Folding and Insertion of Transmembrane Helices at the ER
Abstract
1. Introduction
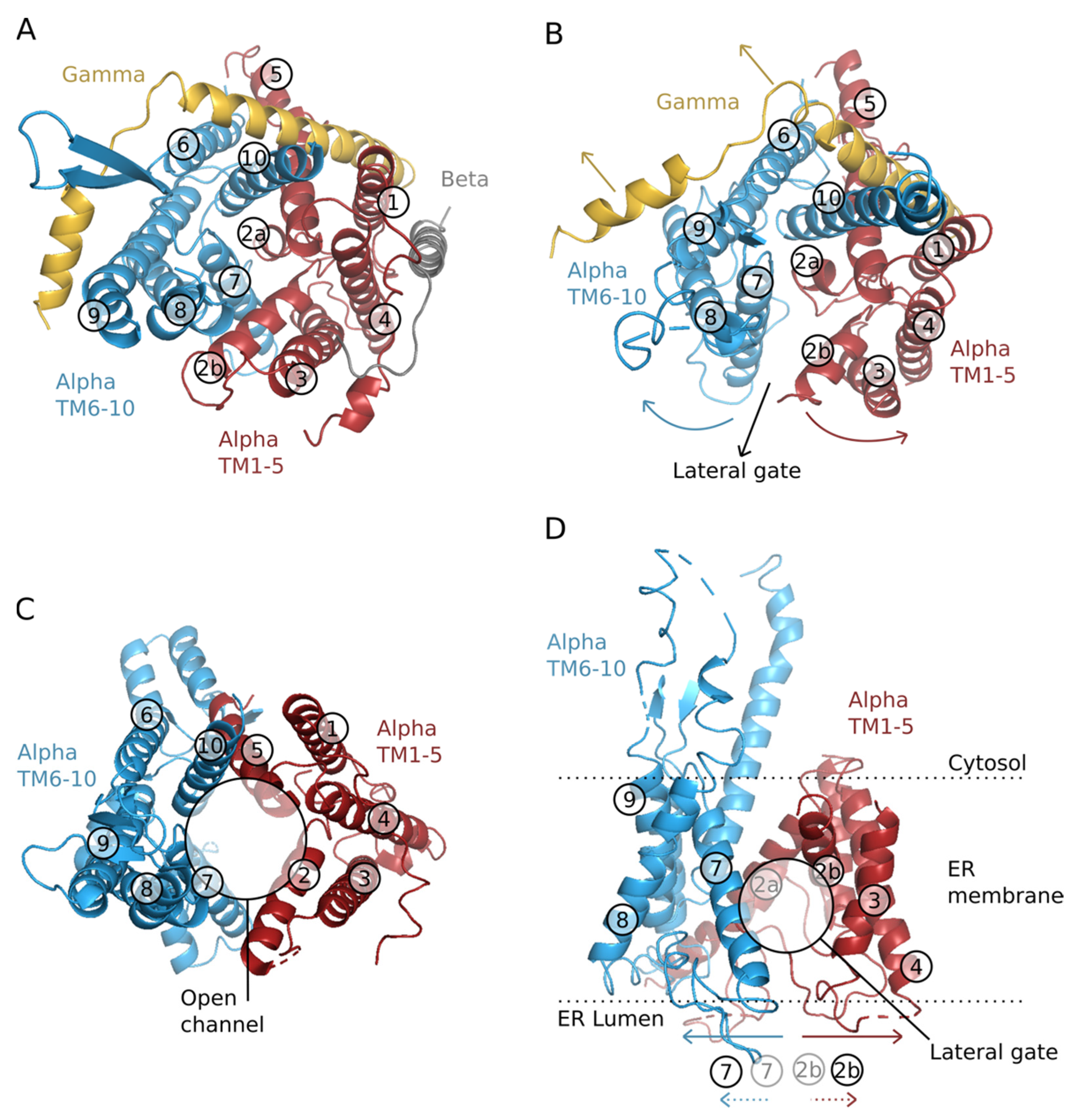
2. Structure-Function of the Translocon
3. Targeting to the Translocon
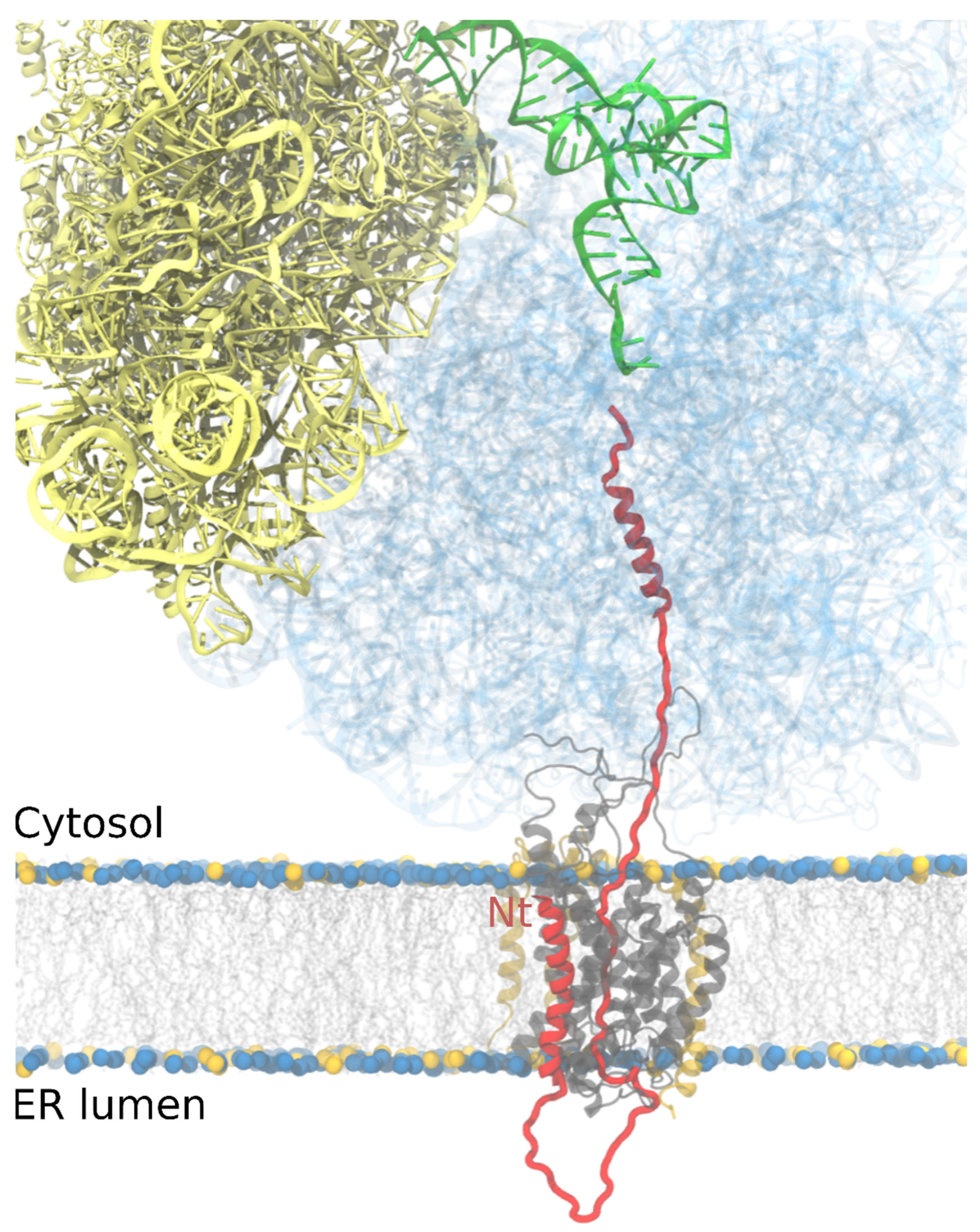
4. Getting across the Membrane
5. Integrating into the Membrane
6. Where Does Folding of TM Helices Occur?
7. Route Into, Through, and Out of the Translocon
8. Exploring the Limits of TM Domain Insertion
9. Concluding Remarks
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
Abbreviations
References
- Ito, K.; Shimokawa-Chiba, N.; Chiba, S. Sec translocon has an insertase-like function in addition to polypeptide conduction through the channel. F1000Research 2019, 8, 2126. [Google Scholar] [CrossRef]
- Farkas, A.; Bohnsack, K.E. Capture and delivery of tail-anchored proteins to the endoplasmic reticulum. J. Cell Biol. 2021, 220, e202105004. [Google Scholar] [CrossRef] [PubMed]
- Mateja, A.; Keenan, R.J. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 2018, 51, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, P.J.; Hegde, R.S. The role of EMC during membrane protein biogenesis. Trends Cell Biol. 2019, 29, 371–384. [Google Scholar] [CrossRef] [PubMed]
- Wideman, J.G. The ubiquitous and ancient ER membrane protein complex (EMC): Tether or not? F1000Research 2015, 4, 624. [Google Scholar] [CrossRef]
- Rapoport, T.A.; Li, L.; Park, E. Structural and mechanistic insights into protein translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390. [Google Scholar] [CrossRef]
- Bano-Polo, M.; Martinez-Garay, C.A.; Grau, B.; Martinez-Gil, L.; Mingarro, I. Membrane insertion and topology of the translocon-associated protein (TRAP) gamma subunit. Biochim. Biophys. Acta Biomembr. 2017, 1859, 903–909. [Google Scholar] [CrossRef]
- Pfeffer, S.; Dudek, J.; Schaffer, M.; Ng, B.G.; Albert, S.; Plitzko, J.M.; Baumeister, W.; Zimmermann, R.; Freeze, H.H.; Engel, B.D.; et al. Dissecting the molecular organization of the translocon-associated protein complex. Nat. Commun. 2017, 8, 14516. [Google Scholar] [CrossRef]
- Sommer, N.; Junne, T.; Kalies, K.U.; Spiess, M.; Hartmann, E. TRAP assists membrane protein topogenesis at the mammalian ER membrane. Biochim. Biophys. Acta 2013, 1833, 3104–3111. [Google Scholar] [CrossRef]
- Braunger, K.; Pfeffer, S.; Shrimal, S.; Gilmore, R.; Berninghausen, O.; Mandon, E.C.; Becker, T.; Forster, F.; Beckmann, R. Structural basis for coupling protein transport and N-glycosylation at the mammalian endoplasmic reticulum. Science 2018, 360, 215–219. [Google Scholar] [CrossRef]
- Gemmer, M.; Forster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340. [Google Scholar] [CrossRef] [PubMed]
- Chitwood, P.J.; Juszkiewicz, S.; Guna, A.; Shao, S.; Hegde, R.S. EMC is required to initiate accurate membrane protein topogenesis. Cell 2018, 175, 1507–1519.e16. [Google Scholar] [CrossRef] [PubMed]
- Van den Berg, B.; Clemons, W.M., Jr.; Collinson, I.; Modis, Y.; Hartmann, E.; Harrison, S.C.; Rapoport, T.A. X-ray structure of a protein-conducting channel. Nature 2004, 427, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Osborne, A.R.; Rapoport, T.A.; van den Berg, B. Protein translocation by the Sec61/SecY channel. Annu. Rev. Cell Dev. Biol. 2005, 21, 529–550. [Google Scholar] [CrossRef]
- Voorhees, R.M.; Hegde, R.S. Toward a structural understanding of co-translational protein translocation. Curr. Opin. Cell Biol. 2016, 41, 91–99. [Google Scholar] [CrossRef]
- Gumbart, J.; Schulten, K. The roles of pore ring and plug in the SecY protein-conducting channel. J. Gen. Physiol. 2008, 132, 709–719. [Google Scholar] [CrossRef]
- Park, E.; Rapoport, T.A. Preserving the membrane barrier for small molecules during bacterial protein translocation. Nature 2011, 473, 239–242. [Google Scholar] [CrossRef] [PubMed]
- Junne, T.; Kocik, L.; Spiess, M. The hydrophobic core of the Sec61 translocon defines the hydrophobicity threshold for membrane integration. Mol. Biol. Cell 2010, 21, 1662–1670. [Google Scholar] [CrossRef]
- Martinez-Gil, L.; Sauri, A.; Marti-Renom, M.A.; Mingarro, I. Membrane protein integration into the endoplasmic reticulum. FEBS J. 2011, 278, 3846–3858. [Google Scholar] [CrossRef]
- Johnson, A.E.; van Waes, M.A. The translocon: A dynamic gateway at the ER membrane. Annu. Rev. Cell Dev. Biol. 1999, 15, 799–842. [Google Scholar] [CrossRef]
- Keenan, R.J.; Freymann, D.M.; Stroud, R.M.; Walter, P. The signal recognition particle. Annu. Rev. Biochem. 2001, 70, 755–775. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Hegde, R.S. Membrane protein insertion at the endoplasmic reticulum. Annu. Rev. Cell Dev. Biol. 2011, 27, 25–56. [Google Scholar] [CrossRef] [PubMed]
- Frauenfeld, J.; Gumbart, J.; Sluis, E.O.; Funes, S.; Gartmann, M.; Beatrix, B.; Mielke, T.; Berninghausen, O.; Becker, T.; Schulten, K.; et al. Cryo-EM structure of the ribosome-SecYE complex in the membrane environment. Nat. Struct. Mol. Biol. 2011, 18, 614–621. [Google Scholar] [CrossRef] [PubMed]
- Dowhan, W.; Vitrac, H.; Bogdanov, M. Lipid-assisted membrane protein folding and topogenesis. Protein J. 2019, 38, 274–288. [Google Scholar] [CrossRef]
- Whitley, P.; Mingarro, I. Stitching proteins into membranes, not sew simple. Biol. Chem. 2014, 395, 1417–1424. [Google Scholar] [CrossRef] [PubMed]
- Higy, M.; Junne, T.; Spiess, M. Topogenesis of membrane proteins at the endoplasmic reticulum. Biochemistry 2004, 43, 12716–12722. [Google Scholar] [CrossRef]
- Sauri, A.; Tamborero, S.; Martinez-Gil, L.; Johnson, A.E.; Mingarro, I. Viral membrane protein topology is dictated by multiple determinants in its sequence. J. Mol. Biol. 2009, 387, 113–128. [Google Scholar] [CrossRef] [PubMed]
- Voorhees, R.M.; Hegde, R.S. Structure of the Sec61 channel opened by a signal sequence. Science 2016, 351, 88–91. [Google Scholar] [CrossRef]
- Mingarro, I.; Nilsson, I.; Whitley, P.; von Heijne, G. Different conformations of nascent polypeptides during translocation across the ER membrane. BMC Cell Biol. 2000, 1, 3. [Google Scholar] [CrossRef]
- Whitley, P.; Nilsson, I.M.; von Heijne, G. A nascent secretory protein may traverse the ribosome/endoplasmic reticulum translocase complex as an extended chain. J. Biol. Chem. 1996, 271, 6241–6244. [Google Scholar] [CrossRef]
- Hessa, T.; Kim, H.; Bihlmaier, K.; Lundin, C.; Boekel, J.; Andersson, H.; Nilsson, I.; White, S.H.; von Heijne, G. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 2005, 433, 377–381. [Google Scholar] [CrossRef] [PubMed]
- Hessa, T.; Meindl-Beinker, N.M.; Bernsel, A.; Kim, H.; Sato, Y.; Lerch-Bader, M.; Nilsson, I.; White, S.H.; von Heijne, G. Molecular code for transmembrane-helix recognition by the Sec61 translocon. Nature 2007, 450, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Devaraneni, P.K.; Conti, B.; Matsumura, Y.; Yang, Z.; Johnson, A.E.; Skach, W.R. Stepwise insertion and inversion of a type II signal anchor sequence in the ribosome-Sec61 translocon complex. Cell 2011, 146, 134–147. [Google Scholar] [CrossRef] [PubMed]
- Ojemalm, K.; Watson, H.R.; Roboti, P.; Cross, B.C.; Warwicker, J.; von Heijne, G.; High, S. Positional editing of transmembrane domains during ion channel assembly. J. Cell Sci. 2013, 126, 464–472. [Google Scholar] [CrossRef]
- Ota, K.; Sakaguchi, M.; von Heijne, G.; Hamasaki, N.; Mihara, K. Forced transmembrane orientation of hydrophilic polypeptide segments in multispanning membrane proteins. Mol. Cell 1998, 2, 495–503. [Google Scholar] [CrossRef]
- Sadlish, H.; Pitonzo, D.; Johnson, A.E.; Skach, W.R. Sequential triage of transmembrane segments by Sec61alpha during biogenesis of a native multispanning membrane protein. Nat. Struct. Mol. Biol. 2005, 12, 870–878. [Google Scholar] [CrossRef]
- Sauri, A.; McCormick, P.J.; Johnson, A.E.; Mingarro, I. Sec61alpha and TRAM are sequentially adjacent to a nascent viral membrane protein during its ER integration. J. Mol. Biol. 2007, 366, 366–374. [Google Scholar] [CrossRef]
- Sauri, A.; Saksena, S.; Salgado, J.; Johnson, A.E.; Mingarro, I. Double-spanning plant viral movement protein integration into the endoplasmic reticulum membrane is signal recognition particle-dependent, translocon-mediated, and concerted. J. Biol. Chem. 2005, 280, 25907–25912. [Google Scholar] [CrossRef]
- Cymer, F.; von Heijne, G.; White, S.H. Mechanisms of integral membrane protein insertion and folding. J. Mol. Biol. 2015, 427, 999–1022. [Google Scholar] [CrossRef]
- White, S.H.; Ladokhin, A.S.; Jayasinghe, S.; Hristova, K. How membranes shape protein structure. J. Biol. Chem. 2001, 276, 32395–32398. [Google Scholar] [CrossRef]
- Wimley, W.C.; Creamer, T.P.; White, S.H. Solvation energies of amino acid side chains and backbone in a family of host-guest pentapeptides. Biochemistry 1996, 35, 5109–5124. [Google Scholar] [CrossRef] [PubMed]
- Wimley, W.C.; White, S.H. Designing transmembrane alpha-helices that insert spontaneously. Biochemistry 2000, 39, 4432–4442. [Google Scholar] [CrossRef] [PubMed]
- Woolhead, C.A.; McCormick, P.J.; Johnson, A.E. Nascent membrane and secretory proteins differ in FRET-detected folding far inside the ribosome and in their exposure to ribosomal proteins. Cell 2004, 116, 725–736. [Google Scholar] [CrossRef]
- Lu, J.; Deutsch, C. Secondary structure formation of a transmembrane segment in Kv channels. Biochemistry 2005, 44, 8230–8243. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Deutsch, C. Folding zones inside the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2005, 12, 1123–1129. [Google Scholar] [CrossRef]
- Bhushan, S.; Gartmann, M.; Halic, M.; Armache, J.P.; Jarasch, A.; Mielke, T.; Berninghausen, O.; Wilson, D.N.; Beckmann, R. alpha-Helical nascent polypeptide chains visualized within distinct regions of the ribosomal exit tunnel. Nat. Struct. Mol. Biol. 2010, 17, 313–317. [Google Scholar] [CrossRef]
- Bano-Polo, M.; Baeza-Delgado, C.; Tamborero, S.; Hazel, A.; Grau, B.; Nilsson, I.; Whitley, P.; Gumbart, J.C.; von Heijne, G.; Mingarro, I. Transmembrane but not soluble helices fold inside the ribosome tunnel. Nat. Commun. 2018, 9, 5246. [Google Scholar] [CrossRef]
- Nilsson, O.B.; Hedman, R.; Marino, J.; Wickles, S.; Bischoff, L.; Johansson, M.; Muller-Lucks, A.; Trovato, F.; Puglisi, J.D.; O’Brien, E.P.; et al. Cotranslational protein folding inside the ribosome exit tunnel. Cell Rep. 2015, 12, 1533–1540. [Google Scholar] [CrossRef]
- Wruck, F.; Tian, P.; Kudva, R.; Best, R.B.; von Heijne, G.; Tans, S.J.; Katranidis, A. The ribosome modulates folding inside the ribosomal exit tunnel. Commun. Biol. 2021, 4, 523. [Google Scholar] [CrossRef]
- Tu, L.; Khanna, P.; Deutsch, C. Transmembrane segments form tertiary hairpins in the folding vestibule of the ribosome. J. Mol. Biol. 2014, 426, 185–198. [Google Scholar] [CrossRef]
- Baars, L.; Wagner, S.; Wickstrom, D.; Klepsch, M.; Ytterberg, A.J.; van Wijk, K.J.; de Gier, J.W. Effects of SecE depletion on the inner and outer membrane proteomes of Escherichia coli. J. Bacteriol. 2008, 190, 3505–3525. [Google Scholar] [CrossRef]
- Berhanu, S.; Ueda, T.; Kuruma, Y. Artificial photosynthetic cell producing energy for protein synthesis. Nat. Commun. 2019, 10, 1325. [Google Scholar] [CrossRef] [PubMed]
- Matsubayashi, H.; Kuruma, Y.; Ueda, T. In vitro synthesis of the E. coli Sec translocon from DNA. Angew Chem. 2014, 53, 7535–7538. [Google Scholar] [CrossRef]
- Eaglesfield, R.; Madsen, M.A.; Sanyal, S.; Reboud, J.; Amtmann, A. Cotranslational recruitment of ribosomes in protocells recreates a translocon-independent mechanism of proteorhodopsin biogenesis. iScience 2021, 24, 102429. [Google Scholar] [CrossRef] [PubMed]
- Doan, K.N.; Grevel, A.; Martensson, C.U.; Ellenrieder, L.; Thornton, N.; Wenz, L.S.; Opalinski, L.; Guiard, B.; Pfanner, N.; Becker, T. The mitochondrial import complex MIM functions as main translocase for alpha-helical outer membrane proteins. Cell Rep. 2020, 31, 107567. [Google Scholar] [CrossRef] [PubMed]
- Glick, B.S.; Von Heijne, G. Saccharomyces cerevisiae mitochondria lack a bacterial-type sec machinery. Protein Sci. 1996, 5, 2651–2652. [Google Scholar] [CrossRef] [PubMed]
- Jia, L.; Dienhart, M.; Schramp, M.; McCauley, M.; Hell, K.; Stuart, R.A. Yeast Oxa1 interacts with mitochondrial ribosomes: The importance of the C-terminal region of Oxa1. EMBO J. 2003, 22, 6438–6447. [Google Scholar] [CrossRef] [PubMed]
- Mulkidjanian, A.Y.; Galperin, M.Y.; Koonin, E.V. Co-evolution of primordial membranes and membrane proteins. Trends Biochem. Sci. 2009, 34, 206–215. [Google Scholar] [CrossRef]
- Cross, B.C.; High, S. Dissecting the physiological role of selective transmembrane-segment retention at the ER translocon. J. Cell Sci. 2009, 122, 1768–1777. [Google Scholar] [CrossRef]
- Park, E.; Rapoport, T.A. Mechanisms of Sec61/SecY-mediated protein translocation across membranes. Annu. Rev. Biophys. 2012, 41, 21–40. [Google Scholar] [CrossRef]
- Voss, N.R.; Gerstein, M.; Steitz, T.A.; Moore, P.B. The geometry of the ribosomal polypeptide exit tunnel. J. Mol. Biol. 2006, 360, 893–906. [Google Scholar] [CrossRef] [PubMed]
- Gumbart, J.; Chipot, C.; Schulten, K. Free energy of nascent-chain folding in the translocon. J. Am. Chem. Soc. 2011, 133, 7602–7607. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Egea, P.F.; Stroud, R.M. Lateral opening of a translocon upon entry of protein suggests the mechanism of insertion into membranes. Proc. Natl. Acad. Sci. USA 2010, 107, 17182–17187. [Google Scholar] [CrossRef] [PubMed]
- Gogala, M.; Becker, T.; Beatrix, B.; Armache, J.P.; Barrio-Garcia, C.; Berninghausen, O.; Beckmann, R. Structures of the Sec61 complex engaged in nascent peptide translocation or membrane insertion. Nature 2014, 506, 107–110. [Google Scholar] [CrossRef] [PubMed]
- Gumbart, J.C.; Teo, I.; Roux, B.; Schulten, K. Reconciling the roles of kinetic and thermodynamic factors in membrane-protein insertion. J. Am. Chem. Soc. 2013, 135, 2291–2297. [Google Scholar] [CrossRef] [PubMed]
- Mackinnon, A.L.; Paavilainen, V.O.; Sharma, A.; Hegde, R.S.; Taunton, J. An allosteric Sec61 inhibitor traps nascent transmembrane helices at the lateral gate. eLife 2014, 3, e01483. [Google Scholar] [CrossRef]
- Niesen, M.J.M.; Zimmer, M.H.; Miller, T.F., 3rd. Dynamics of co-translational membrane protein integration and translocation via the sec translocon. J. Am. Chem. Soc. 2020, 142, 5449–5460. [Google Scholar] [CrossRef]
- Park, E.; Menetret, J.F.; Gumbart, J.C.; Ludtke, S.J.; Li, W.; Whynot, A.; Rapoport, T.A.; Akey, C.W. Structure of the SecY channel during initiation of protein translocation. Nature 2014, 506, 102–106. [Google Scholar] [CrossRef]
- Mercier, E.; Wang, X.; Maiti, M.; Wintermeyer, W.; Rodnina, M.V. Lateral gate dynamics of the bacterial translocon during cotranslational membrane protein insertion. Proc. Natl. Acad. Sci. USA 2021, 118, e2100474118. [Google Scholar] [CrossRef]
- Ge, Y.; Draycheva, A.; Bornemann, T.; Rodnina, M.V.; Wintermeyer, W. Lateral opening of the bacterial translocon on ribosome binding and signal peptide insertion. Nat. Commun. 2014, 5, 5263. [Google Scholar] [CrossRef]
- Pfeffer, S.; Burbaum, L.; Unverdorben, P.; Pech, M.; Chen, Y.; Zimmermann, R.; Beckmann, R.; Forster, F. Structure of the native Sec61 protein-conducting channel. Nat. Commun. 2015, 6, 8403. [Google Scholar] [CrossRef] [PubMed]
- Shurtleff, M.J.; Itzhak, D.N.; Hussmann, J.A.; Schirle Oakdale, N.T.; Costa, E.A.; Jonikas, M.; Weibezahn, J.; Popova, K.D.; Jan, C.H.; Sinitcyn, P.; et al. The ER membrane protein complex interacts cotranslationally to enable biogenesis of multipass membrane proteins. eLife 2018, 7, e37018. [Google Scholar] [CrossRef] [PubMed]
- Bowie, J.U. Solving the membrane protein folding problem. Nature 2005, 438, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Nicolaus, F.; Metola, A.; Mermans, D.; Liljenstrom, A.; Krc, A.; Abdullahi, S.M.; Zimmer, M.; Miller Iii, T.F.; von Heijne, G. Residue-by-residue analysis of cotranslational membrane protein integration in vivo. eLife 2021, 10, e64302. [Google Scholar] [CrossRef] [PubMed]
- Gumbart, J.C.; Chipot, C. Decrypting protein insertion through the translocon with free-energy calculations. Biochim. Biophys. Acta 2016, 1858, 1663–1671. [Google Scholar] [CrossRef]
- Zhang, B.; Miller, T.F., 3rd. Hydrophobically stabilized open state for the lateral gate of the Sec translocon. Proc. Natl. Acad. Sci. USA 2010, 107, 5399–5404. [Google Scholar] [CrossRef]
- Duong, F.; Wickner, W. Sec-dependent membrane protein biogenesis: SecYEG, preprotein hydrophobicity and translocation kinetics control the stop-transfer function. EMBO J. 1998, 17, 696–705. [Google Scholar] [CrossRef]
- Trueman, S.F.; Mandon, E.C.; Gilmore, R. Translocation channel gating kinetics balances protein translocation efficiency with signal sequence recognition fidelity. Mol. Biol. Cell 2011, 22, 2983–2993. [Google Scholar] [CrossRef]
- Rapp, M.; Granseth, E.; Seppala, S.; von Heijne, G. Identification and evolution of dual-topology membrane proteins. Nat. Struct. Mol. Biol. 2006, 13, 112–116. [Google Scholar] [CrossRef]
- Seppala, S.; Slusky, J.S.; Lloris-Garcera, P.; Rapp, M.; von Heijne, G. Control of membrane protein topology by a single C-terminal residue. Science 2010, 328, 1698–1700. [Google Scholar] [CrossRef]
- Seurig, M.; Ek, M.; von Heijne, G.; Fluman, N. Dynamic membrane topology in an unassembled membrane protein. Nat. Chem. Biol. 2019, 15, 945–948. [Google Scholar] [CrossRef] [PubMed]
- Van Lehn, R.C.; Zhang, B.; Miller, T.F., 3rd. Regulation of multispanning membrane protein topology via post-translational annealing. eLife 2015, 4, e08697. [Google Scholar] [CrossRef]
- Baeza-Delgado, C.; von Heijne, G.; Marti-Renom, M.A.; Mingarro, I. Biological insertion of computationally designed short transmembrane segments. Sci. Rep. 2016, 6, 23397. [Google Scholar] [CrossRef] [PubMed]
- Andersen, O.S.; Koeppe, R.E., 2nd. Bilayer thickness and membrane protein function: An energetic perspective. Annu. Rev. Biophys. Biomol. Struct. 2007, 36, 107–130. [Google Scholar] [CrossRef] [PubMed]
- Mouritsen, O.G.; Bloom, M. Mattress model of lipid-protein interactions in membranes. Biophys. J. 1984, 46, 141–153. [Google Scholar] [CrossRef]
- Baeza-Delgado, C.; Marti-Renom, M.A.; Mingarro, I. Structure-based statistical analysis of transmembrane helices. Eur. Biophys. J. 2013, 42, 199–207. [Google Scholar] [CrossRef] [PubMed]
- Chen, A.; Moy, V.T. Cross-linking of cell surface receptors enhances cooperativity of molecular adhesion. Biophys. J. 2000, 78, 2814–2820. [Google Scholar] [CrossRef][Green Version]
- Mitra, K.; Ubarretxena-Belandia, I.; Taguchi, T.; Warren, G.; Engelman, D.M. Modulation of the bilayer thickness of exocytic pathway membranes by membrane proteins rather than cholesterol. Proc. Natl. Acad. Sci. USA 2004, 101, 4083–4088. [Google Scholar] [CrossRef]
- Rawicz, W.; Olbrich, K.C.; McIntosh, T.; Needham, D.; Evans, E. Effect of chain length and unsaturation on elasticity of lipid bilayers. Biophys. J. 2000, 79, 328–339. [Google Scholar] [CrossRef]
- Killian, J.A. Hydrophobic mismatch between proteins and lipids in membranes. Biochim. Biophys. Acta 1998, 1376, 401–415. [Google Scholar] [CrossRef]
- Krishnakumar, S.S.; London, E. Effect of sequence hydrophobicity and bilayer width upon the minimum length required for the formation of transmembrane helices in membranes. J. Mol. Biol. 2007, 374, 671–687. [Google Scholar] [CrossRef]
- Jaud, S.; Fernandez-Vidal, M.; Nilsson, I.; Meindl-Beinker, N.M.; Hubner, N.C.; Tobias, D.J.; von Heijne, G.; White, S.H. Insertion of short transmembrane helices by the Sec61 translocon. Proc. Natl. Acad. Sci. USA 2009, 106, 11588–11593. [Google Scholar] [CrossRef]
- Ulmschneider, M.B.; Ulmschneider, J.P.; Schiller, N.; Wallace, B.A.; von Heijne, G.; White, S.H. Spontaneous transmembrane helix insertion thermodynamically mimics translocon-guided insertion. Nat. Commun. 2014, 5, 4863. [Google Scholar] [CrossRef] [PubMed]
- Grau, B.; Javanainen, M.; Garcia-Murria, M.J.; Kulig, W.; Vattulainen, I.; Mingarro, I.; Martinez-Gil, L. The role of hydrophobic matching on transmembrane helix packing in cells. Cell Stress 2017, 1, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Bano-Polo, M.; Martinez-Gil, L.; Wallner, B.; Nieva, J.L.; Elofsson, A.; Mingarro, I. Charge pair interactions in transmembrane helices and turn propensity of the connecting sequence promote helical hairpin insertion. J. Mol. Biol. 2013, 425, 830–840. [Google Scholar] [CrossRef] [PubMed]
- Monne, M.; Hermansson, M.; von Heijne, G. A turn propensity scale for transmembrane helices. J. Mol. Biol. 1999, 288, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Bano-Polo, M.; Baeza-Delgado, C.; Orzaez, M.; Marti-Renom, M.A.; Abad, C.; Mingarro, I. Polar/Ionizable residues in transmembrane segments: Effects on helix-helix packing. PLoS ONE 2012, 7, e44263. [Google Scholar] [CrossRef]
- MacCallum, J.L.; Bennett, W.F.; Tieleman, D.P. Distribution of amino acids in a lipid bilayer from computer simulations. Biophys. J. 2008, 94, 3393–3404. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Gil, L.; Perez-Gil, J.; Mingarro, I. The surfactant peptide KL4 sequence is inserted with a transmembrane orientation into the endoplasmic reticulum membrane. Biophys. J. 2008, 95, L36–L38. [Google Scholar] [CrossRef]
- Garcia-Saez, A.J.; Coraiola, M.; Dalla Serra, M.; Mingarro, I.; Menestrina, G.; Salgado, J. Peptides derived from apoptotic Bax and Bid reproduce the poration activity of the parent full-length proteins. Biophys. J. 2005, 88, 3976–3990. [Google Scholar] [CrossRef]
- Garcia-Saez, A.J.; Coraiola, M.; Serra, M.D.; Mingarro, I.; Muller, P.; Salgado, J. Peptides corresponding to helices 5 and 6 of Bax can independently form large lipid pores. FEBS J. 2006, 273, 971–981. [Google Scholar] [CrossRef]
- Koneru, J.K.; Prakashchand, D.D.; Dube, N.; Ghosh, P.; Mondal, J. Spontaneous transmembrane pore formation by short-chain synthetic peptide. Biophys. J. 2021, 120, 4557–4574. [Google Scholar] [CrossRef]
- Choma, C.; Gratkowski, H.; Lear, J.D.; DeGrado, W.F. Asparagine-mediated self-association of a model transmembrane helix. Nat. Struct. Biol. 2000, 7, 161–166. [Google Scholar] [CrossRef]
- Zhou, F.X.; Cocco, M.J.; Russ, W.P.; Brunger, A.T.; Engelman, D.M. Interhelical hydrogen bonding drives strong interactions in membrane proteins. Nat. Struct. Biol. 2000, 7, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.X.; Merianos, H.J.; Brunger, A.T.; Engelman, D.M. Polar residues drive association of polyleucine transmembrane helices. Proc. Natl. Acad. Sci. USA 2001, 98, 2250–2255. [Google Scholar] [CrossRef]
- Almada, J.C.; Bortolotti, A.; Ruysschaert, J.M.; de Mendoza, D.; Inda, M.E.; Cybulski, L.E. Interhelical H-bonds modulate the activity of a polytopic transmembrane kinase. Biomolecules 2021, 11, 938. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Mitra, N.; Gratkowski, H.; Vilaire, G.; Litvinov, R.; Nagasami, C.; Weisel, J.W.; Lear, J.D.; DeGrado, W.F.; Bennett, J.S. Activation of integrin alphaIIbbeta3 by modulation of transmembrane helix associations. Science 2003, 300, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sato, Y.; Hessa, T.; von Heijne, G.; Lee, J.K.; Kodama, I.; Sakaguchi, M.; Uozumi, N. Contribution of hydrophobic and electrostatic interactions to the membrane integration of the Shaker K+ channel voltage sensor domain. Proc. Natl. Acad. Sci. USA 2007, 104, 8263–8268. [Google Scholar] [CrossRef]
- Hou, B.; Lin, P.J.; Johnson, A.E. Membrane protein TM segments are retained at the translocon during integration until the nascent chain cues FRET-detected release into bulk lipid. Mol. Cell 2012, 48, 398–408. [Google Scholar] [CrossRef]
- Russ, W.P.; Engelman, D.M. The GxxxG motif: A framework for transmembrane helix-helix association. J. Mol. Biol. 2000, 296, 911–919. [Google Scholar] [CrossRef]
- Lu, Y.; Turnbull, I.R.; Bragin, A.; Carveth, K.; Verkman, A.S.; Skach, W.R. Reorientation of aquaporin-1 topology during maturation in the endoplasmic reticulum. Mol. Biol. Cell 2000, 11, 2973–2985. [Google Scholar] [CrossRef] [PubMed]
- Skach, W.R. Cellular mechanisms of membrane protein folding. Nat. Struct. Mol. Biol. 2009, 16, 606–612. [Google Scholar] [CrossRef] [PubMed]
- Tamborero, S.; Vilar, M.; Martinez-Gil, L.; Johnson, A.E.; Mingarro, I. Membrane insertion and topology of the translocating chain-associating membrane protein (TRAM). J. Mol. Biol. 2011, 406, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Pleiner, T.; Tomaleri, G.P.; Januszyk, K.; Inglis, A.J.; Hazu, M.; Voorhees, R.M. Structural basis for membrane insertion by the human ER membrane protein complex. Science 2020, 369, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Gumbart, J.; Trabuco, L.G.; Schreiner, E.; Villa, E.; Schulten, K. Regulation of the protein-conducting channel by a bound ribosome. Structure 2009, 17, 1453–1464. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Whitley, P.; Grau, B.; Gumbart, J.C.; Martínez-Gil, L.; Mingarro, I. Folding and Insertion of Transmembrane Helices at the ER. Int. J. Mol. Sci. 2021, 22, 12778. https://doi.org/10.3390/ijms222312778
Whitley P, Grau B, Gumbart JC, Martínez-Gil L, Mingarro I. Folding and Insertion of Transmembrane Helices at the ER. International Journal of Molecular Sciences. 2021; 22(23):12778. https://doi.org/10.3390/ijms222312778
Chicago/Turabian StyleWhitley, Paul, Brayan Grau, James C. Gumbart, Luis Martínez-Gil, and Ismael Mingarro. 2021. "Folding and Insertion of Transmembrane Helices at the ER" International Journal of Molecular Sciences 22, no. 23: 12778. https://doi.org/10.3390/ijms222312778
APA StyleWhitley, P., Grau, B., Gumbart, J. C., Martínez-Gil, L., & Mingarro, I. (2021). Folding and Insertion of Transmembrane Helices at the ER. International Journal of Molecular Sciences, 22(23), 12778. https://doi.org/10.3390/ijms222312778





