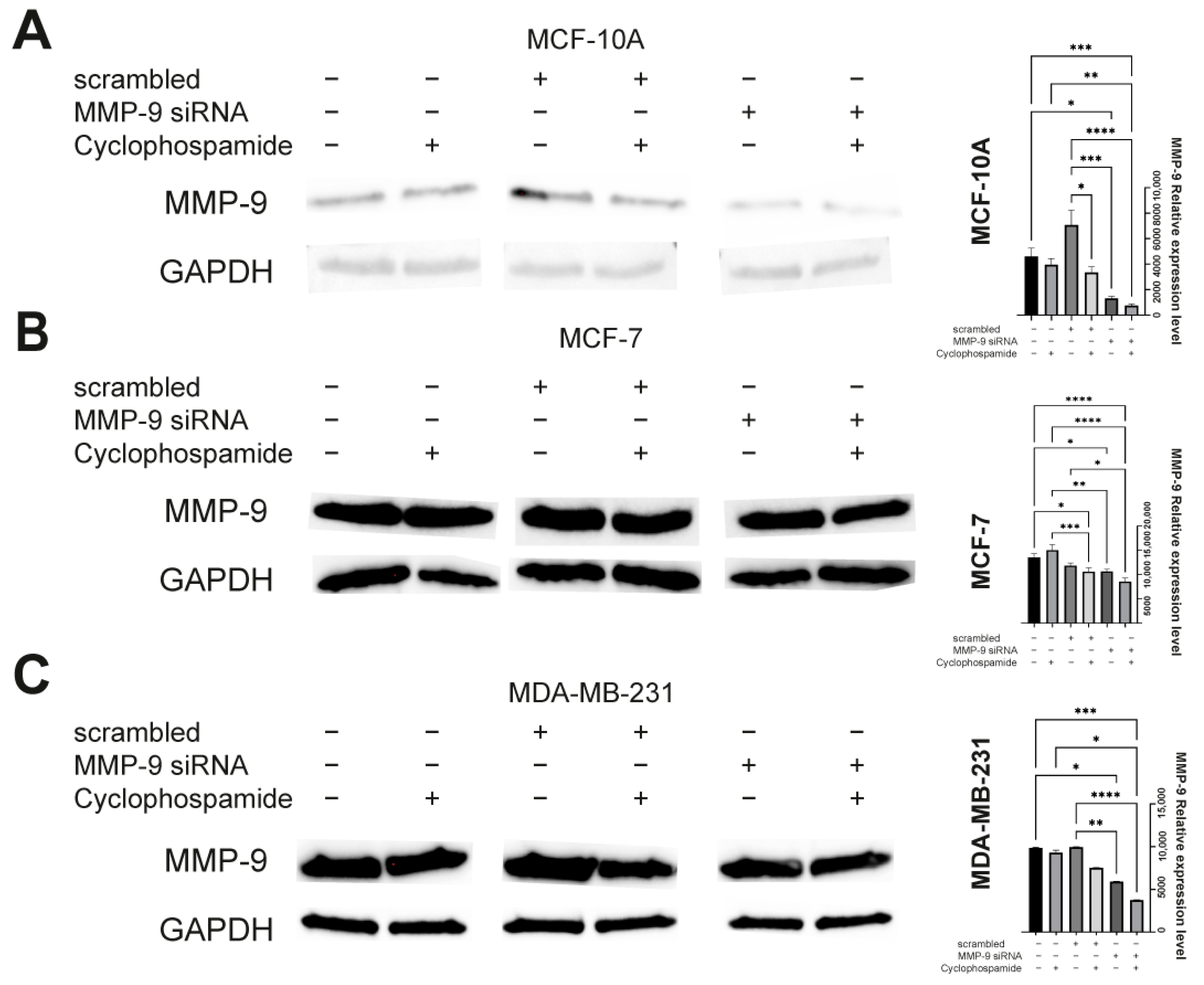
The cells were transfected by electroporation using nucleofection technique according to the manufacturer’s instruction and optimization techniques. Briefly, the cells were transferred to 24-well plate delivered with Nucleofection Kit (Amaxa, SF Cell Line 4D-Nucleofector X Kit L, cat. No. V4XC-2024, Lonza, Basel, Switzerland) (Amaxa, SE Cell Line 4D-Nucleofector X Kit L, cat. No. V4XC-1024, Lonza, Basel, Switzerland). The wells containing cells were supplemented with Nucleofector solutions and MMP-9 siRNA (cat. No. sc-29400, Santa Cruz, Dallas, TX, USA). The plate was covered with Dipping Electrode Array, put into Nucleofector and electroporated using dedicated impulse program. The programs were dependent on the type of the cell and defined by manufacturer (MCF-7: EN-130, MDA-MB-231: CH-125, MCF-10A: T-024). As a negative control the control siRNA (cat. No. sc-37007, Santa Cruz, Dallas, TX, USA) was used. Semi-quantitative analysis of post-translational MMP-9 expression was analyzed by Western blot according to the methodology described below.
4.3. TCGA, GTEx and CPTAC Analysis
To assess the expression of MMP-9 and KRT19 (CK19) and their clinical relevance in breast cancer we analyzed the TCGA, GTEx, and CPTAC data. The TCGA and GTEx mRNA expression data were retrieved via Xena Functional Genomics Explorer [
36] and CPTAC protein data were obtained from the UALCAN database [
37]. The cutpoints for the analysis were calculated using Evaluate Cutpoint R package [
38]. The statistical analysis for RNA-seq data was performed in GraphPad Prism 9 and CPTAC statistical analysis was taken directly from the database.
4.4. MTT Assay
Cells were seeded in 24-well plates, cultured for 24 h, and treated with CP for another 24 h. The increasing drug doses used in the studies were: 0.1 mM, 0.5 mM, 1 mM, 2 mM, 2.5 mM, 5 mM, 8 mM, 10 mM. A stock solution of CP was prepared by dissolving the powder in 100% DMSO to a final 182.7 mM concentration. The dilutions were prepared in a complete growth medium immediately before use. The MTT salt was dissolved (50 mg/mL) in phosphate-buffered saline (PBS, cat. No. 21-040-CV, Corning, New York, NY, USA), diluted 1:9 in phenol red-free DMEM (Corning, New York, NY, USA) right before the experiment and 500 μL were added to each well for 3 h (incubator). The resulting purple formazan crystals were dissolved with dimethyl sulfoxide solution (DMSO, cat. No. 210520182, CHEMPUR, Piekary Śląskie, Poland). The results of the MTT assay have been documented using BioTEK 800Ts fluorescence microplate reader (BioTEK, Bad Friedrichshall, Germany) at 570 nm wavelength.
4.5. Cell Death Analysis
The cell death (apoptosis, necrosis) was analyzed by flow cytometry using Guava® easyCyte™ 6HT-2L system and InCyte Software (version 3.3, Merck KGaA, Darmstadt, Germany). The cells were seeded in the 12-well plates. The next day cells were treated with a 5 mM concentration of the CP and incubated for 24 h. Then, the cells were detached. The cell suspension was centrifuged at 300 RCF for 5 min at room temperature (RT). The pellet was resuspended in 200 µL of Annexin Binding Buffer (ABB, cat. No. V13246, Thermo Fisher Scientific, Waltham, MA, USA). The cells were stained using propidium iodide solution (cat. No. P1304MP, Thermo Fisher Scientific, Waltham, MA, USA) and Annexin V Alexa Fluor 488 (cat. No. A13201, Thermo Fisher Scientific, Waltham, MA, USA) following the manufacturer’s instructions (15 min, RT, dark). Next, the cell suspension was transferred to a 96-well plate (cat. No. 0030601106, Eppendorf, Hamburg, Germany) for the cytometric analysis.
4.6. Cell Cycle Analysis
The cell cycle was analyzed using flow cytometry. MCF-7 and MDA-MB-231 cells were cultured into 12-well plates. After reaching 70% confluence, the cells were treated with CP and incubated for 24 h. Then, the cells were detached, centrifuged (300 RCF, 5 min, RT), and fixed in 1 mL 70% ethanol (cat. No. PA-06-396480427-25L, Pol-Aura, Różnowo, Poland) for 24 h at −20 °C. The fixed cells were centrifuged (500 RCF, 7 min, RT) to remove ethanol. The pellet was washed with PBS. Next, the cells were suspended in 200 μL FxCycle PI/RNase Staining Solution (cat. No. F10797, Thermo Fisher Scientific, Waltham, MA, USA) and incubated according to the Manufacturer’s instruction (30 min, RT, dark). The cells were placed in a 96-well plate and analyzed using Guava® easyCyte™ 6HT-2L system and InCyte Software (version 3.3, Merck KGaA, Darmstadt, Germany).
4.7. Immunoblotting Technique
Western blot was used to assess the efficiency of transfection with MMP-9 siRNA and to measure CK19, vimentin and N-cadherin protein level. The cells used in the experiment were transfected using the protocol described above and seeded in T-24 culture bottles. After 24 h, the cells were treated with the CP solution and incubated for another 24 h. Control cells were cultured under the same conditions but without CP addition. The lysis mixture contained RIPA buffer (cat. No. 20-188, Merck KGaA, Merck KGaA, Darmstadt, Germany) and Halt protease inhibitor cocktail (cat. No. 78438, Thermo Scientific, Waltham, MA, USA). The lysis was carried out for 10 min at 4 °C. Next, the samples were centrifuged (10.000 RCF, 10 min, 4 °C). The supernatant was used for the quantitative determination of the concentration of proteins in samples by the Pierce™ BCA Protein Assay Kit (cat. No. 23225, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s recommendations. The quantitative measurement of protein resulted in a standard curve, which allowed to calculate the volume of each sample and standardize the amount of protein taken for analysis. The samples were prepared by mixing appropriate volumes of cell lysates, NuPage Sample Reducing Agent (cat. No. NP0009, Thermo Fisher Scientific, Waltham, MA, USA), Tris-Glycine SDS Sample Buffer (cat. No. LC2676, Thermo Fisher Scientific, Waltham, MA, USA), and distilled water. The heat denaturation of the samples was carried out at 85 °C for 3 min. The denatured samples were placed in the polyacrylamide gel (Novex™ WedgeWell™ 10–20% Tris-Glycine Gel 1.0 × 10 well, XP10200BOX, Thermo Fisher Scientific, Waltham, MA, USA). PageRuler™ Prestained Protein Ladder, 10 to 180 kDa (cat. No. 26616, Thermo Fisher Scientific, Waltham, MA, USA) was used to assess the protein molecular weight. The electrophoretic separation was carried out for 30 min at 225 V in Tris-Glycine SDS Running Buffer (cat. No. LC2675-5, Thermo Fisher Scientific, Waltham, MA, USA). The next step was dry transfer onto a nitrocellulose membrane using the iBlot Dry Western Blotting System (7 min, 10 V). Transfer efficiency was checked by staining the nitrocellulose membrane with Ponceau S solution (cat. No. P7170, Merck KGaA, Merck KGaA, Darmstadt, Germany). The membranes were incubated in an enhancer solution for 20 min. Primary (anti- MMP-9: cat. No. sc-21733, Santa Cruz, Dallas, TX, USA; anti-CK9: cat. No. MA5-12663, Thermo Fisher Scientific, Waltham, MA, USA; anti-vimentin: cat. No. MA5-16409, Thermo Fisher Scientific, Waltham, MA, USA; anti-N-cadherin: cat. No. 33-3900, Thermo Fisher Scientific, Waltham, MA, USA, and secondary antibodies HRP (cat. No. 31460, cat. No. 31430, Thermo Fisher Scientific, Waltham, MA, USA) were diluted in the iBind Flex Solution Kit (cat. No. SLF2020, Thermo Fisher Scientific, Waltham, MA, USA) according to the manufacturer’s instructions. The membranes were applied onto iBind Flex Cards (cat. No. SLF2010, Thermo Fisher Scientific, Waltham, MA, USA) and placed in the iBind Automated Western System (Thermo Fisher Scientific, Waltham, MA, USA) for 24 h at 4 °C. Finally, the results were visualized using enhanced chemiluminescence (ECL, cat. No. 32109, Thermo Fisher Scientific, Waltham, MA, USA) detection system and documented by the ChemiDoc MP Imaging System (Bio-Rad, Hercules, CA, USA ). The intensity of the bands was measured in ImageLab (Bio-Rad, Hercules, CA, USA).
4.8. Immunofluorescent Staining
The cells were seeded into 12-well plates and grown on sterile ⌀18 mm glass coverslips. After 24 h, the cells were treated by the 5 mM CP concentration for another 24 h (incubator). Coverslips were fixed in 4% paraformaldehyde (PFA, cat. No. 28906, Thermo Fisher Scientific, Waltham, MA, USA; 20 min, RT). All further steps were preceded by washing the cells three times with PBS (5 min, RT). The fixed cells were permeabilized with the 0.25% Triton X-100 in PBS (10 min, RT). To block non-specific interactions 5% BSA (cat. No. 9048-46-8, Merck KGaA, Merck KGaA, Darmstadt, Germany) in PBS was used (20 min, RT). The next step was incubation with CK19, vimentin and N-cadherin primary antibodies (1:100 in BSA, 1 h, RT). Then, the cells were incubated with secondary antibodies (Alexa Fluor 594, Thermo Fisher Scientific, Waltham, MA, USA, 1:200 in PBS, 1 h, RT, dark). Alexa Fluor 488 phalloidin (cat. No. A12379,) was used for the labeling of F-actin (1:20 in PBS, 20 min, RT, dark). Cell nuclei were stained with DAPI (1:100 in distilled water, 10 min, RT, dark). The coverslips were mounted with Aqua Poly/Mount (cat. No. 18606-5, Polysciences) and photographed using the C1 confocal microscope (Nikon, Tokyo, Japan) and 100× oil immersion objective. The images were collected at the brightest signals of protein using Nikon EZ-C1 software (Nikon, Tokyo, Japan). Laser power, gains, and pixel dwell time were kept constant for all repeats for each protein.
To quantitatively confirm the fluorescence level of the selected proteins, analogous labeling steps were performed on the suspension of transfected and non-transfected untreated and treated MCF-7 and MDA-MB-231 cells. The staining results were measured by the microcapillary flow cytometry system and analyzed using FlowJo software (version 10.07, BD Biosciences, Frankin Lakes, NJ, USA).
4.9. Transwell Cell Migration and Invasion Assays
The migration and invasion of the non-tumorigenic cells and mammary gland cancer cell lines were assessed using cell culture inserts (cat. No. 140620, Thermo Fisher Scientific, Waltham, MA, USA). The inserts were put into a 24-well plate. Control and treated cells were transfected and non-transfected cells were plated on inserts at a density of 105 cells per well in a culture medium supplemented with 1% FBS. Under the inserts, a medium with 20% FBS was added to attract the cells to pass through the insert pores. For the invasion assay Matrigel (cat. No. 354277, Corning, New York, NY, USA) was prepared by diluting the stock solution in the 1:4 ratio in PBS, poured onto the inner surface of the insert, and polymerized for 24 h. The control and treated cells were plated on the surface of the Matrigel at a density analogous to the cell migration assay. The experimental assessment of the invasive potential of the cells was carried out for 24 h in standard culture conditions (95% humidity, 5% CO2, and 37 °C).
In both experiments, transwells were fixed with a 3.7% paraformaldehyde solution. The cells were permeabilized using the 100% ice-cold methanol (621990426, Avantor, Toruń, Poland) solution (20 min, RT). The next step was staining with the 0.4% crystal violet (cat. No. C0775, Thermo Fisher Scientific, Waltham, MA, USA) solution (15 min, RT). Noninvasive/nonmigrating cells were removed from the inner insert surfaces using the cotton swabs. The outer surface of the insert membrane was photographed using a light microscope with a CCD digital camera (DS-5Mc-U1). The percentage values of invasive and migrating cells were calculated using ImageJ software (NIH).
4.10. Clonogenic Assay
Transfected and non-transfected MCF-7 and MDA-MB-231 cells were treated with 5 mM concentration of the CP for 24 h or left untreated in the case of control cells. Then, the cells were seeded into Petri dishes at a density of 1 × 103 cells and cultured for 14 days in standard conditions (37 °C and 5% CO2). The resulting colonies were fixed with 4% paraformaldehyde in PBS (20 min, RT), stained using a 0.4% crystal violet in PBS (15 min, RT), and washed with PBS. The results were documented using ChemiDoc MP Imaging System. The number of colonies was measured by ImageJ software (NIH).
4.11. Wound Healing
The transfected and non-transfected cells of both lines were seeded and grown in 6-well plates until reaching 100% confluency in standard cell culture conditions. The 100 μL sterile pipette tips were used to scratch the surface in the center of each well. The cells were treated with 5 mM CP or left untreated in the case of control cells. Experiment was performed in the inverted motorized microscope (Zeiss) equipped with an incubation system for live-cell imaging (Pecon) containing EC Plan-Neofluar 10×/0.30 Ph1 air objective, Axiocam 503 mono camera, and ZEN 2 software. The wound surface and other parameters were measured using ImageJ software.
4.12. Statistical Analysis
All procedures were performed in at least three independent replicates. In the case of significantly different results, they were discarded, and the number of repetitions increased accordingly. The obtained results were analyzed with the GraphPadPrism 9 Software. The significance was assumed at p > 0.05. The data were compared using the Kruskal–Wallis test with the exception of the apoptosis data (Kruskal–Wallis test with Dunn’s multiple comparisons test), the MTT assay (Wilcoxon test), and the cell cycle data (two-way ANOVA).