Abstract
Pseudomonas is characterized by its great capacity to colonize different ecological niches, but also by its antimicrobial resistance and pathogenicity, causing human, animal, or plant diseases. Raw and undercooked food is a potential carrier of foodborne disease. The aim of this study was to determine the occurrence of Pseudomonas spp. among raw vegetables, analysing their antimicrobial resistance, virulence, and molecular typing. A total of 163 Pseudomonas spp. isolates (12 different species) were recovered from 77 of the 145 analysed samples (53.1%) and were classified into 139 different pulsed-field gel electrophoresis patterns. Low antimicrobial resistance levels, but one multidrug-resistant isolate, were found. Among the 37 recovered P. aeruginosa strains, 28 sequence-types and nine serotypes were detected. Eleven OprD patterns and an insertion sequence (ISPa1635) truncating the oprD gene of one imipenem-resistant strain were found. Ten virulotypes were observed, including four exoU-positive and thirty-one exoS-positive strains. The lasR gene was absent in three ST155 strains and was truncated by different insertion sequences (ISPre2, IS1411, and ISPst7) in other three strains. High biofilm, motility, pigment, elastase, and rhamnolipid production were detected. Our study demonstrated a low occurrence of P. aeruginosa (18%) and low antimicrobial resistance, but a high number of virulence-related traits in these P. aeruginosa strains, highlighting their pathological importance.
1. Introduction
Vegetables and fresh fruit are important products in a healthy diet. In recent years, the search for a good lifestyle has led to an increased consumption of fresh products. Nevertheless, vegetables have become increasingly recognised as potential carriers of foodborne diseases due to various contamination sources, such as dust, soil, manure, irrigation water, or wild animal faeces [1,2]. Moreover, fresh vegetables which are grown close to the soil, are often consumed raw, exposing consumers to the risk of infection [3]. Most foodborne diseases are not reported, and sometimes outbreaks may affect a wide number of people. Thus, there is a special interest to know the epidemiology and spread of foodborne pathogens that are adaptable to different environmental conditions [4]. Additionally, bacteria can develop antimicrobial resistance due to spontaneous mutations or acquisition of resistance mechanisms by horizontal gene transfer. Antimicrobial resistant isolates can also spread antimicrobial resistance genes to other commensal and pathogenic bacteria [5]. The role of food in human exposure to antimicrobial resistant bacteria, as well as a reservoir of resistance genes, is becoming a growing food safety issue [3].
Pseudomonas is a non-fermenting Gram-negative bacterium that colonizes different niches, due to their metabolic capacity and broad potential for adaptation to different conditions [6]. This genus includes a wide variety of species, Pseudomonas aeruginosa being the most important one. This species is a major opportunistic human pathogen with increasing medical and veterinary importance. The significance of P. aeruginosa is marked by its great resistance to antimicrobials and antiseptics and the presence of multiple virulence factors [6]. P. aeruginosa uses its big arsenal of pathogenicity factors (including adhesins and secretion toxins, effector proteins, proteases, elastases and pigments) to interfere with host defences. The type 3 secretion system (T3SS) is a major virulence weapon that contributes to cytotoxicity and acute infections, injecting potent exotoxins called effectors (ExoU, ExoS, ExoY and ExoT) into cytoplasm of the host cell due to its syringe form [6,7]. The ExoU effector is associated with an increased risk of early clinical mortality [8,9]. Furthermore, it has been demonstrated that the predominance and persistence of this species in food and on surfaces of food processing plants is related to its ability to form biofilm. Most of these virulence factors are under the control of a cell density recognition mechanism called Quorum-Sensing (QS) [10,11]. P. aeruginosa possesses two well-defined and interrelated QS systems, las and rhl, which are used to regulate gene expression through the production and secretion of autoinducers, by lasI and rhlI genes, activating LasR and RhlR regulators, respectively [10].
Previous studies have given information about the detection of Pseudomonas spp. in food of animal or vegetable origin [1,3,12,13], but there are little data about their antimicrobial resistance and virulence phenotype. For this reason, the purpose of this study was to analyse and characterise the Pseudomonas spp. population in food vegetables, at both genotypic and phenotypic levels.
2. Results
2.1. Isolates of Pseudomonas spp.
In total, 163 Pseudomonas spp. were isolated from 77 vegetal samples (prevalence of 53.1%), belonging to (number of positive samples): lettuce (18), chard (14), potato (11), green bean (10), cucumber (9), zucchini (8) and onion/leek (7) (Table S1). Regarding each sample type, the highest prevalence was detected in lettuce (90%) and chard (70%) samples, and the lowest one was among the onion/leek samples (26.3%).
The 163 isolates were classified into twelve different Pseudomonas species. P. putida (51 isolates), P. aeruginosa (50 isolates) and P. mendocina (32 isolates) were the most abundant ones and were isolated from 46, 26 and 29 samples, respectively (Table 1). Thirty samples harboured more than one different Pseudomonas species (Table S1).

Table 1.
Description of the twelve different Pseudomonas species isolated from raw vegetables, including the number of isolates and the samples where they were recovered.
2.2. Antimicrobial Susceptibility Testing
The 163 Pseudomonas spp. isolates showed the following resistance percentages to: aztreonam (42%), imipenem (1.8%), meropenem (1.8%), doripenem (1.8%), piperacillin (0.6%), and ceftazidime (0.6%). The isolates showed susceptibility to the remaining seven antibiotics tested. Fifty-four per cent of the isolates were susceptible to all antibiotics tested (Table S1). Conversely, one P. fluorescens isolate (Ps876), recovered from a zucchini collected in an orchard, showed a multidrug-resistance phenotype, showing resistance to imipenem, meropenem, doripenem, ceftazidime and aztreonam (Table S1).
None of the 163 isolates showed class A carbapenemase, metallo-beta-lactamase (MBL) or extended spectrum beta-lactamase (ESBL) phenotypes, whereas the AmpC inducible phenotype was detected in 31% of the isolates, which included all P. aeruginosa isolates and the multidrug resistant P. fluorescens isolate (Ps876) (Table S1).
2.3. Molecular Typing
One hundred and thirty-nine different pulsed-field gel electrophoresis (PFGE) patterns were detected among the 163 Pseudomonas spp. (Table S1), and regarding the 50 P. aeruginosa isolates, 36 PFGE patterns were observed (Table 2). Indistinguishable patterns were only detected among isolates from the same sample, selecting one strain for next steps; except for two P. aeruginosa isolates that showed the same PFGE pattern, but they were recovered from two different chard samples (Table 2 and Table S1). One strain with different PFGE pattern and species per sample were included in further studies. Additionally, three P. putida isolates from the same cucumber were included because they showed the same PFGE profiles but different resistance phenotypes. After these criteria, 142 Pseudomonas spp. strains were chosen: 105 strains corresponding to Pseudomonas non-aeruginosa species and 37 P. aeruginosa strains (Table S1).

Table 2.
Genotypic and phenotypic characteristics of the 37 P. aeruginosa strains recovered from food vegetables.
Twenty-eight different sequence types (ST) were determined among the 37 P. aeruginosa strains using Multilocus sequence typing (MLST) method (Table 2). Eight of them (ST2416, ST2427-ST2432 and ST2448) were first described in this study and named by the MLST database. Seven ST were repeated more than once: ST155, ST274, ST982, ST1226, ST1228, ST2416 and ST2432 (Table 2 and Figure 1). The P. aeruginosa strains were distributed into three clusters (Cluster I, II and III) when a phylogenetic tree based on the MLST was obtained (Figure 1). Cluster I was an outlier and only included the Ps760 strain (ST2448). Cluster II included the new high-risk clone ST155, whereas the cluster III possessed the intercontinental clones, ST253, ST274 and ST395. The new ST were distributed between both clusters.
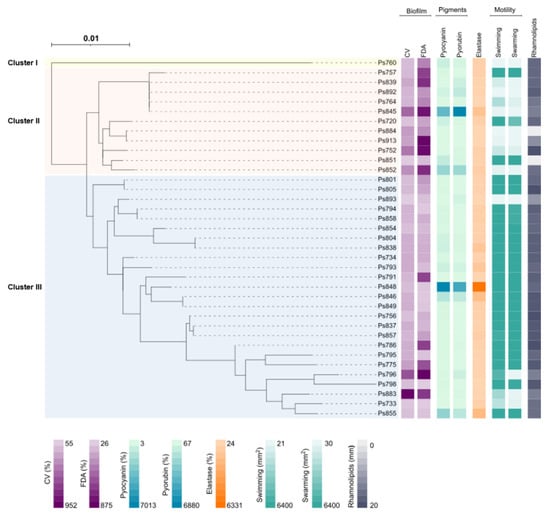
Figure 1.
Maximum-likelihood phylogenetic tree based on the sequence type (MLST) and phenotypic characteristics of the 37 P. aeruginosa strains analysed in this study. The phylogenetic tree was obtained using the IQTREE v.1.6.1 [14] software. The iTol v.4 [15] program was used to visualise the phylogenetic tree and to perform the eight heatmaps. In order: biofilm biomass production (CV), bacterial metabolic activity inside the biofilm (FDA), pyocyanin and pyorubin production, elastase activity, swimming and swarming motility and rhamnolipids production. Legend values belong to the minimum and maximum data for each phenotypic assay (Table 2). The three clusters (I, II and III) were marked with different colours: yellow (Cluster I), red (Cluster II) and blue (Cluster III).
2.4. Serotyping
Nine different serotypes were identified in these 37 P. aeruginosa strains, in addition to five non-agglutinable (Ps798, Ps839, Ps884, Ps892 and Ps913) and one poly-agglutinable (Ps796) strains. Serotype O:6 was the most predominant (40.5%), followed by O:5 (10.8%) and O:1 (8.1%) (Table 2). Serotypes O:2, O:7, O:10, O:12; O:13, O:14 and O:15 were not found in this study. Strains belonging to the same ST showed the same serotype, except for ST155 strains that were non-agglutinable or O:6 serotype (Table 2).
2.5. Characterisation of Porin OprD
The oprD gene was amplified in all P. aeruginosa strains, and eleven amino acidic OprD profiles were distinguished (Table 2 and Table S2). Only two strains had the same pattern as P. aeruginosa PAO1 (pattern A, wild type), and pattern B was the most frequently detected (23 strains). The deletion of two amino acids in the region from amino acid 372 to 383 of the loop 7 (loop L7-short), which encodes a protein OprD of 441 amino acids, was identified in 24 strains (64.8%) (patterns B and E) (Table 2 and Table S2). Regarding the two imipenem-resistant strains, pattern B was observed in Ps839 strain, and the Ps884 strain showed the oprD gene truncated by the insertion sequence ISPa1635 at nucleotide position 561 (pattern K). This insertion sequence belongs to the IS4 family, and this is the first description of ISPa1635 truncating oprD gene. Thus, the sequence was included in GenBank with the accession number MH050332.
A total of 11 P. aeruginosa strains (one per pattern) with different protein OprD profiles were selected to study their outer membrane proteins by SDS-PAGE. The OprD band was detected in all tested strains except in pattern K, which corresponded to the strain with ISPa1635 element truncating the oprD gene.
2.6. Virulence Patterns
The presence of virulence and QS genes was investigated in the 37 P. aeruginosa strains, and ten different virulence patterns were obtained (Table 2 and Table S3). Regarding the T3SS, exoU gene was detected in 4 P. aeruginosa strains and exoS in 31 strains. Neither exoU nor exoS genes were amplified in two strains (pattern VII). Moreover, exoA and exoY genes did not amplify in one and three strains, respectively. The exoT, lasA, lasB, aprA, rhlAB, rhlC, rhlI, and rhlR genes were detected in all strains, whereas exlA gene was absent in all of them. The lasI and lasR genes, involved in the QS system, were not amplified in three strains (pattern IV) that belonged to ST155 (Ps764, Ps839, Ps892). In the remaining ST155 strain (Ps845), as well as in two more strains (Ps796, Ps852), the lasR amplicon sized higher than 2,000 bp, resulting in the first description of three insertion sequences (IS1411, ISPre2, and ISPst7, respectively) and truncating this gene (Table 2 and Figure 2). All lasR sequences were submitted in GenBank (accession number): IS1411 (MH050330), ISPre2 (MH050329), and ISPst7 (MH050331). Finally, the lasR gene of Ps893 strain sized lower than expected, 647 instead of 720 bp, due to the presence of a deletion at the beginning of the gene (Figure 2).
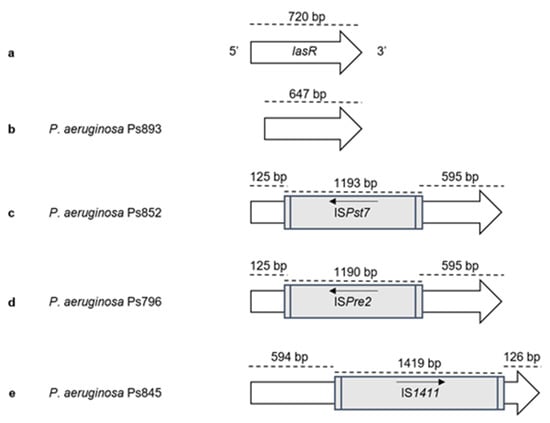
Figure 2.
Representation of truncated lasR genes found among our P. aeruginosa strains. The position and orientation of the gene and insertion sequences are indicated by arrows: (a) lasR gene (PA1430) of the control strain P. aeruginosa PAO1 (NCBI accession number NC_002516); (b) Ps893 strain, lasR size is 647 bp due to the presence of a deletion at the beginning of the gene; (c) Ps852 strain, lasR truncated at position 125 bp by the insertion sequence ISPst7 (1193 bp) (GenBank accession number MH050331); (d) Ps796 strain, lasR truncated at position 125 bp by the insertion sequence ISPre2 (1190 bp) (GenBank accession number MH050329); (e) Ps845 strain, lasR truncated at position 595 bp by the insertion sequence IS1411 (1419 bp) (GenBank accession number MH050330).
2.7. Biofilm Quantification
Table 2 and Figure 1 summarise the biofilm biomass production (CV) and the bacterial metabolic activity inside the biofilm (FDA) of the 37 P. aeruginosa strains. The 92% of P. aeruginosa strains displayed higher values for biomass biofilm production than P. aeruginosa PAO1. Ps883 and Ps796 strains showed the highest percentages (952% and 600%, respectively), whereas Ps893, Ps851 and Ps733 exhibited the lowest ones (56, 65 and 90%, respectively). For FDA assay, 73% of strains showed more metabolic activity than the control strain P. aeruginosa PAO1. Ps752, Ps796, Ps845, Ps852 and Ps913 were the highest producers (>700%), and Ps798 the lowest producer (26.2%). Likewise, strains that had an absent or truncated lasR gene showed high levels of biomass production and bacterial metabolic activity inside the biofilm, except for Ps893 that showed low levels in comparison with reference P. aeruginosa PAO1 strain.
2.8. Motility
The different swarming and swimming patterns detected among the 37 P. aeruginosa strains are included in Figure S1. Analysing the swimming results, many strains belonging to the Cluster III showed higher motility than the remaining ones (Table 2 and Figure 1). In fact, 23 of 37 strains (62.1%) covered the entire Petri dish surface (5363.7 to 6400 mm2) in swimming and swarming. Ps893 and Ps884 strains described the lowest swimming (21.6 and 25.2 mm2) and swarming (30.0 and 38.3 mm2) values (Table 2 and Figure 1). Ps796 showed a high value of swimming (5363.7 mm2) but showed a low swarming value (97.2 mm2).
P. aeruginosa strains having an absent or truncated lasR gene showed medium and low levels of motility, except the Ps796 swimming (Table 2 and Figure 1). Ps892 (absent gene) and Ps893 (truncated gene) showed the lowest motility values. Considering the strains which carried an insertion sequence truncating the lasR gene (patterns IIa, IIb and VI), the swarming levels were lower than the swimming motility ones.
2.9. Elastase and Pigment Production
Results for quantification of pyorubin and pyocyanin production as well as elastase activity are summarised in Table 2 and Figure 1.
For pyorubin assay, 70.2% of strains showed high levels of production in comparison with P. aeruginosa PAO1. Besides, Ps845, Ps848 and Ps852 showed the highest levels of pyorubin. Conversely, 27% of strains were the lowest pyorubin producers, highlighting Ps733, Ps775 and Ps854 strains. For the pyocyanin values, 62% showed high levels of production, being Ps845 and Ps848 the most producers. Conversely, fourteen strains showed low levels in comparison with reference strain, and Ps883, Ps884 and Ps913 strains were the weakest producers. The four strains with an absent or truncated lasR gene showed low levels of pyocyanin production.
2.10. Rhamnolipids Detection
Figure S2 shows some of the results obtained with the rhamnolipid assays in P. aeruginosa strains. The 94.6% of the strains showed halos ≥11 mm (the minimum value observed), many of them (10 strains) with halos ranging 19–20 mm of diameter (Table 2 and Figure 1). Ps851 and Ps884 did not produce rhamnolipids.
3. Discussion
Food and the environment have been described as reservoirs of bacteria harbouring antimicrobial resistance genes that could be transferred or mobilised into human pathogens. Moreover, the extensive use and even the misuse of antimicrobial agents in clinic, animal production, and agriculture could be a way to select and disseminate these resistant human pathogens [5], and among them Pseudomonas genus. Some reports showed different clinical cases caused by environmental Pseudomonas, such as P. mendocina, P. monteilii or P. putida [16,17,18,19], although P. aeruginosa is the most important pathogenic bacterium. In our study, the 53.1% of fresh vegetables were positive for Pseudomonas spp., and lettuce and chard were the most frequently contaminated vegetables, as well as in previous studies [12,13]. According to our results and previous reports [1,20,21], the vegetables cultivated in contact with the soil may be contaminated more easily with Pseudomonas coming from soil, fertilizers, manure or water used for irrigation. In contrast, the lowest presence of Pseudomonas spp. was among onion samples. This fact could be due to the layered structure of the onion and/or the bioactive compounds present in onions [22,23,24]. Previous studies revealed that fresh onions, even onion wastes, exhibited high antimicrobial activity against bacteria such as Escherichia coli, P. fluorescens and Bacillus cereus, among others.
Antibiotic susceptibility testing revealed that all Pseudomonas spp. were susceptible to aminoglycosides and fluoroquinolones, and they showed low resistance rates for aztreonam and carbapenems. Only two imipenem-resistant P. aeruginosa strains were detected, and none of them were an MBL producer. The imipenem-resistance of Ps884 strain was associated with the loss of function of its OprD porin due to the truncation of the oprD gene by the insertion sequence ISPa1635. The inactivation of this porin gene by insertion sequences has been deeply studied in clinical strains [25,26,27], but this is the first time that the ISPa1635 has been identified in a P. aeruginosa from food origin and truncating oprD gene. Conversely, the imipenem-resistant Ps839 strain showed the same amino acid changes detected in the OprD porin as those reported in carbapenem-susceptible P. aeruginosa isolates [28,29]. Thus, other resistance mechanisms such as active efflux pumps or AmpC hyperproduction could be involved in that phenotype.
Considering P. aeruginosa QS genes, rhlI and rhlR genes were amplified in all P. aeruginosa strains. However, lasI and lasR genes were not detected in three strains; the other three strains showed insertion sequences truncating the lasR gene (IS1411, ISPst7 and ISPre2), and one strain showed a short lasR gene leading the possibility of losing the QS function. Several reports have mentioned the frequency of lasI and lasR mutations, or the lack of lasR gene among clinical and environmental isolates to favour their adaptation or persistence [30,31,32]. They have also demonstrated that a mutation in lasR does not lead to virulence factors loss, due to the regulation mediated by the rhl system, taking control of the phenazines production or rhamnolipids synthesis [10]. In addition, there are two other QS mechanisms, the Pseudomonas quinolone signal (PQS) and the integrated quorum sensing (IQS) mechanism, able to replace, in many cases, the LasR function [6,10]. Regarding our phenotypic results, it is important to remark that a high percentage of analysed P. aeruginosa showed high levels of biofilm, pigments and rhamnolipids production, and elastase activity even those with an absent or truncated lasR gene; however, in these cases, the strains were not as mobile as the remaining strains. Nevertheless, the hypothesis of the action of other QS mechanisms could demonstrate the pathological importance of these P. aeruginosa strains.
In P. aeruginosa, the T3SS mechanism contributes to cytotoxicity and acute infections [6,7]. The exoU gene was detected in four strains, all of them situated in the same branch of the MLST cluster, including the ST253 [33] and the new one ST2427, and belonging to O:11 serotype or poly-agglutinable, as other reports [34]. Usually, the exoU gene is described in clinical isolates, but it has also been detected in environmental strains [29,35].
The pathogenicity and host adaptation of P. aeruginosa is associated with its worldwide dissemination and specific sequence types. Among all sequence types detected, unlike other reports regarding clinical isolates, the most important “high-risk clones” ST111, ST175 and ST235, were not found. However, intercontinental clones disseminated worldwide, newest high-risk clones, such as ST155 and ST244 [36,37], as well as the epidemic clone ST274 circulating in Spain [38,39], were detected. All of those, including ST252, ST253, and ST395 epidemic clones were also previously observed in clinical animal and environmental samples [36,40,41,42]. However, none of the epidemic clones showed the same pathogen phenotype. In this case, the low motility but high biofilm production that possessed the STs from the second cluster, where ST155 clone was included. Conversely, the P. aeruginosa strains belonging to the third cluster exhibited higher motility, where high-risk clones ST253, ST274 and ST395 were englobed. However, there are some exceptions, such as P. aeruginosa strains with lacked or truncated lasR gene, Ps852 (ST267), Ps893 (ST395) or Ps764, Ps839, Ps845 and Ps892 (ST155). Further studies are needed to delve into the relationship between the genotype and phenotype of these epidemic clones because, to the best of our knowledge, this is the first time the analysis of this relationship is described in environmental samples.
4. Materials and Methods
4.1. Bacterial Isolates
One hundred and forty-five samples of raw vegetables were recovered from orchards (82 samples) and little markets (63 samples) of different areas of La Rioja region (Spain), during 2015. Samples were divided as follows: 20 lettuces (Lactuca sativa), 22 cucumbers (Cucumis sativus), 20 zucchinis (Cucurbita pepo), 23 onions/leeks (Allium cepa/Allium ampeloprasum var. porrum), 20 potatoes (Solanum tuberosum), 20 green beans (Phaseolus vulgaris) and 20 chards (Beta vulgaris var. cicla).
A quantity of 30–35 g of each sample was enriched in 100 mL of Tryptose Soy Broth (Becton Dickinson, Franklin Lake, NJ, USA), and homogenised in a stomacher. A suspension volume (40 mL) was incubated at 37 °C during 24 h in agitation. Then, 100 µL of this suspension was streaked onto Cetrimide-agar plates (Becton Dickinson, Le Pont de Claix, France) and incubated at 42 °C during 24–48 h. Two or three different colonies per plate, presumptive of being Pseudomonas, were selected, identified by classical biochemical methods (Triple Sugar Iron and oxidase tests), and confirmed by PCR amplification and sequencing of 16S rRNA fragment [28] and by Matrix-Assisted Laser Desorption/Ionization Time-of-Flight (MALDI-TOF) mass spectrometry (Bruker, Billerica, MA, USA).
4.2. Antimicrobial Susceptibility Testing
Antimicrobial susceptibility testing was performed by disc diffusion method following the Clinical and Laboratory Standards Institute guidelines [43]. Thirteen antipseudomonal agents were tested (disc concentration), including piperacillin-tazobactam (100/10 µg), piperacillin (100 µg), aztreonam (30 µg), cefepime (30 µg), ceftazidime (30 µg), imipenem (10 µg), meropenem (10 µg), doripenem (10 µg), tobramycin (10 µg), gentamicin (10 µg), amikacin (30 µg), netilmicin (30 µg), and ciprofloxacin (5 µg). ESBL, MBL, class A carbapenemase and inducible AmpC phenotypes were determined by double-disc synergy tests [29].
4.3. Molecular Typing
The clonal relationship among the recovered isolates was determined by PFGE with SpeI restriction enzyme [28]. PFGE patterns were analysed by the Java program GelJ using the Dice coefficient [44].
MLST for P. aeruginosa was performed by PCR and sequencing [29,45]. Allelic profiles and sequence types (STs) were assigned according to the PubMLST database (http://pubmlst.org/paeruginosa/ accessed on April 2016). A maximum-likelihood phylogenetic tree, relating the sequence types of the P. aeruginosa strains, was performed using IQTREE v.1.6.1 [14], and visualised with iTol v.4 [15].
4.4. Serotyping
P. aeruginosa strains were serotyped by slide agglutination according to the International Antigenic Typing Scheme (IATS), using 16 type O monovalent antisera specific for P. aeruginosa (O:1 to O:16) following the manufacturer’s protocol (Bio-Rad, Temse, Belgium).
4.5. Characterisation of Porin OprD
Amino acid changes of the porin OprD were analysed by PCR and sequencing in all P. aeruginosa strains [29]. The mutations were determined by comparison with the sequence of the control strain P. aeruginosa PAO1 (GenBank accession number AE004091).
The outer membrane proteins (OMPs) of selected strains were stained with Coomassie Brilliant Blue and were visualised by SDS-PAGE (Sodium Dodecyl Sulphate-PolyAcrylamide Gel Electrophoresis) in a Bio-Rad Mini-Protean II apparatus (Bio-Rad, Temse, Belgium) as previously described [46]. P. aeruginosa PAO1 and PAO1 lacking-OprD were included as control strains.
4.6. Detection of Virulence Marker Genes
The presence of exoU, exoS, exoY, exoT, exlA, exoA, lasA, lasB, aprA, rhlAB, rhlC, rhlI, rhlR, lasI, and lasR virulence and QS genes was analysed by PCR in P. aeruginosa strains [47].
4.7. Biofilm Quantification
The analysis of the total biofilm biomass was performed by crystal violet (CV) staining, and the bacterial metabolic activity inside the biofilm structure by fluorescein diacetate (FDA) assay among P. aeruginosa strains [47]. Both methods were carried out in flat-bottom microtiter 96-well plates after 24 h of bacterial incubation in Müeller–Hinton broth at 37 °C. For CV assay, 66% acetic acid and 10% CV were used, and the FDA working solution concentration was 0.1 mg/mL. Measures were performed using a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany). All assays were carried out in triplicate, including P. aeruginosa PAO1 as control.
4.8. Motility
Swarming and swimming motilities were studied in P. aeruginosa strains [47], placing 4 µL of bacterial suspension (1 × 109 cells in Luria–Bertani (LB) broth) on the middle of 0.5% (swarming) and 0.3% (swimming) LB agar plates and subsequent incubation at 37 °C overnight. The plates were imaged with Chemi Doc system (Bio-Rad, Temse, Belgium), and processed with Image Lab software (version 6.0.1, Bio-Rad). The entire plate area was 6400 mm2. All assays were performed in triplicate, including P. aeruginosa PAO1 as a control strain.
4.9. Elastase and Pigment Production
Bacteria were grown overnight in LB broth at 37 °C with shaking at 120 rpm. After centrifugation, 900 μL of supernatant was used to determine the elastase activity in P. aeruginosa strains by the Elastin Congo Red assay as previously described [48].
The chloroform extract method was used to quantify pyocyanin and pyorubin phenazines by measuring the absorbance of the corresponding solutions: the organic phase at 520 for pyocyanin, and the aqueous phase at 525 nm for pyorubin, using a POLARstar Omega microplate reader (BMG Labtech, Ortenberg, Germany) [49], and including P. aeruginosa PAO1 as control.
4.10. Rhamnolipids Detection
The detection of P. aeruginosa biosurfactant producers was carried out by the Cetyl Trimethylammonium Bromide–Methylene Blue (CTAB-MB) agar plates method [50,51,52]. One colony of each P. aeruginosa strain studied was inoculated in 3 mL of mineral salt medium (MSM) broth and was incubated at 35 °C and 130 rpm during 48 h. Following previous procedures, shallow wells of 6 mm were cut on the CTAB-MB agar plate surface. Twenty microliters of the inoculum were added into each well. The plates were incubated for 24–48 h at 35 °C, and then stored in the fridge for at least 24 h, to intensify the blue colour of the plates to facilitate the recognition of the rhamnolipids production. The halo was measured (mm). P. aeruginosa PAO1 was used as a positive control.
5. Conclusions
Pseudomonas strains contaminating fresh vegetables were found in this work, especially in lettuce and chard. A variety of different Pseudomonas species, including pathogenic to humans, such as P. aeruginosa, P. mendocina, P. monteilii or P. putida were detected. P. aeruginosa was recovered from 26 (17.9 %) of the vegetable samples and belonged to many different clones, comprising some international clones. Moreover, these strains showed low resistance to antibiotics but high presence of virulence-related traits, as high biofilm, pigments and rhamnolipids production. P. aeruginosa is an opportunistic human pathogen, and the food chain might be a source of transmission to humans. Identifying the natural reservoirs of this important pathogen and elucidating its molecular biology are crucial tasks in the pursuit of minimising its transmission. For all these reasons, the application of proper hygiene practices along the food production/supply chain is essential, not only for vegetable workers, but also for consumers.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/ijms222312626/s1.
Author Contributions
Conceptualization, L.R.-R., C.T., and Y.S.; Data curation, L.R.-R. and Y.S.; Formal analysis, L.R.-R., B.R.-B., C.L., M.L., G.C. and Y.S.; Funding acquisition, Y.S.; Investigation, L.R.-R., B.R.-B., C.L., M.L., G.C. and Y.S.; Supervision, C.T. and Y.S.; Writing—original draft, L.R.-R., B.R.-B., C.L., M.L., C.T. and Y.S.; Writing—review and editing, L.R.-R., B.R.-B., C.L., M.L., G.C., C.T. and Y.S. All authors have read and agreed to the published version of the manuscript.
Funding
Lidia Ruiz-Roldán and Gabriela Chichón had a predoctoral fellowship from the Consejería de Industria, Innovación y Empleo, Gobierno de La Rioja, Spain. Carmen Lozano was supported by a Sara Borrell Postdoctoral Contract (CD15/00125), from the Instituto de Salud Carlos III of Spain. This work was supported by the Instituto de Salud Carlos III of Spain (ISCIII) (projects FIS PI12/01276 and PI16/01381) (Co-funded by European Regional Development Fund (FEDER) “A way to make Europe”).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All datasets are available. New sequences of lasR genes truncated by insertion sequence elements were submitted in GenBank (accession number): IS1411 (MH050330), ISPre2 (MH050329), and ISPst7 (MH050331). The oprD gene truncated by the insertion sequence ISPa1635 was included in GenBank with the accession number MH050332.
Acknowledgments
This publication made use of the Pseudomonas aeruginosa MLST website (Jolley, K.A., Bray, J.E., Maiden, M.C.J. Open-access bacterial population genomics: BIGSdb software, the PubMLST.org website and their applications. Wellcome Open Res. 2018, 3, 124. doi: 10.12688/wellcomeopenres.14826.1). Part of this study was presented at the 27th European Congress of Clinical Microbiology and Infectious Diseases (ECCMID) (Abstract Nº 6532, Vienna, Austria, 22–25 April 2017), the 16th International Congress on Pseudomonas (Nº P114, Liverpool, UK, 5–9 September 2017) and the 28th ECCMID (Abstract Nº 975, Madrid, Spain, 21–24 April 2018).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Hölzel, C.S.; Tetens, J.L.; Schwaiger, K. Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: A need for quantitative risk assessment. Foodborne Pathog. Dis. 2018, 15, 671–688. [Google Scholar] [CrossRef]
- Jung, D.; Rubin, J.E. Identification of antimicrobial resistant bacteria from plant-based food products imported into Canada. Int. J. Food Microbiol. 2020, 319, 108509. [Google Scholar] [CrossRef]
- Iseppi, R.; de Niederhäusern, S.; Bondi, M.; Messi, P.; Sabia, C. Extended-Spectrum β-lactamase, AmpC, and MBL-producing Gram-Negative bacteria on fresh vegetables and ready-to-eat salads sold in local markets. Microb. Drug Resist. 2018, 24, 1156–1164. [Google Scholar] [CrossRef]
- Prieto, M.; Colin, P.; Fernández-Escámez, P.; Alvarez-Ordóñez, A. Epidemiology, detection, and control of foodborne microbial pathogens. Biomed. Res. Int. 2015, 2015, 617417. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; Ning, J.; Ahmed, S.; Huang, J.; Ullah, R.; An, B.; Hao, H.; Dai, M.; Huang, L.; Wang, X.; et al. Selection and dissemination of antimicrobial resistance in Agri-food production. Antimicrob. Resist. Infect. Control 2019, 8, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Moradali, M.F.; Ghods, S.; Rehm, B.H.A. Pseudomonas aeruginosa lifestyle: A paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 2017, 7, 39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pena, R.T.; Blasco, L.; Ambroa, A.; González-Pedrajo, B.; Fernández-García, L.; López, M.; Bleriot, I.; Bou, G.; García-Contreras, R.; Wood, T.K.; et al. Relationship between Quorum Sensing and Secretion Systems. Front. Microbiol. 2019, 10, 1100. [Google Scholar] [CrossRef] [Green Version]
- Foulkes, D.M.; McLean, K.; Haneef, A.S.; Fernig, D.G.; Winstanley, C.; Berry, N.; Kaye, S.B. Pseudomonas aeruginosa toxin ExoU as a therapeutic target in the treatment of bacterial infections. Microorganisms 2019, 7, 707. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tümmler, B.; Klockgether, J. Recent advances in understanding Pseudomonas aeruginosa as a pathogen. F1000Research 2017, 6, 1261. [Google Scholar] [CrossRef]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar] [CrossRef] [Green Version]
- Quintieri, L.; Fanelli, F.; Caputo, L. Antibiotic resistant Pseudomonas spp. spoilers in fresh dairy products: An underestimated risk and the control strategies. Foods 2019, 8, 372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allydice-Francis, K.; Brown, P.D. Diversity of antimicrobial resistance and virulence determinants in Pseudomonas aeruginosa associated with fresh vegetables. Int. J. Microbiol. 2012, 2012, 426241. [Google Scholar] [CrossRef] [Green Version]
- Estepa, V.; Rojo-Bezares, B.; Torres, C.; Sáenz, Y. Genetic lineages and antimicrobial resistance in Pseudomonas spp. isolates recovered from food samples. Foodborne Pathog. Dis. 2015, 12, 486–491. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.-T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A Fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaerts, P.; Bouchahrouf, W.; Lissoir, B.; Denis, O.; Glupczynski, Y. IMP-13-producing Pseudomonas monteilii recovered in a hospital environment. J. Antimicrob. Chemother. 2011, 66, 2434–2435. [Google Scholar] [CrossRef] [Green Version]
- Gani, M.; Rao, S.; Miller, M.; Scoular, S. Pseudomonas mendocina bacteremia: A case study and review of literature. Am. J. Case Rep. 2019, 20, 453–458. [Google Scholar] [CrossRef]
- Kim, S.E.; Park, S.-H.; Park, H.B.; Park, K.-H.; Kim, S.-H.; Jung, S.-I.; Shin, J.-H.; Jang, H.-C.; Kang, S.J. Nosocomial Pseudomonas putida Bacteremia: High Rates of Carbapenem Resistance and Mortality. Chonnam Med. J. 2012, 48, 91. [Google Scholar] [CrossRef] [Green Version]
- Yoshino, Y.; Kitazawa, T.; Kamimura, M.; Tatsuno, K.; Yotsuyanagi, H.; Ota, Y. Pseudomonas putida bacteremia in adult patients: Five case reports and a review of the literature. J. Infect. Chemother. 2011, 17, 278–282. [Google Scholar] [CrossRef]
- Crone, S.; Vives-Flórez, M.; Kvich, L.; Saunders, A.M.; Malone, M.; Nicolaisen, M.H.; Martínez-García, E.; Rojas-Acosta, C.; Catalina Gomez-Puerto, M.; Calum, H.; et al. The environmental occurrence of Pseudomonas aeruginosa. APMIS 2020, 128, 220–231. [Google Scholar] [CrossRef]
- Schroth, M.N.; Cho, J.J.; Green, S.K.; Kominos, S.D. Epidemiology of Pseudomonas aeruginosa in agricultural areas. J. Med. Microbiol. 2018, 67, 1191–1201. [Google Scholar] [CrossRef]
- Sharma, K.; Mahato, N.; Lee, Y.R. Systematic study on active compounds as antibacterial and antibiofilm agent in aging onions. J. Food Drug Anal. 2018, 26, 518–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramos, F.A.; Takaishi, Y.; Shirotori, M.; Kawaguchi, Y.; Tsuchiya, K.; Shibata, H.; Higuti, T.; Tadokoro, T.; Takeuchi, M. Antibacterial and antioxidant activities of quercetin oxidation products from yellow onion (Allium cepa) skin. J. Agric. Food Chem. 2006, 54, 3551–3557. [Google Scholar] [CrossRef] [PubMed]
- Santas, J.; Almajano, M.P.; Carbó, R. Antimicrobial and antioxidant activity of crude onion (Allium cepa, L.) extracts. Int. J. Food Sci. Tech. 2010, 45, 403–409. [Google Scholar] [CrossRef]
- Bocharova, Y.; Savinova, T.; Shagin, D.A.; Shelenkov, A.A.; Mayanskiy, N.A.; Chebotar, I.V. Inactivation of the oprD porin gene by a novel insertion sequence ISPa195 associated with large deletion in a carbapenem-resistant Pseudomonas aeruginosa clinical isolate. J. Glob. Antimicrob. Resist. 2019, 17, 309–311. [Google Scholar] [CrossRef] [PubMed]
- Fournier, D.; Richardot, C.; Müller, E.; Robert-Nicoud, M.; Llanes, C.; Plésiat, P.; Jeannot, K. Complexity of resistance mechanisms to imipenem in intensive care unit strains of Pseudomonas aeruginosa. J. Antimicrob. Chemother. 2013, 68, 1772–1780. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Ba, Z.; Wu, G.; Wang, W.; Lin, S.; Yang, H. Insertion sequence ISRP10 inactivation of the oprD gene in imipenem-resistant Pseudomonas aeruginosa clinical isolates. Int. J. Antimicrob. Agents 2016, 47, 375–379. [Google Scholar] [CrossRef]
- Estepa, V.; Rojo-Bezares, B.; Torres, C.; Sáenz, Y. Faecal carriage of Pseudomonas aeruginosa in healthy humans: Antimicrobial susceptibility and global genetic lineages. FEMS Microbiol. Ecol. 2014, 89, 15–19. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Roldán, L.; Bellés, A.; Bueno, J.; Azcona-Gutiérrez, J.M.; Rojo-Bezares, B.; Torres, C.; Castillo, F.J.; Sáenz, Y.; Seral, C. Pseudomonas aeruginosa isolates from Spanish children: Occurrence in faecal samples, antimicrobial resistance, virulence, and molecular typing. Biomed Res. Int. 2018, 2018, 8060178. [Google Scholar] [CrossRef] [Green Version]
- Feltner, J.B.; Wolter, D.J.; Pope, C.E.; Groleau, M.; Smalley, N.E.; Greenberg, E.P. LasR variant cystic fibrosis isolates reveal an adaptable Quorum-Sensing hierarchy in Pseudomonas aeruginosa. Am. Soc. Microbiol. 2016, 7, e01513–e01516. [Google Scholar] [CrossRef] [Green Version]
- Lee, D.J.; Jo, A.R.; Jang, M.C.; Nam, J.; Choi, H.J.; Choi, G.-W.; Sung, H.Y.; Bae, H.; Ku, Y.-G.; Chi, Y.-T. Analysis of two Quorum Sensing-deficient isolates of Pseudomonas aeruginosa. Microb. Pathog. 2018, 119, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Morales, E.; González-Valdez, A.; Servín-González, L.; Soberón-Chávez, G. Pseudomonas aeruginosa Quorum-Sensing response in the absence of functional LasR and LasI proteins: The case of strain 148, a virulent dolphin isolate. FEMS Microbiol. Lett. 2017, 364. [Google Scholar] [CrossRef] [PubMed]
- Sawa, T.; Momiyama, K.; Mihara, T.; Kainuma, A.; Kinoshita, M.; Moriyama, K. Molecular epidemiology of clinically high-risk Pseudomonas aeruginosa strains: Practical overview. Microbiol. Inmunol. 2020, 64, 331–344. [Google Scholar] [CrossRef]
- Berthelot, P.; Attree, I.; Plésiat, P.; Chabert, J.; de Bentzmann, S.; Pozzetto, B.; Grattard, F. Genotypic and phenotypic analysis of Type III Secretion System in a cohort of Pseudomonas aeruginosa bacteremia isolates: Evidence for a possible association between O serotypes and exo genes. J. Infect. Dis. 2003, 188, 512–518. [Google Scholar] [CrossRef] [Green Version]
- Petit, S.M.C.; Lavenir, R.; Colinon-Dupuich, C.; Boukerb, A.M.; Cholley, P.; Bertrand, X.; Freney, J.; Doléans-Jordheim, A.; Nazaret, S.; Laurent, F.; et al. Lagooning of wastewaters favors dissemination of clinically relevant Pseudomonas aeruginosa. Res. Microbiol. 2013, 164, 856–866. [Google Scholar] [CrossRef]
- Cholley, P.; Ka, R.; Guyeux, C.; Thouverez, M.; Guessennd, N.; Ghebremedhin, B.; Frank, T.; Bertrand, X.; Hocquet, D. Population structure of clinical Pseudomonas aeruginosa from West and Central African countries. PLoS ONE 2014, 9, e107008. [Google Scholar] [CrossRef] [Green Version]
- Kidd, T.J.; Ritchie, S.R.; Ramsay, K.A.; Grimwood, K.; Bell, S.C.; Rainey, P.B. Pseudomonas aeruginosa exhibits frequent recombination, but only a limited association between genotype and ecological setting. PLoS ONE 2012, 7, e44199. [Google Scholar] [CrossRef] [Green Version]
- Del Barrio-Tofiño, E.; López-Causapé, C.; Cabot, G.; Rivera, A.; Benito, N.; Segura, C.; Montero, M.M.; Sorlí, L.; Tubau, F.; Gómez-Zorrilla, S.; et al. Genomics and Susceptibility Profiles of Extensively Drug-Resistant Pseudomonas aeruginosa Isolates from Spain. Antimicrob. Agents Chemother. 2017, 61, e01589-17. [Google Scholar] [CrossRef] [Green Version]
- Ocampo-Sosa, A.A.; Fernández-Martínez, M.; Cabot, G.; Peña, C.; Tubau, F.; Oliver, A.; Martínez-Martínez, L. Draft genome sequence of the quorum-sensing and biofilm-producing Pseudomonas aeruginosa strain Pae221, belonging to the epidemic high-risk clone sequence type 274. Genome Announc. 2015, 3, e01343-14. [Google Scholar] [CrossRef] [Green Version]
- Freschi, L.; Bertelli, C.; Jeukens, J.; Moore, M.P.; Kukavica-Ibrulj, I.; Emond-Rheault, J.-G.; Hamel, J.; Fothergill, J.L.; Tucker, N.P.; McClean, S.; et al. Genomic characterisation of an international Pseudomonas aeruginosa reference panel indicates that the two major groups draw upon distinct mobile gene pools. FEMS Microbiol. Lett. 2018, 365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haenni, M.; Hocquet, D.; Ponsin, C.; Cholley, P.; Guyeux, C.; Madec, J.-Y.; Bertrand, X. Population structure and antimicrobial susceptibility of Pseudomonas aeruginosa from animal infections in France. BMC Vet. Res. 2015, 11, 9. [Google Scholar] [CrossRef] [Green Version]
- Petitjean, M.; Martak, D.; Silvant, A.; Bertrand, X.; Valot, B.; Hocquet, D. Genomic characterization of a local epidemic Pseudomonas aeruginosa reveals specific features of the widespread clone 1–10. Microb. Genom. 2017, 3, e000129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing, 26th ed.; CLSI supplement M100S; Clinical and Laboratory Standards Institute: Wayne, PA, USA, 2016; ISBN 1-56238-923-8. [Google Scholar]
- Heras, J.; Domínguez, C.; Mata, E.; Pascual, V.; Lozano, C.; Torres, C.; Zarazaga, M. GelJ-a tool for analyzing DNA fingerprint gel images. BMC Bioinform. 2015, 16, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Curran, B.; Jonas, D.; Grundmann, H.; Pitt, T.; Dowson, C.G. Development of a multilocus sequence typing scheme for the opportunistic pathogen Pseudomonas aeruginosa. J. Clin. Microbiol. 2004, 42, 5644–5649. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Martínez, L.; López-Jiménez, L.; Fusté, E.; Vinuesa, T.; Martínez, J.P.; Viñas, M. Class 1 integrons in environmental and clinical isolates of Pseudomonas aeruginosa. Int. J. Antimicrob. Agents 2011, 38, 398–402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruiz-Roldán, L.; Rojo-Bezares, B.; de Toro, M.; López, M.; Toledano, P.; Lozano, C.; Chichón, G.; Alvarez-Erviti, L.; Torres, C.; Sáenz, Y. Antimicrobial resistance and virulence of Pseudomonas spp. among healthy animals: Concern about exolysin ExlA detection. Sci. Rep. 2020, 10, 11667. [Google Scholar] [CrossRef]
- Glessner, A.; Smith, R.S.; Iglewski, B.H.; Robinson, J.B. Roles of Pseudomonas aeruginosa las and rhl quorum-sensing systems in control of twitching motility. J. Bacteriol. 1997, 179, 5756–5767. [Google Scholar] [CrossRef] [Green Version]
- Anantharajah, A.; Buyck, J.M.; Sundin, C.; Tulkens, P.M.; Mingeot-Leclercq, M.P.; Van Bambeke, F. Salicylidene acylhydrazides and hydroxyquinolines act as inhibitors of type three secretion systems in Pseudomonas aeruginosa by distinct mechanisms. Antimicrob. Agents Chemother. 2017, 61, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gunther IV, N.W.; Nuñez, A.; Fett, W.; Solaiman, D.K.Y. Production of rhamnolipids by Pseudomonas chlororaphis, a nonpathogenic bacterium. Appl. Environ. Microbiol. 2005, 71, 2288–2293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pinzon, N.M.; Ju, L.-K. Improved detection of rhamnolipid production using agar plates containing methylene blue and cetyl trimethylammonium bromide. Biotechnol. Lett. 2009, 31, 1583–1588. [Google Scholar] [CrossRef]
- Siegmund, I.; Wagner, F. New method for detecting rhamnolipids excreted by Pseudomonas species during growth on mineral agar. Biotechnol. Tech. 1991, 5, 265–268. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).