Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy
Abstract
:1. Brief History of Apoptosis
2. Apoptosis Induction and Regulation
2.1. Intrinsic Pathway
2.2. Extrinsic Pathway
2.2.1. Tumor Necrosis Factor Receptor (TNFR)
2.2.2. TNF-Related Apoptosis-Inducing Ligand (TRAIL)
2.2.3. Fas Receptor
3. Apoptosis in Human Diseases
4. Autophagy
5. Molecular Basis of Autophagy
6. Different Types of Autophagy
6.1. Macroautophagy
6.1.1. Initiation
6.1.2. Nucleation
6.1.3. Elongation
6.1.4. Maturation
6.1.5. Fusion
6.2. Microautophagy
6.3. Chaperone-Mediated Autophagy
7. Autophagy in Health and Disease
8. Crosstalk between Apoptosis and Autophagy
9. Conclusions
10. Materials and Methods
10.1. Developmental Apoptosis
10.2. Electron Microscopy
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trent, C.; Tsuing, N.; Horvitz, H.R. Egg-laying defective mutants of the nematode Caenorhabditis elegans. Genetics 1983, 104, 619–647. [Google Scholar] [CrossRef] [PubMed]
- Esposti, M.D. Apoptosis: Who was first? Cell Death Differ. 1998, 5, 719. [Google Scholar] [CrossRef]
- André, N. Hippocrates of Cos and apoptosis. Lancet 2003, 361, 1306. [Google Scholar] [CrossRef]
- Fernández-Flores, A.; Aguilera, B.; Yau, P.; Oliva, H. An old meaning of the word apoptosis. Lancet 2002, 359, 1072. [Google Scholar] [CrossRef]
- Diamantis, A.; Magiorkinis, E.; Sakorafas, G.H.; Androutsos, G. A Brief History of Apoptosis: From Ancient to Modern Times. Oncol. Res. Treat. 2008, 31, 702–706. [Google Scholar] [CrossRef]
- Gerschenson, L.E.; Geske, F.J. Virchow and apoptosis. Am. J. Pathol. 2001, 158, 1543. [Google Scholar] [CrossRef]
- Judah, J.D.; Ahmed, K.; McLean, A.E. Pathogenesis of cell necrosis. Fed. Proc. 1965, 24, 1217–1221. [Google Scholar] [PubMed]
- Trump, B.F.; Goldblatt, P.J.; Stowell, R.E. Studies on Necrosis of Mouse Liver in Vitro. Ultrastructural Alterations in the Mitochondria of Hepatic Parenchymal Cells. Lab. Investig. 1965, 14, 343–371. [Google Scholar] [PubMed]
- Stoica, B.A.; Faden, A.I. Programmed Neuronal Cell Death Mechanisms in CNS Injury. In Acute Neuronal Injury; Fujikawa, D.G., Ed.; Springer: Boston, MA, USA, 2010; pp. 169–200. [Google Scholar]
- Klion, F.M.; Schaffner, F. The ultrastructure of acidophilic “Councilman-like” bodies in the liver. Am. J. Pathol. 1966, 48, 755–767. [Google Scholar] [PubMed]
- Kerr, J.F. An electron-microscope study of liver cell necrosis due to heliotrine. J. Pathol. 1969, 97, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F.; Searle, J. A suggested explanation for the paradoxically slow growth rate of basal-cell carcinomas that contain numerous mitotic figures. J. Pathol. 1972, 107, 41–44. [Google Scholar] [CrossRef] [PubMed]
- Kerr, J.F. Shrinkage necrosis: A distinct mode of cellular death. J. Pathol. 1971, 105, 13–20. [Google Scholar] [CrossRef]
- Saunders, J.W., Jr. Death in embryonic systems. Science 1966, 154, 604–612. [Google Scholar] [CrossRef]
- Webster, D.A.; Gross, J. Studies on possible mechanisms of programmed cell death in the chick embryo. Dev. Biol. 1970, 22, 157–184. [Google Scholar] [CrossRef]
- Kerr, J.F. A histochemical study of hypertrophy and ischaemic injury of rat liver with special reference to changes in lysosomes. J. Pathol. Bacteriol. 1965, 90, 419–435. [Google Scholar] [CrossRef]
- Kerr, J.F. History of the events leading to the formulation of the apoptosis concept. Toxicology 2002, 181–182, 471–474. [Google Scholar] [CrossRef]
- Haanen, C.; Vermes, I. Apoptosis and inflammation. Mediat. Inflamm. 1995, 4, 5–15. [Google Scholar] [CrossRef]
- Crawford, A.M.; Kerr, J.F.; Currie, A.R. The relationship of acute mesodermal cell death to the teratogenic effects of 7-OHM-12-MBA in the foetal rat. Br. J. Cancer 1972, 26, 498–503. [Google Scholar] [CrossRef] [Green Version]
- Wyllie, A.H.; Kerr, J.F.; Currie, A.R. Cell death in the normal neonatal rat adrenal cortex. J. Pathol. 1973, 111, 255–261. [Google Scholar] [CrossRef]
- Wyllie, A.H.; Kerr, J.F.; Macaskill, I.A.; Currie, A.R. Adrenocortical cell deletion: The role of ACTH. J. Pathol. 1973, 111, 85–94. [Google Scholar] [CrossRef]
- Kerr, J.F. An electron microscopic study of liver cell necrosis due to albitocin. Pathology 1970, 2, 251–259. [Google Scholar] [CrossRef] [PubMed]
- Kress, M.; May, E.; Cassingena, R.; May, P. Simian virus 40-transformed cells express new species of proteins precipitable by anti-simian virus 40 tumor serum. J. Virol. 1979, 31, 472–483. [Google Scholar] [CrossRef] [Green Version]
- Lane, D.P.; Crawford, L.V. T antigen is bound to a host protein in SV40-transformed cells. Nature 1979, 278, 261–263. [Google Scholar] [CrossRef] [PubMed]
- Linzer, D.I.; Levine, A.J. Characterization of a 54K dalton cellular SV40 tumor antigen present in SV40-transformed cells and uninfected embryonal carcinoma cells. Cell 1979, 17, 43–52. [Google Scholar] [CrossRef]
- Melero, J.A.; Stitt, D.T.; Mangel, W.F.; Carroll, R.B. Identification of new polypeptide species (48–55K) immunoprecipitable by antiserum to purified large T antigen and present in SV40-infected and -transformed cells. Virology 1979, 93, 466–480. [Google Scholar] [CrossRef]
- Smith, A.E.; Smith, R.; Paucha, E. Characterization of different tumor antigens present in cells transformed by simian virus 40. Cell 1979, 18, 335–346. [Google Scholar] [CrossRef]
- Rotter, V.; Witte, O.N.; Coffman, R.; Baltimore, D. Abelson murine leukemia virus-induced tumors elicit antibodies against a host cell protein, P50. J. Virol. 1980, 36, 547–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeLeo, A.B.; Jay, G.; Appella, E.; Dubois, G.C.; Law, L.W.; Old, L.J. Detection of a transformation-related antigen in chemically induced sarcomas and other transformed cells of the mouse. Proc. Natl. Acad. Sci. USA 1979, 76, 2420–2424. [Google Scholar] [CrossRef] [Green Version]
- Levine, A.J.; Oren, M. The first 30 years of p53: Growing ever more complex. Nat. Rev. Cancer 2009, 9, 749–758. [Google Scholar] [CrossRef] [Green Version]
- Baker, S.J.; Vogelstein, B. p53: A tumor suppressor hiding in plain sight. J. Mol. Cell Biol. 2019, 11, 536–538. [Google Scholar] [CrossRef]
- Baker, S.J.; Fearon, E.R.; Nigro, J.M.; Hamilton, S.R.; Preisinger, A.C.; Jessup, J.M.; van Tuinen, P.; Ledbetter, D.H.; Barker, D.F.; Nakamura, Y.; et al. Chromosome 17 deletions and p53 gene mutations in colorectal carcinomas. Science 1989, 244, 217–221. [Google Scholar] [CrossRef] [PubMed]
- Finlay, C.A.; Hinds, P.W.; Levine, A.J. The p53 proto-oncogene can act as a suppressor of transformation. Cell 1989, 57, 1083–1093. [Google Scholar] [CrossRef]
- Sulston, J.E.; Horvitz, H.R. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans. Dev. Biol. 1977, 56, 110–156. [Google Scholar] [CrossRef]
- Sulston, J.E.; Schierenberg, E.; White, J.G.; Thomson, J.N. The embryonic cell lineage of the nematode Caenorhabditis elegans. Dev. Biol. 1983, 100, 64–119. [Google Scholar] [CrossRef]
- Ellis, H.M.; Horvitz, H.R. Genetic control of programmed cell death in the nematode C. elegans. Cell 1986, 44, 817–829. [Google Scholar] [CrossRef]
- Hedgecock, E.M.; Thomson, J.N. A gene required for nuclear and mitochondrial attachment in the nematode Caenorhabditis elegans. Cell 1982, 30, 321–330. [Google Scholar] [CrossRef]
- Yuan, J.; Horvitz, H.R. The Caenorhabditis elegans cell death gene ced-4 encodes a novel protein and is expressed during the period of extensive programmed cell death. Development 1992, 116, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Hengartner, M.O.; Ellis, R.E.; Horvitz, H.R. Caenorhabditis elegans gene ced-9 protects cells from programmed cell death. Nature 1992, 356, 494–499. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Finger, L.R.; Yunis, J.; Nowell, P.C.; Croce, C.M. Cloning of the chromosome breakpoint of neoplastic B cells with the t(14;18) chromosome translocation. Science 1984, 226, 1097–1099. [Google Scholar] [CrossRef]
- Cleary, M.L.; Smith, S.D.; Sklar, J. Cloning and structural analysis of cDNAs for bcl-2 and a hybrid bcl-2/immunoglobulin transcript resulting from the t(14;18) translocation. Cell 1986, 47, 19–28. [Google Scholar] [CrossRef]
- Tsujimoto, Y.; Croce, C.M. Analysis of the structure, transcripts, and protein products of bcl-2, the gene involved in human follicular lymphoma. Proc. Natl.Acad. Sci. USA 1986, 83, 5214–5218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vaux, D.L.; Cory, S.; Adams, J.M. Bcl-2 gene promotes haemopoietic cell survival and cooperates with c-myc to immortalize pre-B cells. Nature 1988, 335, 440–442. [Google Scholar] [CrossRef] [PubMed]
- Reed, J.C. Bcl-2-family proteins and hematologic malignancies: History and future prospects. Blood 2008, 111, 3322–3330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delbridge, A.R.; Grabow, S.; Strasser, A.; Vaux, D.L. Thirty years of BCL-2: Translating cell death discoveries into novel cancer therapies. Nat. Rev. Cancer 2016, 16, 99–109. [Google Scholar] [CrossRef] [PubMed]
- McDonnell, T.J.; Deane, N.; Platt, F.M.; Nunez, G.; Jaeger, U.; McKearn, J.P.; Korsmeyer, S.J. bcl-2-immunoglobulin transgenic mice demonstrate extended B cell survival and follicular lymphoproliferation. Cell 1989, 57, 79–88. [Google Scholar] [CrossRef]
- Vaux, D.L.; Weissman, I.L.; Kim, S.K. Prevention of programmed cell death in Caenorhabditis elegans by human bcl-2. Science 1992, 258, 1955–1957. [Google Scholar] [CrossRef]
- Yuan, J.; Shaham, S.; Ledoux, S.; Ellis, H.M.; Horvitz, H.R. The C. elegans cell death gene ced-3 encodes a protein similar to mammalian interleukin-1 beta-converting enzyme. Cell 1993, 75, 641–652. [Google Scholar] [CrossRef]
- Miura, M.; Zhu, H.; Rotello, R.; Hartwieg, E.A.; Yuan, J. Induction of apoptosis in fibroblasts by IL-1 beta-converting enzyme, a mammalian homolog of the C. elegans cell death gene ced-3. Cell 1993, 75, 653–660. [Google Scholar] [CrossRef]
- Cerretti, D.P.; Kozlosky, C.J.; Mosley, B.; Nelson, N.; Van Ness, K.; Greenstreet, T.A.; March, C.J.; Kronheim, S.R.; Druck, T.; Cannizzaro, L.A.; et al. Molecular cloning of the interleukin-1 beta converting enzyme. Science 1992, 256, 97–100. [Google Scholar] [CrossRef] [PubMed]
- Thornberry, N.A.; Bull, H.G.; Calaycay, J.R.; Chapman, K.T.; Howard, A.D.; Kostura, M.J.; Miller, D.K.; Molineaux, S.M.; Weidner, J.R.; Aunins, J.; et al. A novel heterodimeric cysteine protease is required for interleukin-1 beta processing in monocytes. Nature 1992, 356, 768–774. [Google Scholar] [CrossRef]
- Alnemri, E.S.; Livingston, D.J.; Nicholson, D.W.; Salvesen, G.; Thornberry, N.A.; Wong, W.W.; Yuan, J. Human ICE/CED-3 protease nomenclature. Cell 1996, 87, 171. [Google Scholar] [CrossRef] [Green Version]
- Wilson, K.P.; Black, J.A.; Thomson, J.A.; Kim, E.E.; Griffith, J.P.; Navia, M.A.; Murcko, M.A.; Chambers, S.P.; Aldape, R.A.; Raybuck, S.A.; et al. Structure and mechanism of interleukin-1 beta converting enzyme. Nature 1994, 370, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Malinowski, J.J.; Grasberger, B.L.; Trakshel, G.; Huston, E.E.; Helaszek, C.T.; Smallwood, A.M.; Ator, M.A.; Banks, T.M.; Brake, P.G.; Ciccarelli, R.B.; et al. Production, purification, and crystallization of human interleukin-1 beta converting enzyme derived from an Escherichia coli expression system. Protein Sci. 1995, 4, 2149–2155. [Google Scholar] [CrossRef]
- Miller, D.K.; Ayala, J.M.; Egger, L.A.; Raju, S.M.; Yamin, T.T.; Ding, G.J.; Gaffney, E.P.; Howard, A.D.; Palyha, O.C.; Rolando, A.M.; et al. Purification and characterization of active human interleukin-1 beta-converting enzyme from THP.1 monocytic cells. J. Biol. Chem. 1993, 268, 18062–18069. [Google Scholar] [CrossRef]
- Kumar, S.; Kinoshita, M.; Noda, M.; Copeland, N.G.; Jenkins, N.A. Induction of apoptosis by the mouse Nedd2 gene, which encodes a protein similar to the product of the Caenorhabditis elegans cell death gene ced-3 and the mammalian IL-1 beta-converting enzyme. Genes Dev. 1994, 8, 1613–1626. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.; Shaham, S.; Horvitz, H.R. The Caenorhabditis elegans cell-death protein CED-3 is a cysteine protease with substrate specificities similar to those of the human CPP32 protease. Genes Dev. 1996, 10, 1073–1083. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes-Alnemri, T.; Litwack, G.; Alnemri, E.S. CPP32, a novel human apoptotic protein with homology to Caenorhabditis elegans cell death protein Ced-3 and mammalian interleukin-1 beta-converting enzyme. J. Biol. Chem. 1994, 269, 30761–30764. [Google Scholar] [CrossRef]
- Wang, L.; Miura, M.; Bergeron, L.; Zhu, H.; Yuan, J. Ich-1, an Ice/ced-3-related gene, encodes both positive and negative regulators of programmed cell death. Cell 1994, 78, 739–750. [Google Scholar] [CrossRef]
- Johnson, A.L.; Bridgham, J.T.; Bergeron, L.; Yuan, J. Characterization of the avian Ich-1 cDNA and expression of Ich-1L mRNA in the hen ovary. Gene 1997, 192, 227–233. [Google Scholar] [CrossRef]
- Tewari, M.; Quan, L.T.; O’Rourke, K.; Desnoyers, S.; Zeng, Z.; Beidler, D.R.; Poirier, G.G.; Salvesen, G.S.; Dixit, V.M. Yama/CPP32β, a mammalian homolog of CED-3, is a CrmA-inhibitable protease that cleaves the death substrate poly(ADP-ribose) polymerase. Cell 1995, 81, 801–809. [Google Scholar] [CrossRef] [Green Version]
- Nicholson, D.W.; Ali, A.; Thornberry, N.A.; Vaillancourt, J.P.; Ding, C.K.; Gallant, M.; Gareau, Y.; Griffin, P.R.; Labelle, M.; Lazebnik, Y.A.; et al. Identification and inhibition of the ICE/CED-3 protease necessary for mammalian apoptosis. Nature 1995, 376, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Walker, N.P.; Talanian, R.V.; Brady, K.D.; Dang, L.C.; Bump, N.J.; Ferenz, C.R.; Franklin, S.; Ghayur, T.; Hackett, M.C.; Hammill, L.D.; et al. Crystal structure of the cysteine protease interleukin-1 beta-converting enzyme: A (p20/p10)2 homodimer. Cell 1994, 78, 343–352. [Google Scholar] [CrossRef]
- Alnemri, E.S.; Fernandes-Alnemri, T.; Litwack, G. Cloning and expression of four novel isoforms of human interleukin-1 beta converting enzyme with different apoptotic activities. J. Biol. Chem. 1995, 270, 4312–4317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes-Alnemri, T.; Litwack, G.; Alnemri, E.S. Mch2, a new member of the apoptotic Ced-3/Ice cysteine protease gene family. Cancer Res. 1995, 55, 2737–2742. [Google Scholar]
- Fernandes-Alnemri, T.; Takahashi, A.; Armstrong, R.; Krebs, J.; Fritz, L.; Tomaselli, K.J.; Wang, L.; Yu, Z.; Croce, C.M.; Salveson, G.; et al. Mch3, a novel human apoptotic cysteine protease highly related to CPP32. Cancer Res. 1995, 55, 6045–6052. [Google Scholar]
- Boise, L.H.; Gonzalez-Garcia, M.; Postema, C.E.; Ding, L.; Lindsten, T.; Turka, L.A.; Mao, X.; Nunez, G.; Thompson, C.B. bcl-x, a bcl-2-related gene that functions as a dominant regulator of apoptotic cell death. Cell 1993, 74, 597–608. [Google Scholar] [CrossRef] [Green Version]
- Kozopas, K.M.; Yang, T.; Buchan, H.L.; Zhou, P.; Craig, R.W. MCL1, a gene expressed in programmed myeloid cell differentiation, has sequence similarity to BCL2. Proc. Natl.Acad. Sci. USA 1993, 90, 3516–3520. [Google Scholar] [CrossRef] [Green Version]
- Gibson, L.; Holmgreen, S.P.; Huang, D.C.; Bernard, O.; Copeland, N.G.; Jenkins, N.A.; Sutherland, G.R.; Baker, E.; Adams, J.M.; Cory, S. bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene 1996, 13, 665–675. [Google Scholar]
- Farrow, S.N.; White, J.H.; Martinou, I.; Raven, T.; Pun, K.T.; Grinham, C.J.; Martinou, J.C.; Brown, R. Cloning of a bcl-2 homologue by interaction with adenovirus E1B 19K. Nature 1995, 374, 731–733. [Google Scholar] [CrossRef] [PubMed]
- Kiefer, M.C.; Brauer, M.J.; Powers, V.C.; Wu, J.J.; Umansky, S.R.; Tomei, L.D.; Barr, P.J. Modulation of apoptosis by the widely distributed Bcl-2 homologue Bak. Nature 1995, 374, 736–739. [Google Scholar] [CrossRef] [PubMed]
- Chittenden, T.; Harrington, E.A.; O’Connor, R.; Flemington, C.; Lutz, R.J.; Evan, G.I.; Guild, B.C. Induction of apoptosis by the Bcl-2 homologue Bak. Nature 1995, 374, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Oltvai, Z.N.; Milliman, C.L.; Korsmeyer, S.J. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell 1993, 74, 609–619. [Google Scholar] [CrossRef]
- Boyd, J.M.; Malstrom, S.; Subramanian, T.; Venkatesh, L.K.; Schaeper, U.; Elangovan, B.; D’Sa-Eipper, C.; Chinnadurai, G. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 1994, 79, 341–351. [Google Scholar] [CrossRef]
- Oda, E.; Ohki, R.; Murasawa, H.; Nemoto, J.; Shibue, T.; Yamashita, T.; Tokino, T.; Taniguchi, T.; Tanaka, N. Noxa, a BH3-only member of the Bcl-2 family and candidate mediator of p53-induced apoptosis. Science 2000, 288, 1053–1058. [Google Scholar] [CrossRef] [PubMed]
- Nakano, K.; Vousden, K.H. PUMA, a novel proapoptotic gene, is induced by p53. Mol. Cell 2001, 7, 683–694. [Google Scholar] [CrossRef]
- Boyd, J.M.; Gallo, G.J.; Elangovan, B.; Houghton, A.B.; Malstrom, S.; Avery, B.J.; Ebb, R.G.; Subramanian, T.; Chittenden, T.; Lutz, R.J.; et al. Bik, a novel death-inducing protein shares a distinct sequence motif with Bcl-2 family proteins and interacts with viral and cellular survival-promoting proteins. Oncogene 1995, 11, 1921–1928. [Google Scholar]
- Wang, K.; Yin, X.M.; Chao, D.T.; Milliman, C.L.; Korsmeyer, S.J. BID: A novel BH3 domain-only death agonist. Genes Dev. 1996, 10, 2859–2869. [Google Scholar] [CrossRef] [Green Version]
- Inohara, N.; Ding, L.; Chen, S.; Nunez, G. harakiri, a novel regulator of cell death, encodes a protein that activates apoptosis and interacts selectively with survival-promoting proteins Bcl-2 and Bcl-X(L). EMBO J. 1997, 16, 1686–1694. [Google Scholar] [CrossRef]
- Yang, E.; Zha, J.; Jockel, J.; Boise, L.H.; Thompson, C.B.; Korsmeyer, S.J. Bad, a heterodimeric partner for Bcl-XL and Bcl-2, displaces Bax and promotes cell death. Cell 1995, 80, 285–291. [Google Scholar] [CrossRef] [Green Version]
- Liang, X.H.; Kleeman, L.K.; Jiang, H.H.; Gordon, G.; Goldman, J.E.; Berry, G.; Herman, B.; Levine, B. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J. Virol. 1998, 72, 8586–8596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kale, J.; Osterlund, E.J.; Andrews, D.W. BCL-2 family proteins: Changing partners in the dance towards death. Cell Death Differ. 2018, 25, 65–80. [Google Scholar] [CrossRef] [Green Version]
- Hengartner, M.O.; Horvitz, H.R.C. elegans cell survival gene ced-9 encodes a functional homolog of the mammalian proto-oncogene bcl-2. Cell 1994, 76, 665–676. [Google Scholar] [CrossRef]
- Hengartner, M.O.; Horvitz, H.R. Programmed cell death in Caenorhabditis elegans. Curr. Opin. Genet. Dev. 1994, 4, 581–586. [Google Scholar] [CrossRef]
- Liu, X.; Kim, C.N.; Yang, J.; Jemmerson, R.; Wang, X. Induction of apoptotic program in cell-free extracts: Requirement for dATP and cytochrome c. Cell 1996, 86, 147–157. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Henzel, W.J.; Liu, X.; Lutschg, A.; Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997, 90, 405–413. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Liu, X.; Bhalla, K.; Kim, C.N.; Ibrado, A.M.; Cai, J.; Peng, T.I.; Jones, D.P.; Wang, X. Prevention of apoptosis by Bcl-2: Release of cytochrome c from mitochondria blocked. Science 1997, 275, 1129–1132. [Google Scholar] [CrossRef]
- Kluck, R.M.; Bossy-Wetzel, E.; Green, D.R.; Newmeyer, D.D. The release of cytochrome c from mitochondria: A primary site for Bcl-2 regulation of apoptosis. Science 1997, 275, 1132–1136. [Google Scholar] [CrossRef] [Green Version]
- Pourkarimi, E.; Greiss, S.; Gartner, A. Evidence that CED-9/Bcl2 and CED-4/Apaf-1 localization is not consistent with the current model for C. elegans apoptosis induction. Cell Death Differ. 2012, 19, 406–415. [Google Scholar] [CrossRef] [Green Version]
- Shaham, S.; Horvitz, H.R. Developing Caenorhabditis elegans neurons may contain both cell-death protective and killer activities. Genes Dev. 1996, 10, 578–591. [Google Scholar] [CrossRef] [Green Version]
- Shaham, S.; Horvitz, H.R. An alternatively spliced C. elegans ced-4 RNA encodes a novel cell death inhibitor. Cell 1996, 86, 201–208. [Google Scholar] [CrossRef] [Green Version]
- Xue, D.; Horvitz, H.R. Caenorhabditis elegans CED-9 protein is a bifunctional cell-death inhibitor. Nature 1997, 390, 305–308. [Google Scholar] [CrossRef]
- Spector, M.S.; Desnoyers, S.; Hoeppner, D.J.; Hengartner, M.O. Interaction between the C. elegans cell-death regulators CED-9 and CED-4. Nature 1997, 385, 653–656. [Google Scholar] [CrossRef] [PubMed]
- Conradt, B.; Horvitz, H.R. The C. elegans protein EGL-1 is required for programmed cell death and interacts with the Bcl-2-like protein CED-9. Cell 1998, 93, 519–529. [Google Scholar] [CrossRef] [Green Version]
- Strasser, A.; Huang, D.C.; Vaux, D.L. The role of the bcl-2/ced-9 gene family in cancer and general implications of defects in cell death control for tumourigenesis and resistance to chemotherapy. Biochim. Biophys. Acta 1997, 1333, F151–F178. [Google Scholar] [CrossRef]
- Moller, G. The Nobel Prize in Physiology or Medicine for 2002. Scand. J. Immunol. 2002, 56, 435. [Google Scholar] [CrossRef] [PubMed]
- Hunter, D.R.; Haworth, R.A.; Southard, J.H. Relationship between configuration, function, and permeability in calcium-treated mitochondria. J. Biol. Chem. 1976, 251, 5069–5077. [Google Scholar] [CrossRef]
- Hunter, D.R.; Haworth, R.A. The Ca2+-induced membrane transition in mitochondria. I. The protective mechanisms. Arch. Biochem. Biophys. 1979, 195, 453–459. [Google Scholar] [CrossRef]
- Haworth, R.A.; Hunter, D.R. The Ca2+-induced membrane transition in mitochondria. II. Nature of the Ca2+ trigger site. Arch. BioChem. Biophys. 1979, 195, 460–467. [Google Scholar] [CrossRef]
- Hunter, D.R.; Haworth, R.A. The Ca2+-induced membrane transition in mitochondria. III. Transitional Ca2+ release. Arch. BioChem. Biophys. 1979, 195, 468–477. [Google Scholar] [CrossRef]
- Brumatti, G.; Salmanidis, M.; Ekert, P.G. Crossing paths: Interactions between the cell death machinery and growth factor survival signals. Cell Mol. Life Sci. 2010, 67, 1619–1630. [Google Scholar] [CrossRef] [PubMed]
- Morris, J.L.; Gillet, G.; Prudent, J.; Popgeorgiev, N. Bcl-2 Family of Proteins in the Control of Mitochondrial Calcium Signalling: An Old Chap with New Roles. Int. J. Mol. Sci. 2021, 22, 3730. [Google Scholar] [CrossRef] [PubMed]
- Pihan, P.; Carreras-Sureda, A.; Hetz, C. BCL-2 family: Integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017, 24, 1478–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roos, W.P.; Thomas, A.D.; Kaina, B. DNA damage and the balance between survival and death in cancer biology. Nat. Rev. Cancer 2016, 16, 20–33. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; De Maria, R.; Kroemer, G.; Galluzzi, L. DNA Damage in Stem Cells. Mol. Cell 2017, 66, 306–319. [Google Scholar] [CrossRef] [Green Version]
- Wunderlich, R.; Ruehle, P.F.; Deloch, L.; Unger, K.; Hess, J.; Zitzelsberger, H.; Lauber, K.; Frey, B.; Gaipl, U.S. Interconnection between DNA damage, senescence, inflammation, and cancer. Front. Biosci. 2017, 22, 348–369. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Tu, H.C.; Ren, D.; Takeuchi, O.; Jeffers, J.R.; Zambetti, G.P.; Hsieh, J.J.; Cheng, E.H. Stepwise activation of BAX and BAK by tBID, BIM, and PUMA initiates mitochondrial apoptosis. Mol. Cell 2009, 36, 487–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cheng, E.H.; Wei, M.C.; Weiler, S.; Flavell, R.A.; Mak, T.W.; Lindsten, T.; Korsmeyer, S.J. BCL-2, BCL-X(L) sequester BH3 domain-only molecules preventing BAX- and BAK-mediated mitochondrial apoptosis. Mol. Cell 2001, 8, 705–711. [Google Scholar] [CrossRef]
- Letai, A.; Bassik, M.C.; Walensky, L.D.; Sorcinelli, M.D.; Weiler, S.; Korsmeyer, S.J. Distinct BH3 domains either sensitize or activate mitochondrial apoptosis, serving as prototype cancer therapeutics. Cancer Cell 2002, 2, 183–192. [Google Scholar] [CrossRef] [Green Version]
- O’Neill, K.L.; Huang, K.; Zhang, J.; Chen, Y.; Luo, X. Inactivation of prosurvival Bcl-2 proteins activates Bax/Bak through the outer mitochondrial membrane. Genes Dev. 2016, 30, 973–988. [Google Scholar] [CrossRef] [Green Version]
- Bleicken, S.; Jeschke, G.; Stegmueller, C.; Salvador-Gallego, R.; Garcia-Saez, A.J.; Bordignon, E. Structural model of active Bax at the membrane. Mol. Cell 2014, 56, 496–505. [Google Scholar] [CrossRef] [Green Version]
- Kuwana, T.; Mackey, M.R.; Perkins, G.; Ellisman, M.H.; Latterich, M.; Schneiter, R.; Green, D.R.; Newmeyer, D.D. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002, 111, 331–342. [Google Scholar] [CrossRef] [Green Version]
- Dadsena, S.; Jenner, A.; Garcia-Saez, A.J. Mitochondrial outer membrane permeabilization at the single molecule level. Cell Mol. Life Sci. 2021, 78, 3777–3790. [Google Scholar] [CrossRef]
- Uren, R.T.; Iyer, S.; Kluck, R.M. Pore formation by dimeric Bak and Bax: An unusual pore? Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372. [Google Scholar] [CrossRef]
- Dadsena, S.; Zollo, C.; Garcia-Saez, A.J. Mechanisms of mitochondrial cell death. BioChem. Soc. Trans. 2021, 49, 663–674. [Google Scholar] [CrossRef]
- Tait, S.W.; Green, D.R. Mitochondria and cell death: Outer membrane permeabilization and beyond. Nat. Rev. Mol. Cell Biol. 2010, 11, 621–632. [Google Scholar] [CrossRef] [PubMed]
- Zamzami, N.; Marchetti, P.; Castedo, M.; Zanin, C.; Vayssiere, J.L.; Petit, P.X.; Kroemer, G. Reduction in mitochondrial potential constitutes an early irreversible step of programmed lymphocyte death in vivo. J. Exp. Med. 1995, 181, 1661–1672. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bratton, S.B.; Walker, G.; Srinivasula, S.M.; Sun, X.M.; Butterworth, M.; Alnemri, E.S.; Cohen, G.M. Recruitment, activation and retention of caspases-9 and -3 by Apaf-1 apoptosome and associated XIAP complexes. EMBO J. 2001, 20, 998–1009. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Shiozaki, E.; Srinivasula, S.M.; Wu, Q.; Datta, P.; Alnemri, E.S.; Shi, Y. Structural basis of caspase-7 inhibition by XIAP. Cell 2001, 104, 769–780. [Google Scholar] [CrossRef]
- Deveraux, Q.L.; Roy, N.; Stennicke, H.R.; Van Arsdale, T.; Zhou, Q.; Srinivasula, S.M.; Alnemri, E.S.; Salvesen, G.S.; Reed, J.C. IAPs block apoptotic events induced by caspase-8 and cytochrome c by direct inhibition of distinct caspases. EMBO J. 1998, 17, 2215–2223. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Park, Y.C.; Rich, R.L.; Segal, D.; Myszka, D.G.; Wu, H. Structural basis of caspase inhibition by XIAP: Differential roles of the linker versus the BIR domain. Cell 2001, 104, 781–790. [Google Scholar] [CrossRef]
- Takahashi, R.; Deveraux, Q.; Tamm, I.; Welsh, K.; Assa-Munt, N.; Salvesen, G.S.; Reed, J.C. A single BIR domain of XIAP sufficient for inhibiting caspases. J. Biol. Chem. 1998, 273, 7787–7790. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.L.; Li, X.M. The IAP family: Endogenous caspase inhibitors with multiple biological activities. Cell Res. 2000, 10, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Du, C.; Fang, M.; Li, Y.; Li, L.; Wang, X. Smac, a mitochondrial protein that promotes cytochrome c-dependent caspase activation by eliminating IAP inhibition. Cell 2000, 102, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Kalkavan, H.; Green, D.R. MOMP, cell suicide as a BCL-2 family business. Cell Death Differ. 2018, 25, 46–55. [Google Scholar] [CrossRef] [PubMed]
- Fesik, S.W.; Shi, Y. Structural biology. Controlling the caspases. Science 2001, 294, 1477–1478. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Sun, C.; Olejniczak, E.T.; Meadows, R.P.; Betz, S.F.; Oost, T.; Herrmann, J.; Wu, J.C.; Fesik, S.W. Structural basis for binding of Smac/DIABLO to the XIAP BIR3 domain. Nature 2000, 408, 1004–1008. [Google Scholar] [CrossRef] [PubMed]
- Shiozaki, E.N.; Shi, Y. Caspases, IAPs and Smac/DIABLO: Mechanisms from structural biology. Trends Biochem. Sci. 2004, 29, 486–494. [Google Scholar] [CrossRef]
- Kesavardhana, S.; Malireddi, R.K.S.; Kanneganti, T.D. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu. Rev. Immunol. 2020, 38, 567–595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verhagen, A.M.; Ekert, P.G.; Pakusch, M.; Silke, J.; Connolly, L.M.; Reid, G.E.; Moritz, R.L.; Simpson, R.J.; Vaux, D.L. Identification of DIABLO, a mammalian protein that promotes apoptosis by binding to and antagonizing IAP proteins. Cell 2000, 102, 43–53. [Google Scholar] [CrossRef] [Green Version]
- Acehan, D.; Jiang, X.; Morgan, D.G.; Heuser, J.E.; Wang, X.; Akey, C.W. Three-dimensional structure of the apoptosome: Implications for assembly, procaspase-9 binding, and activation. Mol. Cell 2002, 9, 423–432. [Google Scholar] [CrossRef]
- Bratton, S.B.; Walker, G.; Roberts, D.L.; Cain, K.; Cohen, G.M. Caspase-3 cleaves Apaf-1 into an approximately 30 kDa fragment that associates with an inappropriately oligomerized and biologically inactive approximately 1.4 MDa apoptosome complex. Cell Death Differ. 2001, 8, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Cain, K. Chemical-induced apoptosis: Formation of the Apaf-1 apoptosome. Drug Metab. Rev. 2003, 35, 337–363. [Google Scholar] [CrossRef] [PubMed]
- Cain, K.; Bratton, S.B.; Langlais, C.; Walker, G.; Brown, D.G.; Sun, X.M.; Cohen, G.M. Apaf-1 oligomerizes into biologically active approximately 700-kDa and inactive approximately 1.4-MDa apoptosome complexes. J. Biol. Chem. 2000, 275, 6067–6070. [Google Scholar] [CrossRef] [Green Version]
- Cain, K.; Brown, D.G.; Langlais, C.; Cohen, G.M. Caspase activation involves the formation of the aposome, a large (approximately 700 kDa) caspase-activating complex. J. Biol. Chem. 1999, 274, 22686–22692. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997, 91, 479–489. [Google Scholar] [CrossRef] [Green Version]
- Shakeri, R.; Kheirollahi, A.; Davoodi, J. Apaf-1: Regulation and function in cell death. Biochimie 2017, 135, 111–125. [Google Scholar] [CrossRef] [PubMed]
- Srinivasula, S.M.; Ahmad, M.; Fernandes-Alnemri, T.; Alnemri, E.S. Autoactivation of procaspase-9 by Apaf-1-mediated oligomerization. Mol. Cell 1998, 1, 949–957. [Google Scholar] [CrossRef]
- Slee, E.A.; Harte, M.T.; Kluck, R.M.; Wolf, B.B.; Casiano, C.A.; Newmeyer, D.D.; Wang, H.G.; Reed, J.C.; Nicholson, D.W.; Alnemri, E.S.; et al. Ordering the cytochrome c-initiated caspase cascade: Hierarchical activation of caspases-2, -3, -6, -7, -8, and -10 in a caspase-9-dependent manner. J. Cell Biol. 1999, 144, 281–292. [Google Scholar] [CrossRef] [PubMed]
- Logue, S.E.; Martin, S.J. Caspase activation cascades in apoptosis. Biochem. Soc. Trans. 2008, 36, 1–9. [Google Scholar] [CrossRef]
- Srinivasula, S.M.; Hegde, R.; Saleh, A.; Datta, P.; Shiozaki, E.; Chai, J.; Lee, R.A.; Robbins, P.D.; Fernandes-Alnemri, T.; Shi, Y.; et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature 2001, 410, 112–116. [Google Scholar] [CrossRef] [PubMed]
- Twiddy, D.; Cain, K. Caspase-9 cleavage, do you need it? BioChem. J. 2007, 405, e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walczak, H.; Krammer, P.H. The CD95 (APO-1/Fas) and the TRAIL (APO-2L) apoptosis systems. Exp. Cell Res. 2000, 256, 58–66. [Google Scholar] [CrossRef] [PubMed]
- Flynn, J.L.; Goldstein, M.M.; Chan, J.; Triebold, K.J.; Pfeffer, K.; Lowenstein, C.J.; Schreiber, R.; Mak, T.W.; Bloom, B.R. Tumor necrosis factor-alpha is required in the protective immune response against Mycobacterium tuberculosis in mice. Immunity 1995, 2, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Giroir, B.P.; Johnson, J.H.; Brown, T.; Allen, G.L.; Beutler, B. The tissue distribution of tumor necrosis factor biosynthesis during endotoxemia. J. Clin. Investig. 1992, 90, 693–698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeffer, K.; Matsuyama, T.; Kundig, T.M.; Wakeham, A.; Kishihara, K.; Shahinian, A.; Wiegmann, K.; Ohashi, P.S.; Kronke, M.; Mak, T.W. Mice deficient for the 55 kd tumor necrosis factor receptor are resistant to endotoxic shock, yet succumb to L. monocytogenes infection. Cell 1993, 73, 457–467. [Google Scholar] [CrossRef]
- Baer, M.; Dillner, A.; Schwartz, R.C.; Sedon, C.; Nedospasov, S.; Johnson, P.F. Tumor necrosis factor alpha transcription in macrophages is attenuated by an autocrine factor that preferentially induces NF-kappaB p50. Mol. Cell Biol. 1998, 18, 5678–5689. [Google Scholar] [CrossRef] [Green Version]
- Kriegler, M.; Perez, C.; DeFay, K.; Albert, I.; Lu, S.D. A novel form of TNF/cachectin is a cell surface cytotoxic transmembrane protein: Ramifications for the complex physiology of TNF. Cell 1988, 53, 45–53. [Google Scholar] [CrossRef]
- Jiang, Y.; Yu, M.; Hu, X.; Han, L.; Yang, K.; Ba, H.; Zhang, Z.; Yin, B.; Yang, X.P.; Li, Z.; et al. STAT1 mediates transmembrane TNF-alpha-induced formation of death-inducing signaling complex and apoptotic signaling via TNFR1. Cell Death Differ. 2017, 24, 660–671. [Google Scholar] [CrossRef] [Green Version]
- Gon, S.; Gatanaga, T.; Sendo, F. Involvement of two types of TNF receptor in TNF-alpha induced neutrophil apoptosis. Microbiol. Immunol. 1996, 40, 463–465. [Google Scholar] [CrossRef]
- Pennica, D.; Nedwin, G.E.; Hayflick, J.S.; Seeburg, P.H.; Derynck, R.; Palladino, M.A.; Kohr, W.J.; Aggarwal, B.B.; Goeddel, D.V. Human tumour necrosis factor: Precursor structure, expression and homology to lymphotoxin. Nature 1984, 312, 724–729. [Google Scholar] [CrossRef]
- Micheau, O.; Tschopp, J. Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 2003, 114, 181–190. [Google Scholar] [CrossRef] [Green Version]
- Kischkel, F.C.; Lawrence, D.A.; Tinel, A.; LeBlanc, H.; Virmani, A.; Schow, P.; Gazdar, A.; Blenis, J.; Arnott, D.; Ashkenazi, A. Death receptor recruitment of endogenous caspase-10 and apoptosis initiation in the absence of caspase-8. J. Biol. Chem. 2001, 276, 46639–46646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Chun, H.J.; Wong, W.; Spencer, D.M.; Lenardo, M.J. Caspase-10 is an initiator caspase in death receptor signaling. Proc. Natl. Acad. Sci. USA 2001, 98, 13884–13888. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Herreweghe, F.; Festjens, N.; Declercq, W.; Vandenabeele, P. Tumor necrosis factor-mediated cell death: To break or to burst, that’s the question. Cell Mol. Life Sci. 2010, 67, 1567–1579. [Google Scholar] [CrossRef]
- Kim, J.W.; Choi, E.J.; Joe, C.O. Activation of death-inducing signaling complex (DISC) by pro-apoptotic C-terminal fragment of RIP. Oncogene 2000, 19, 4491–4499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider-Brachert, W.; Tchikov, V.; Neumeyer, J.; Jakob, M.; Winoto-Morbach, S.; Held-Feindt, J.; Heinrich, M.; Merkel, O.; Ehrenschwender, M.; Adam, D.; et al. Compartmentalization of TNF receptor 1 signaling: Internalized TNF receptosomes as death signaling vesicles. Immunity 2004, 21, 415–428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dohrman, A.; Kataoka, T.; Cuenin, S.; Russell, J.Q.; Tschopp, J.; Budd, R.C. Cellular FLIP (long form) regulates CD8+ T cell activation through caspase-8-dependent NF-kappa B activation. J. Immunol. 2005, 174, 5270–5278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuda, I.; Matsuo, K.; Matsushita, Y.; Haruna, Y.; Niwa, M.; Kataoka, T. The C-terminal domain of the long form of cellular FLICE-inhibitory protein (c-FLIPL) inhibits the interaction of the caspase 8 prodomain with the receptor-interacting protein 1 (RIP1) death domain and regulates caspase 8-dependent nuclear factor kappaB (NF-kappaB) activation. J. Biol. Chem. 2014, 289, 3876–3887. [Google Scholar] [CrossRef] [Green Version]
- Seol, D.-W.; Li, J.; Seol, M.-H.; Park, S.-Y.; Talanian, R.V.; Billiar, T.R. Signaling Events Triggered by Tumor Necrosis Factor-related Apoptosis-inducing Ligand (TRAIL): Caspase-8 Is Required for TRAIL-induced Apoptosis. Cancer Res. 2001, 61, 1138. [Google Scholar]
- Eskes, R.; Desagher, S.; Antonsson, B.; Martinou, J.C. Bid induces the oligomerization and insertion of Bax into the outer mitochondrial membrane. Mol. Cell Biol. 2000, 20, 929–935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gross, A.; Yin, X.M.; Wang, K.; Wei, M.C.; Jockel, J.; Milliman, C.; Erdjument-Bromage, H.; Tempst, P.; Korsmeyer, S.J. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 1999, 274, 1156–1163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kamada, S.; Washida, M.; Hasegawa, J.; Kusano, H.; Funahashi, Y.; Tsujimoto, Y. Involvement of caspase-4(-like) protease in Fas-mediated apoptotic pathway. Oncogene 1997, 15, 285–290. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhu, H.; Xu, C.J.; Yuan, J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998, 94, 491–501. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Budihardjo, I.; Zou, H.; Slaughter, C.; Wang, X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998, 94, 481–490. [Google Scholar] [CrossRef] [Green Version]
- Srinivasula, S.M.; Ahmad, M.; Fernandes-Alnemri, T.; Litwack, G.; Alnemri, E.S. Molecular ordering of the Fas-apoptotic pathway: The Fas/APO-1 protease Mch5 is a CrmA-inhibitable protease that activates multiple Ced-3/ICE-like cysteine proteases. Proc. Natl. Acad. Sci. USA 1996, 93, 14486–14491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, M.C.; Lindsten, T.; Mootha, V.K.; Weiler, S.; Gross, A.; Ashiya, M.; Thompson, C.B.; Korsmeyer, S.J. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000, 14, 2060–2071. [Google Scholar]
- Hennino, A.; Berard, M.; Casamayor-Palleja, M.; Krammer, P.H.; Defrance, T. Regulation of the Fas death pathway by FLICE-inhibitory protein in primary human B cells. J. Immunol. 2000, 165, 3023–3030. [Google Scholar] [CrossRef] [Green Version]
- Irmler, M.; Thome, M.; Hahne, M.; Schneider, P.; Hofmann, K.; Steiner, V.; Bodmer, J.L.; Schroter, M.; Burns, K.; Mattmann, C.; et al. Inhibition of death receptor signals by cellular FLIP. Nature 1997, 388, 190–195. [Google Scholar] [CrossRef]
- Krueger, A.; Schmitz, I.; Baumann, S.; Krammer, P.H.; Kirchhoff, S. Cellular FLICE-inhibitory protein splice variants inhibit different steps of caspase-8 activation at the CD95 death-inducing signaling complex. J. Biol. Chem. 2001, 276, 20633–20640. [Google Scholar] [CrossRef] [Green Version]
- Lavrik, I.N.; Krammer, P.H. Regulation of CD95/Fas signaling at the DISC. Cell Death Differ. 2012, 19, 36–41. [Google Scholar] [CrossRef]
- Scaffidi, C.; Schmitz, I.; Krammer, P.H.; Peter, M.E. The role of c-FLIP in modulation of CD95-induced apoptosis. J. Biol. Chem. 1999, 274, 1541–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.; Webster, J.D.; Varfolomeev, E.; Kwon, Y.C.; Cheng, J.H.; Zhang, J.; Dugger, D.L.; Wickliffe, K.E.; Maltzman, A.; Sujatha-Bhaskar, S.; et al. RIP1 inhibition blocks inflammatory diseases but not tumor growth or metastases. Cell Death Differ. 2020, 27, 161–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Bidere, N.; Staudt, D.; Cubre, A.; Orenstein, J.; Chan, F.K.; Lenardo, M. Competitive control of independent programs of tumor necrosis factor receptor-induced cell death by TRADD and RIP1. Mol. Cell Biol. 2006, 26, 3505–3513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Zhang, M.; Huang, X.; Liang, W.; Li, G.; Lu, X.; Li, Y.; Pan, H.; Shi, L.; Zhu, H.; et al. Ubiquitination of RIPK1 regulates its activation mediated by TNFR1 and TLRs signaling in distinct manners. Nat. Commun. 2020, 11, 6364. [Google Scholar] [CrossRef]
- O’Donnell, M.A.; Legarda-Addison, D.; Skountzos, P.; Yeh, W.C.; Ting, A.T. Ubiquitination of RIP1 regulates an NF-kappaB-independent cell-death switch in TNF signaling. Curr. Biol. 2007, 17, 418–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smyth, P.; Sessler, T.; Scott, C.J.; Longley, D.B. FLIP(L): The pseudo-caspase. FEBS J. 2020, 287, 4246–4260. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, P.M.; Eby, M.; Jasmin, A.; Bookwalter, A.; Murray, J.; Hood, L. Death receptor 5, a new member of the TNFR family, and DR4 induce FADD-dependent apoptosis and activate the NF-kappaB pathway. Immunity 1997, 7, 821–830. [Google Scholar] [CrossRef] [Green Version]
- MacFarlane, M.; Ahmad, M.; Srinivasula, S.M.; Fernandes-Alnemri, T.; Cohen, G.M.; Alnemri, E.S. Identification and molecular cloning of two novel receptors for the cytotoxic ligand TRAIL. J. Biol. Chem. 1997, 272, 25417–25420. [Google Scholar] [CrossRef] [Green Version]
- Pan, G.; O’Rourke, K.; Chinnaiyan, A.M.; Gentz, R.; Ebner, R.; Ni, J.; Dixit, V.M. The receptor for the cytotoxic ligand TRAIL. Science 1997, 276, 111–113. [Google Scholar] [CrossRef]
- Walczak, H.; Degli-Esposti, M.A.; Johnson, R.S.; Smolak, P.J.; Waugh, J.Y.; Boiani, N.; Timour, M.S.; Gerhart, M.J.; Schooley, K.A.; Smith, C.A.; et al. TRAIL-R2: A novel apoptosis-mediating receptor for TRAIL. EMBO J. 1997, 16, 5386–5397. [Google Scholar] [CrossRef]
- Wu, G.S.; Burns, T.F.; McDonald, E.R., 3rd; Jiang, W.; Meng, R.; Krantz, I.D.; Kao, G.; Gan, D.D.; Zhou, J.Y.; Muschel, R.; et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat. Genet. 1997, 17, 141–143. [Google Scholar] [CrossRef] [PubMed]
- Schneider, P.; Thome, M.; Burns, K.; Bodmer, J.L.; Hofmann, K.; Kataoka, T.; Holler, N.; Tschopp, J. TRAIL receptors 1 (DR4) and 2 (DR5) signal FADD-dependent apoptosis and activate NF-kappaB. Immunity 1997, 7, 831–836. [Google Scholar] [CrossRef] [Green Version]
- Suliman, A.; Lam, A.; Datta, R.; Srivastava, R.K. Intracellular mechanisms of TRAIL: Apoptosis through mitochondrial-dependent and -independent pathways. Oncogene 2001, 20, 2122–2133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wajant, H. The Fas signaling pathway: More than a paradigm. Science 2002, 296, 1635–1636. [Google Scholar] [CrossRef] [PubMed]
- Itoh, N.; Nagata, S. A novel protein domain required for apoptosis. Mutational analysis of human Fas antigen. J. Biol. Chem. 1993, 268, 10932–10937. [Google Scholar] [CrossRef]
- Itoh, N.; Yonehara, S.; Ishii, A.; Yonehara, M.; Mizushima, S.; Sameshima, M.; Hase, A.; Seto, Y.; Nagata, S. The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 1991, 66, 233–243. [Google Scholar] [CrossRef]
- Oehm, A.; Behrmann, I.; Falk, W.; Pawlita, M.; Maier, G.; Klas, C.; Li-Weber, M.; Richards, S.; Dhein, J.; Trauth, B.C.; et al. Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor superfamily. Sequence identity with the Fas antigen. J. Biol. Chem. 1992, 267, 10709–10715. [Google Scholar] [CrossRef]
- Schneider, P.; Bodmer, J.L.; Holler, N.; Mattmann, C.; Scuderi, P.; Terskikh, A.; Peitsch, M.C.; Tschopp, J. Characterization of Fas (Apo-1, CD95)-Fas ligand interaction. J. Biol. Chem. 1997, 272, 18827–18833. [Google Scholar] [CrossRef] [Green Version]
- Suda, T.; Takahashi, T.; Golstein, P.; Nagata, S. Molecular cloning and expression of the Fas ligand, a novel member of the tumor necrosis factor family. Cell 1993, 75, 1169–1178. [Google Scholar] [CrossRef]
- Vincenz, C.; Dixit, V.M. Fas-associated death domain protein interleukin-1beta-converting enzyme 2 (FLICE2), an ICE/Ced-3 homologue, is proximally involved in CD95- and p55-mediated death signaling. J. Biol. Chem. 1997, 272, 6578–6583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muzio, M.; Chinnaiyan, A.M.; Kischkel, F.C.; O’Rourke, K.; Shevchenko, A.; Ni, J.; Scaffidi, C.; Bretz, J.D.; Zhang, M.; Gentz, R.; et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell 1996, 85, 817–827. [Google Scholar] [CrossRef] [Green Version]
- Boldin, M.P.; Goncharov, T.M.; Goltsev, Y.V.; Wallach, D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 1996, 85, 803–815. [Google Scholar] [CrossRef] [Green Version]
- Memon, S.A.; Hou, J.; Moreno, M.B.; Zacharchuk, C.M. Apoptosis induced by a chimeric Fas/FLICE receptor: Lack of requirement for Fas- or FADD-binding proteins. J. Immunol. 1998, 160, 2046–2049. [Google Scholar] [PubMed]
- Medema, J.P.; Scaffidi, C.; Kischkel, F.C.; Shevchenko, A.; Mann, M.; Krammer, P.H.; Peter, M.E. FLICE is activated by association with the CD95 death-inducing signaling complex (DISC). EMBO J. 1997, 16, 2794–2804. [Google Scholar] [CrossRef] [PubMed]
- Muzio, M.; Stockwell, B.R.; Stennicke, H.R.; Salvesen, G.S.; Dixit, V.M. An induced proximity model for caspase-8 activation. J. Biol. Chem. 1998, 273, 2926–2930. [Google Scholar] [CrossRef] [Green Version]
- Vanden Berghe, T.; van Loo, G.; Saelens, X.; Van Gurp, M.; Brouckaert, G.; Kalai, M.; Declercq, W.; Vandenabeele, P. Differential signaling to apoptotic and necrotic cell death by Fas-associated death domain protein FADD. J. Biol. Chem. 2004, 279, 7925–7933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamkanfi, M.; Declercq, W.; Kalai, M.; Saelens, X.; Vandenabeele, P. Alice in caspase land. A phylogenetic analysis of caspases from worm to man. Cell Death Differ. 2002, 9, 358–361. [Google Scholar] [CrossRef]
- Julien, O.; Wells, J.A. Caspases and their substrates. Cell Death Differ. 2017, 24, 1380–1389. [Google Scholar] [CrossRef] [PubMed]
- Salvesen, G.S.; Ashkenazi, A. Snapshot: Caspases. Cell 2011, 147, 476. [Google Scholar] [CrossRef] [Green Version]
- Gray, D.C.; Mahrus, S.; Wells, J.A. Activation of specific apoptotic caspases with an engineered small-molecule-activated protease. Cell 2010, 142, 637–646. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coleman, M.L.; Sahai, E.A.; Yeo, M.; Bosch, M.; Dewar, A.; Olson, M.F. Membrane blebbing during apoptosis results from caspase-mediated activation of ROCK I. Nat. Cell Biol. 2001, 3, 339–345. [Google Scholar] [CrossRef]
- Kawane, K.; Nagata, S. Nucleases in programmed cell death. Methods Enzymol. 2008, 442, 271–287. [Google Scholar] [CrossRef] [PubMed]
- Leverrier, Y.; Ridley, A.J. Apoptosis: Caspases orchestrate the ROCK ‘n’ bleb. Nat. Cell Biol. 2001, 3, E91–E93. [Google Scholar] [CrossRef] [PubMed]
- Martin, S.J.; Finucane, D.M.; Amarante-Mendes, G.P.; O’Brien, G.A.; Green, D.R. Phosphatidylserine externalization during CD95-induced apoptosis of cells and cytoplasts requires ICE/CED-3 protease activity. J. Biol. Chem. 1996, 271, 28753–28756. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nagata, S. DNA degradation in development and programmed cell death. Annu Rev. Immunol. 2005, 23, 853–875. [Google Scholar] [CrossRef] [PubMed]
- Naito, M.; Nagashima, K.; Mashima, T.; Tsuruo, T. Phosphatidylserine externalization is a downstream event of interleukin-1 beta-converting enzyme family protease activation during apoptosis. Blood 1997, 89, 2060–2066. [Google Scholar] [CrossRef]
- Santavanond, J.P.; Rutter, S.F.; Atkin-Smith, G.K.; Poon, I.K.H. Apoptotic Bodies: Mechanism of Formation, Isolation and Functional Relevance. Subcell Biochem. 2021, 97, 61–88. [Google Scholar] [CrossRef]
- Sebbagh, M.; Renvoize, C.; Hamelin, J.; Riche, N.; Bertoglio, J.; Breard, J. Caspase-3-mediated cleavage of ROCK I induces MLC phosphorylation and apoptotic membrane blebbing. Nat. Cell Biol. 2001, 3, 346–352. [Google Scholar] [CrossRef]
- Vanags, D.M.; Porn-Ares, M.I.; Coppola, S.; Burgess, D.H.; Orrenius, S. Protease involvement in fodrin cleavage and phosphatidylserine exposure in apoptosis. J. Biol. Chem. 1996, 271, 31075–31085. [Google Scholar] [CrossRef] [Green Version]
- Shi, Y. Caspase activation: Revisiting the induced proximity model. Cell 2004, 117, 855–858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cande, C.; Cohen, I.; Daugas, E.; Ravagnan, L.; Larochette, N.; Zamzami, N.; Kroemer, G. Apoptosis-inducing factor (AIF): A novel caspase-independent death effector released from mitochondria. Biochimie 2002, 84, 215–222. [Google Scholar] [CrossRef]
- Susin, S.A.; Lorenzo, H.K.; Zamzami, N.; Marzo, I.; Snow, B.E.; Brothers, G.M.; Mangion, J.; Jacotot, E.; Costantini, P.; Loeffler, M.; et al. Molecular characterization of mitochondrial apoptosis-inducing factor. Nature 1999, 397, 441–446. [Google Scholar] [CrossRef]
- Ziegler, D.S.; Kung, A.L. Therapeutic targeting of apoptosis pathways in cancer. Curr. Opin. Oncol. 2008, 20, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Inuzuka, H.; Shaik, S.; Onoyama, I.; Gao, D.; Tseng, A.; Maser, R.S.; Zhai, B.; Wan, L.; Gutierrez, A.; Lau, A.W.; et al. SCF(FBW7) regulates cellular apoptosis by targeting MCL1 for ubiquitylation and destruction. Nature 2011, 471, 104–109. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Wan, P.; Xiang, Q.; Huang, S.; Huang, S.; Wang, J.; Wu, K.; Wu, J. E3 Ubiquitin Ligase ASB17 Promotes Apoptosis by Ubiquitylating and Degrading BCLW and MCL1. Biology 2021, 10, 234. [Google Scholar] [CrossRef]
- Slomp, A.; Moesbergen, L.M.; Eldering, E.; Kersten, M.J.; Minnema, M.C.; Peperzak, V. Phosphatase PP2A enhances MCL-1 protein half-life in multiple myeloma cells. Cell Death Dis. 2021, 12, 229. [Google Scholar] [CrossRef] [PubMed]
- Edlich, F.; Banerjee, S.; Suzuki, M.; Cleland, M.M.; Arnoult, D.; Wang, C.; Neutzner, A.; Tjandra, N.; Youle, R.J. Bcl-x(L) retrotranslocates Bax from the mitochondria into the cytosol. Cell 2011, 145, 104–116. [Google Scholar] [CrossRef] [Green Version]
- Todt, F.; Cakir, Z.; Reichenbach, F.; Emschermann, F.; Lauterwasser, J.; Kaiser, A.; Ichim, G.; Tait, S.W.; Frank, S.; Langer, H.F.; et al. Differential retrotranslocation of mitochondrial Bax and Bak. EMBO J. 2015, 34, 67–80. [Google Scholar] [CrossRef] [Green Version]
- Campbell, K.J.; Tait, S.W.G. Targeting BCL-2 regulated apoptosis in cancer. Open Biol. 2018, 8, 180002. [Google Scholar] [CrossRef]
- Liu, Y.; Hernandez, A.M.; Shibata, D.; Cortopassi, G.A. BCL2 translocation frequency rises with age in humans. Proc. Natl.Acad. Sci. USA 1994, 91, 8910–8914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strasser, A.; Harris, A.W.; Cory, S. E mu-bcl-2 transgene facilitates spontaneous transformation of early pre-B and immunoglobulin-secreting cells but not T cells. Oncogene 1993, 8, 1–9. [Google Scholar]
- Soond, S.M.; Savvateeva, L.V.; Makarov, V.A.; Gorokhovets, N.V.; Townsend, P.A.; Zamyatnin, A.A., Jr. Cathepsin S Cleaves BAX as a Novel and Therapeutically Important Regulatory Mechanism for Apoptosis. Pharmaceutics 2021, 13, 339. [Google Scholar] [CrossRef]
- Lowe, S.W.; Schmitt, E.M.; Smith, S.W.; Osborne, B.A.; Jacks, T. p53 is required for radiation-induced apoptosis in mouse thymocytes. Nature 1993, 362, 847–849. [Google Scholar] [CrossRef] [PubMed]
- Fields, S.; Jang, S.K. Presence of a potent transcription activating sequence in the p53 protein. Science 1990, 249, 1046–1049. [Google Scholar] [CrossRef]
- Stewart-Ornstein, J.; Iwamoto, Y.; Miller, M.A.; Prytyskach, M.A.; Ferretti, S.; Holzer, P.; Kallen, J.; Furet, P.; Jambhekar, A.; Forrester, W.C.; et al. p53 dynamics vary between tissues and are linked with radiation sensitivity. Nat. Commun. 2021, 12, 898. [Google Scholar] [CrossRef] [PubMed]
- Miyashita, T.; Reed, J.C. Tumor suppressor p53 is a direct transcriptional activator of the human bax gene. Cell 1995, 80, 293–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanz, G.; Singh, M.; Peuget, S.; Selivanova, G. Inhibition of p53 inhibitors: Progress, challenges and perspectives. J. Mol. Cell Biol. 2019, 11, 586–599. [Google Scholar] [CrossRef] [Green Version]
- Bremer, E. Targeting of the tumor necrosis factor receptor superfamily for cancer immunotherapy. ISRN Oncol. 2013, 2013, 371854. [Google Scholar] [CrossRef] [PubMed]
- Ukrainskaya, V.M.; Stepanov, A.V.; Glagoleva, I.S.; Knorre, V.D.; Belogurov, A.A.J.; Gabibov, A.G. Death Receptors: New Opportunities in Cancer Therapy. Acta Nat. 2017, 9, 55–63. [Google Scholar] [CrossRef]
- Masum, A.A.; Yokoi, K.; Hisamatsu, Y.; Naito, K.; Shashni, B.; Aoki, S. Design and synthesis of a luminescent iridium complex-peptide hybrid (IPH) that detects cancer cells and induces their apoptosis. Bioorg. Med. Chem. 2018, 26, 4804–4816. [Google Scholar] [CrossRef] [PubMed]
- Masum, A.A.; Hisamatsu, Y.; Yokoi, K.; Aoki, S. Luminescent Iridium Complex-Peptide Hybrids (IPHs) for Therapeutics of Cancer: Design and Synthesis of IPHs for Detection of Cancer Cells and Induction of Their Necrosis-Type Cell Death. Bioinorg. Chem. Appl. 2018, 2018, 7578965. [Google Scholar] [CrossRef] [Green Version]
- Waldman, A.D.; Fritz, J.M.; Lenardo, M.J. A guide to cancer immunotherapy: From T cell basic science to clinical practice. Nat. Rev. Immunol. 2020, 20, 651–668. [Google Scholar] [CrossRef]
- Eguchi, K. Apoptosis in autoimmune diseases. Intern. Med. 2001, 40, 275–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Zhang, Y.; Guan, Z.; Li, H.; Ye, M.; Chen, X.; Shen, J.; Zhou, Y.; Shi, Z.L.; Zhou, P.; et al. SARS-CoV-2 triggers inflammatory responses and cell death through caspase-8 activation. Signal Transduct. Target 2020, 5, 235. [Google Scholar] [CrossRef] [PubMed]
- Ivanisenko, N.V.; Seyrek, K.; Kolchanov, N.A.; Ivanisenko, V.A.; Lavrik, I.N. The role of death domain proteins in host response upon SARS-CoV-2 infection: Modulation of programmed cell death and translational applications. Cell Death Discov. 2020, 6, 101. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Shuai, H.; Hou, Y.; Zhang, X.; Wen, L.; Huang, X.; Hu, B.; Yang, D.; Wang, Y.; Yoon, C.; et al. Targeting highly pathogenic coronavirus-induced apoptosis reduces viral pathogenesis and disease severity. Sci. Adv. 2021, 7, eabf8577. [Google Scholar] [CrossRef] [PubMed]
- Schweichel, J.U.; Merker, H.J. The morphology of various types of cell death in prenatal tissues. Teratology 1973, 7, 253–266. [Google Scholar] [CrossRef]
- Clarke, P.G. Developmental cell death: Morphological diversity and multiple mechanisms. Anat. Embryol. (Berl.) 1990, 181, 195–213. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541. [Google Scholar] [CrossRef]
- Porter, K.R.; Claude, A.; Fullam, E.F. A Study of Tissue Culture Cells by Electron Microscopy: Methods and Preliminary Observations. J. Exp. Med. 1945, 81, 233–246. [Google Scholar] [CrossRef]
- Clark, S.L., Jr. Cellular differentiation in the kidneys of newborn mice studies with the electron microscope. J. Biophys. Biochem. Cytol. 1957, 3, 349–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashford, T.P.; Porter, K.R. Cytoplasmic components in hepatic cell lysosomes. J. Cell Biol. 1962, 12, 198–202. [Google Scholar] [CrossRef] [PubMed]
- De Duve, C.; Wattiaux, R. Functions of lysosomes. Annu. Rev. Physiol. 1966, 28, 435–492. [Google Scholar] [CrossRef] [PubMed]
- Essner, E.; Novikoff, A.B. Human hepatocellular pigments and lysosomes. J. UltraStruct. Res. 1960, 3, 374–391. [Google Scholar] [CrossRef]
- Essner, E.; Novikoff, A.B. Localization of acid phosphatase activity in hepatic lysosomes by means of electron microscopy. J. Biophys. Biochem. Cytol. 1961, 9, 773–784. [Google Scholar] [CrossRef]
- Novikoff, A.B.; Beaufay, H.; De Duve, C. Electron microscopy of lysosomerich fractions from rat liver. J. Biophys. Biochem. Cytol. 1956, 2, 179–184. [Google Scholar] [CrossRef] [Green Version]
- Novikoff, A.B. The proximal tubule cell in experimental hydronephrosis. J. Biophys. Biochem. Cytol. 1959, 6, 136–138. [Google Scholar] [CrossRef]
- Porter, K.R.; Novikoff, A.B. The 1974 nobel prize for physiology or medicine. Science 1974, 186, 516–520. [Google Scholar] [CrossRef]
- Focusing on autophagy. Nat. Cell Biol. 2010, 12, 813. [CrossRef] [Green Version]
- Seglen, P.O. Lysosomes: Their Role in Protein Breakdown; Glaumann, H., Ballard, F.J., Eds.; Academic Press: Cambridge, MA, USA, 1987; pp. 371–414. [Google Scholar]
- Pfeifer, U. Inhibition by insulin of the physiological autophagic breakdown of cell organelles. Acta Biol. Med. Ger. 1977, 36, 1691–1694. [Google Scholar]
- Mitchener, J.S.; Shelburne, J.D.; Bradford, W.D.; Hawkins, H.K. Cellular autophagocytosis induced by deprivation of serum and amino acids in HeLa cells. Am. J. Pathol. 1976, 83, 485–492. [Google Scholar]
- Mortimore, G.E.; Mondon, C.E. Inhibition by insulin of valine turnover in liver. Evidence for a general control of proteolysis. J. Biol. Chem. 1970, 245, 2375–2383. [Google Scholar] [CrossRef]
- Mortimore, G.E.; Schworer, C.M. Induction of autophagy by amino-acid deprivation in perfused rat liver. Nature 1977, 270, 174–176. [Google Scholar] [CrossRef]
- Neely, A.N.; Cox, J.R.; Fortney, J.A.; Schworer, C.M.; Mortimore, G.E. Alterations of lysosomal size and density during rat liver perfusion. Suppression by insulin and amino acids. J. Biol. Chem. 1977, 252, 6948–6954. [Google Scholar] [CrossRef]
- Woodside, K.H.; Mortimore, G.E. Suppression of protein turnover by amino acids in the perfused rat liver. J. Biol. Chem. 1972, 247, 6474–6481. [Google Scholar] [CrossRef]
- Noda, T.; Ohsumi, Y. Tor, a phosphatidylinositol kinase homologue, controls autophagy in yeast. J. Biol. Chem. 1998, 273, 3963–3966. [Google Scholar] [CrossRef] [Green Version]
- Ebato, C.; Uchida, T.; Arakawa, M.; Komatsu, M.; Ueno, T.; Komiya, K.; Azuma, K.; Hirose, T.; Tanaka, K.; Kominami, E.; et al. Autophagy is important in islet homeostasis and compensatory increase of beta cell mass in response to high-fat diet. Cell Metab. 2008, 8, 325–332. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.S.; Chung, K.W.; Won Kim, J.; Kim, J.; Komatsu, M.; Tanaka, K.; Nguyen, Y.H.; Kang, T.M.; Yoon, K.H.; Kim, J.W.; et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008, 8, 318–324. [Google Scholar] [CrossRef] [Green Version]
- Riahi, Y.; Wikstrom, J.D.; Bachar-Wikstrom, E.; Polin, N.; Zucker, H.; Lee, M.S.; Quan, W.; Haataja, L.; Liu, M.; Arvan, P.; et al. Autophagy is a major regulator of beta cell insulin homeostasis. Diabetologia 2016, 59, 1480–1491. [Google Scholar] [CrossRef] [Green Version]
- Ji, J.; Petropavlovskaia, M.; Khatchadourian, A.; Patapas, J.; Makhlin, J.; Rosenberg, L.; Maysinger, D. Type 2 diabetes is associated with suppression of autophagy and lipid accumulation in beta-cells. J. Cell Mol. Med. 2019, 23, 2890–2900. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, R.; Kaushik, S.; Wang, Y.; Xiang, Y.; Novak, I.; Komatsu, M.; Tanaka, K.; Cuervo, A.M.; Czaja, M.J. Autophagy regulates lipid metabolism. Nature 2009, 458, 1131–1135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.; Yao, T.P. Quality control autophagy: A joint effort of ubiquitin, protein deacetylase and actin cytoskeleton. Autophagy 2010, 6, 555–557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sehgal, S.N.; Baker, H.; Vezina, C. Rapamycin (AY-22,989), a new antifungal antibiotic. II. Fermentation, isolation and characterization. J. Antibiot. 1975, 28, 727–732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vezina, C.; Kudelski, A.; Sehgal, S.N. Rapamycin (AY-22,989), a new antifungal antibiotic. I. Taxonomy of the producing streptomycete and isolation of the active principle. J. Antibiot. 1975, 28, 721–726. [Google Scholar] [CrossRef] [PubMed]
- Sabatini, D.M.; Erdjument-Bromage, H.; Lui, M.; Tempst, P.; Snyder, S.H. RAFT1: A mammalian protein that binds to FKBP12 in a rapamycin-dependent fashion and is homologous to yeast TORs. Cell 1994, 78, 35–43. [Google Scholar] [CrossRef]
- Helliwell, S.B.; Wagner, P.; Kunz, J.; Deuter-Reinhard, M.; Henriquez, R.; Hall, M.N. TOR1 and TOR2 are structurally and functionally similar but not identical phosphatidylinositol kinase homologues in yeast. Mol. Biol. Cell 1994, 5, 105–118. [Google Scholar] [CrossRef] [Green Version]
- Kunz, J.; Henriquez, R.; Schneider, U.; Deuter-Reinhard, M.; Movva, N.R.; Hall, M.N. Target of rapamycin in yeast, TOR2, is an essential phosphatidylinositol kinase homolog required for G1 progression. Cell 1993, 73, 585–596. [Google Scholar] [CrossRef]
- Cafferkey, R.; Young, P.R.; McLaughlin, M.M.; Bergsma, D.J.; Koltin, Y.; Sathe, G.M.; Faucette, L.; Eng, W.K.; Johnson, R.K.; Livi, G.P. Dominant missense mutations in a novel yeast protein related to mammalian phosphatidylinositol 3-kinase and VPS34 abrogate rapamycin cytotoxicity. Mol. Cell Biol. 1993, 13, 6012–6023. [Google Scholar] [CrossRef] [Green Version]
- Heitman, J.; Movva, N.R.; Hall, M.N. Targets for cell cycle arrest by the immunosuppressant rapamycin in yeast. Science 1991, 253, 905–909. [Google Scholar] [CrossRef]
- Heitman, J.; Movva, N.R.; Hiestand, P.C.; Hall, M.N. FK 506-binding protein proline rotamase is a target for the immunosuppressive agent FK 506 in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 1991, 88, 1948–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koltin, Y.; Faucette, L.; Bergsma, D.J.; Levy, M.A.; Cafferkey, R.; Koser, P.L.; Johnson, R.K.; Livi, G.P. Rapamycin sensitivity in Saccharomyces cerevisiae is mediated by a peptidyl-prolyl cis-trans isomerase related to human FK506-binding protein. Mol. Cell Biol. 1991, 11, 1718–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiederrecht, G.; Brizuela, L.; Elliston, K.; Sigal, N.H.; Siekierka, J.J. FKB1 encodes a nonessential FK 506-binding protein in Saccharomyces cerevisiae and contains regions suggesting homology to the cyclophilins. Proc. Natl. Acad. Sci. USA 1991, 88, 1029–1033. [Google Scholar] [CrossRef] [Green Version]
- Laplante, M.; Sabatini, D.M. mTOR signaling in growth control and disease. Cell 2012, 149, 274–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blommaart, E.F.; Luiken, J.J.; Blommaart, P.J.; van Woerkom, G.M.; Meijer, A.J. Phosphorylation of ribosomal protein S6 is inhibitory for autophagy in isolated rat hepatocytes. J. Biol. Chem. 1995, 270, 2320–2326. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blommaart, E.F.; Krause, U.; Schellens, J.P.; Vreeling-Sindelarova, H.; Meijer, A.J. The phosphatidylinositol 3-kinase inhibitors wortmannin and LY294002 inhibit autophagy in isolated rat hepatocytes. Eur. J. Biochem. 1997, 243, 240–246. [Google Scholar] [CrossRef] [Green Version]
- mTOR: The Master Regulator. Cell 2012, 149, 955–957. [CrossRef] [Green Version]
- Morita, M.; Gravel, S.P.; Hulea, L.; Larsson, O.; Pollak, M.; St-Pierre, J.; Topisirovic, I. mTOR coordinates protein synthesis, mitochondrial activity and proliferation. Cell Cycle 2015, 14, 473–480. [Google Scholar] [CrossRef] [Green Version]
- Saxton, R.A.; Sabatini, D.M. mTOR Signaling in Growth, Metabolism, and Disease. Cell 2017, 168, 960–976. [Google Scholar] [CrossRef] [Green Version]
- Duong, T.; Rasmussen, N.R.; Ballato, E.; Mote, F.S.; Reiner, D.J. The Rheb-TORC1 signaling axis functions as a developmental checkpoint. Development 2020, 147, dev181727. [Google Scholar] [CrossRef]
- Long, X.; Spycher, C.; Han, Z.S.; Rose, A.M.; Muller, F.; Avruch, J. TOR deficiency in C. elegans causes developmental arrest and intestinal atrophy by inhibition of mRNA translation. Curr. Biol. 2002, 12, 1448–1461. [Google Scholar] [CrossRef] [Green Version]
- Vellai, T.; Takacs-Vellai, K.; Zhang, Y.; Kovacs, A.L.; Orosz, L.; Muller, F. Genetics: Influence of TOR kinase on lifespan in C. elegans. Nature 2003, 426, 620. [Google Scholar] [CrossRef]
- Baugh, L.R.; Hu, P.J. Starvation Responses Throughout the Caenorhabditis elegans Life Cycle. Genetics 2020, 216, 837–878. [Google Scholar] [CrossRef] [PubMed]
- Tissenbaum, H.A. DAF-16: FOXO in the Context of C. elegans. Curr. Top. Dev. Biol. 2018, 127, 1–21. [Google Scholar] [CrossRef] [PubMed]
- Dall, K.B.; Faergeman, N.J. Metabolic regulation of lifespan from a C. elegans perspective. Genes Nutr. 2019, 14, 25. [Google Scholar] [CrossRef] [PubMed]
- Palmisano, N.J.; Melendez, A. Autophagy in C. elegans development. Dev. Biol. 2019, 447, 103–125. [Google Scholar] [CrossRef]
- Takeshige, K.; Baba, M.; Tsuboi, S.; Noda, T.; Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 1992, 119, 301–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukada, M.; Ohsumi, Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 1993, 333, 169–174. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Cregg, J.M.; Dunn, W.A., Jr.; Emr, S.D.; Sakai, Y.; Sandoval, I.V.; Sibirny, A.; Subramani, S.; Thumm, M.; Veenhuis, M.; et al. A unified nomenclature for yeast autophagy-related genes. Dev. Cell 2003, 5, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J. Look people, “Atg” is an abbreviation for “autophagy-related.” That’s it. Autophagy 2012, 8, 1281–1282. [Google Scholar] [CrossRef] [Green Version]
- Levine, B.; Klionsky, D.J. Autophagy wins the 2016 Nobel Prize in Physiology or Medicine: Breakthroughs in baker’s yeast fuel advances in biomedical research. Proc. Natl. Acad. Sci. USA 2017, 114, 201–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aita, V.M.; Liang, X.H.; Murty, V.V.; Pincus, D.L.; Yu, W.; Cayanis, E.; Kalachikov, S.; Gilliam, T.C.; Levine, B. Cloning and genomic organization of beclin 1, a candidate tumor suppressor gene on chromosome 17q21. Genomics 1999, 59, 59–65. [Google Scholar] [CrossRef]
- Liang, X.H.; Jackson, S.; Seaman, M.; Brown, K.; Kempkes, B.; Hibshoosh, H.; Levine, B. Induction of autophagy and inhibition of tumorigenesis by beclin 1. Nature 1999, 402, 672–676. [Google Scholar] [CrossRef]
- Qu, X.; Yu, J.; Bhagat, G.; Furuya, N.; Hibshoosh, H.; Troxel, A.; Rosen, J.; Eskelinen, E.L.; Mizushima, N.; Ohsumi, Y.; et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J. Clin. Investig. 2003, 112, 1809–1820. [Google Scholar] [CrossRef] [Green Version]
- Yue, Z.; Jin, S.; Yang, C.; Levine, A.J.; Heintz, N. Beclin 1, an autophagy gene essential for early embryonic development, is a haploinsufficient tumor suppressor. Proc. Natl. Acad. Sci. USA 2003, 100, 15077–15082. [Google Scholar] [CrossRef] [Green Version]
- Pattingre, S.; Tassa, A.; Qu, X.; Garuti, R.; Liang, X.H.; Mizushima, N.; Packer, M.; Schneider, M.D.; Levine, B. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell 2005, 122, 927–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meléndez, A.; Levine, B. Autophagy in C. elegans. WormBook 2009, 1–26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Palmisano, N.J.; Melendez, A. Detection of Autophagy in Caenorhabditis elegans. Cold Spring Harb Protoc 2016, 2016, pdb-top070466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melendez, A.; Talloczy, Z.; Seaman, M.; Eskelinen, E.L.; Hall, D.H.; Levine, B. Autophagy genes are essential for dauer development and life-span extension in C. elegans. Science 2003, 301, 1387–1391. [Google Scholar] [CrossRef] [Green Version]
- Takacs-Vellai, K.; Vellai, T.; Puoti, A.; Passannante, M.; Wicky, C.; Streit, A.; Kovacs, A.L.; Muller, F. Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr. Biol. 2005, 15, 1513–1517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Søreng, K.; Neufeld, T.P.; Simonsen, A. Chapter One—Membrane Trafficking in Autophagy. In International Review of Cell and Molecular Biology; Galluzzi, L., Ed.; Academic Press: Cambridge, MA, USA, 2018; Volume 336, pp. 1–92. [Google Scholar]
- Nakatogawa, H. Mechanisms governing autophagosome biogenesis. Nat. Rev. Mol. Cell Biol. 2020, 21, 439–458. [Google Scholar] [CrossRef]
- Suzuki, H.; Osawa, T.; Fujioka, Y.; Noda, N.N. Structural biology of the core autophagy machinery. Curr. Opin. Struct. Biol. 2017, 43, 10–17. [Google Scholar] [CrossRef] [Green Version]
- Gross, A.S.; Graef, M. Mechanisms of Autophagy in Metabolic Stress Response. J. Mol. Biol. 2020, 432, 28–52. [Google Scholar] [CrossRef]
- Corona Velazquez, A.F.; Jackson, W.T. So Many Roads: The Multifaceted Regulation of Autophagy Induction. Mol. Cell Biol. 2018, 38, e00303-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matsuura, A.; Tsukada, M.; Wada, Y.; Ohsumi, Y. Apg1p, a novel protein kinase required for the autophagic process in Saccharomyces cerevisiae. Gene 1997, 192, 245–250. [Google Scholar] [CrossRef]
- Chew, L.H.; Lu, S.; Liu, X.; Li, F.K.; Yu, A.Y.; Klionsky, D.J.; Dong, M.Q.; Yip, C.K. Molecular interactions of the Saccharomyces cerevisiae Atg1 complex provide insights into assembly and regulatory mechanisms. Autophagy 2015, 11, 891–905. [Google Scholar] [CrossRef] [Green Version]
- Kamada, Y.; Funakoshi, T.; Shintani, T.; Nagano, K.; Ohsumi, M.; Ohsumi, Y. Tor-mediated induction of autophagy via an Apg1 protein kinase complex. J. Cell Biol. 2000, 150, 1507–1513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, J.; Kuroyanagi, H.; Kuroiwa, A.; Matsuda, Y.; Tokumitsu, H.; Tomoda, T.; Shirasawa, T.; Muramatsu, M. Identification of mouse ULK1, a novel protein kinase structurally related to C. elegans UNC-51. Biochem. Biophys. Res. Commun. 1998, 246, 222–227. [Google Scholar] [CrossRef]
- Kuroyanagi, H.; Yan, J.; Seki, N.; Yamanouchi, Y.; Suzuki, Y.; Takano, T.; Muramatsu, M.; Shirasawa, T. Human ULK1, a novel serine/threonine kinase related to UNC-51 kinase of Caenorhabditis elegans: cDNA cloning, expression, and chromosomal assignment. Genomics 1998, 51, 76–85. [Google Scholar] [CrossRef]
- Yan, J.; Kuroyanagi, H.; Tomemori, T.; Okazaki, N.; Asato, K.; Matsuda, Y.; Suzuki, Y.; Ohshima, Y.; Mitani, S.; Masuho, Y.; et al. Mouse ULK2, a novel member of the UNC-51-like protein kinases: Unique features of functional domains. Oncogene 1999, 18, 5850–5859. [Google Scholar] [CrossRef] [Green Version]
- Cheong, H.; Nair, U.; Geng, J.; Klionsky, D.J. The Atg1 kinase complex is involved in the regulation of protein recruitment to initiate sequestering vesicle formation for nonspecific autophagy in Saccharomyces cerevisiae. Mol. Biol. Cell 2008, 19, 668–681. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kawamata, T.; Kamada, Y.; Kabeya, Y.; Sekito, T.; Ohsumi, Y. Organization of the pre-autophagosomal structure responsible for autophagosome formation. Mol. Biol. Cell 2008, 19, 2039–2050. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Itakura, E.; Mizushima, N. Characterization of autophagosome formation site by a hierarchical analysis of mammalian Atg proteins. Autophagy 2010, 6, 764–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koyama-Honda, I.; Itakura, E.; Fujiwara, T.K.; Mizushima, N. Temporal analysis of recruitment of mammalian ATG proteins to the autophagosome formation site. Autophagy 2013, 9, 1491–1499. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Hara, T.; Kaizuka, T.; Kishi, C.; Takamura, A.; Miura, Y.; Iemura, S.; Natsume, T.; Takehana, K.; Yamada, N.; et al. Nutrient-dependent mTORC1 association with the ULK1-Atg13-FIP200 complex required for autophagy. Mol. Biol. Cell 2009, 20, 1981–1991. [Google Scholar] [CrossRef] [Green Version]
- Jung, C.H.; Jun, C.B.; Ro, S.H.; Kim, Y.M.; Otto, N.M.; Cao, J.; Kundu, M.; Kim, D.H. ULK-Atg13-FIP200 complexes mediate mTOR signaling to the autophagy machinery. Mol. Biol. Cell 2009, 20, 1992–2003. [Google Scholar] [CrossRef] [Green Version]
- Hara, T.; Takamura, A.; Kishi, C.; Iemura, S.; Natsume, T.; Guan, J.L.; Mizushima, N. FIP200, a ULK-interacting protein, is required for autophagosome formation in mammalian cells. J. Cell Biol. 2008, 181, 497–510. [Google Scholar] [CrossRef] [Green Version]
- Hosokawa, N.; Sasaki, T.; Iemura, S.; Natsume, T.; Hara, T.; Mizushima, N. Atg101, a novel mammalian autophagy protein interacting with Atg13. Autophagy 2009, 5, 973–979. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.; Kundu, M.; Viollet, B.; Guan, K.L. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 2011, 13, 132–141. [Google Scholar] [CrossRef] [Green Version]
- Puente, C.; Hendrickson, R.C.; Jiang, X. Nutrient-regulated Phosphorylation of ATG13 Inhibits Starvation-induced Autophagy. J. Biol. Chem. 2016, 291, 6026–6035. [Google Scholar] [CrossRef] [Green Version]
- Dunlop, E.A.; Hunt, D.K.; Acosta-Jaquez, H.A.; Fingar, D.C.; Tee, A.R. ULK1 inhibits mTORC1 signaling, promotes multisite Raptor phosphorylation and hinders substrate binding. Autophagy 2011, 7, 737–747. [Google Scholar] [CrossRef]
- Loffler, A.S.; Alers, S.; Dieterle, A.M.; Keppeler, H.; Franz-Wachtel, M.; Kundu, M.; Campbell, D.G.; Wesselborg, S.; Alessi, D.R.; Stork, B. Ulk1-mediated phosphorylation of AMPK constitutes a negative regulatory feedback loop. Autophagy 2011, 7, 696–706. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sadria, M.; Layton, A.T. Interactions among mTORC, AMPK and SIRT: A computational model for cell energy balance and metabolism. Cell Commun. Signal. 2021, 19, 57. [Google Scholar] [CrossRef] [PubMed]
- Tooze, S.A.; Yoshimori, T. The origin of the autophagosomal membrane. Nat. Cell Biol. 2010, 12, 831–835. [Google Scholar] [CrossRef] [PubMed]
- Gomez-Sanchez, R.; Tooze, S.A.; Reggiori, F. Membrane supply and remodeling during autophagosome biogenesis. Curr. Opin. Cell Biol. 2021, 71, 112–119. [Google Scholar] [CrossRef]
- Rao, Y.; Perna, M.G.; Hofmann, B.; Beier, V.; Wollert, T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat. Commun. 2016, 7, 10338. [Google Scholar] [CrossRef]
- Hollenstein, D.M.; Kraft, C. Autophagosomes are formed at a distinct cellular structure. Curr. Opin. Cell Biol. 2020, 65, 50–57. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef] [Green Version]
- Kern, A.; Dikic, I.; Behl, C. The integration of autophagy and cellular trafficking pathways via RAB GAPs. Autophagy 2015, 11, 2393–2397. [Google Scholar] [CrossRef] [PubMed]
- Ao, X.; Zou, L.; Wu, Y. Regulation of autophagy by the Rab GTPase network. Cell Death Differ. 2014, 21, 348–358. [Google Scholar] [CrossRef] [Green Version]
- Papinski, D.; Schuschnig, M.; Reiter, W.; Wilhelm, L.; Barnes, C.A.; Maiolica, A.; Hansmann, I.; Pfaffenwimmer, T.; Kijanska, M.; Stoffel, I.; et al. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol. Cell 2014, 53, 471–483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Russell, R.C.; Tian, Y.; Yuan, H.; Park, H.W.; Chang, Y.Y.; Kim, J.; Kim, H.; Neufeld, T.P.; Dillin, A.; Guan, K.L. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat. Cell Biol. 2013, 15, 741–750. [Google Scholar] [CrossRef] [Green Version]
- Fogel, A.I.; Dlouhy, B.J.; Wang, C.; Ryu, S.W.; Neutzner, A.; Hasson, S.A.; Sideris, D.P.; Abeliovich, H.; Youle, R.J. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol. Cell Biol. 2013, 33, 3675–3688. [Google Scholar] [CrossRef] [Green Version]
- Park, J.M.; Seo, M.; Jung, C.H.; Grunwald, D.; Stone, M.; Otto, N.M.; Toso, E.; Ahn, Y.; Kyba, M.; Griffin, T.J.; et al. ULK1 phosphorylates Ser30 of BECN1 in association with ATG14 to stimulate autophagy induction. Autophagy 2018, 14, 584–597. [Google Scholar] [CrossRef] [Green Version]
- Simonsen, A.; Tooze, S.A. Coordination of membrane events during autophagy by multiple class III PI3-kinase complexes. J. Cell Biol. 2009, 186, 773–782. [Google Scholar] [CrossRef]
- Petiot, A.; Ogier-Denis, E.; Blommaart, E.F.; Meijer, A.J.; Codogno, P. Distinct classes of phosphatidylinositol 3’-kinases are involved in signaling pathways that control macroautophagy in HT-29 cells. J. Biol. Chem. 2000, 275, 992–998. [Google Scholar] [CrossRef] [Green Version]
- Arico, S.; Petiot, A.; Bauvy, C.; Dubbelhuis, P.F.; Meijer, A.J.; Codogno, P.; Ogier-Denis, E. The tumor suppressor PTEN positively regulates macroautophagy by inhibiting the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2001, 276, 35243–35246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jakobi, A.J.; Huber, S.T.; Mortensen, S.A.; Schultz, S.W.; Palara, A.; Kuhm, T.; Shrestha, B.K.; Lamark, T.; Hagen, W.J.H.; Wilmanns, M.; et al. Structural basis of p62/SQSTM1 helical filaments and their role in cellular cargo uptake. Nat. Commun. 2020, 11, 440. [Google Scholar] [CrossRef] [Green Version]
- Lamark, T.; Svenning, S.; Johansen, T. Regulation of selective autophagy: The p62/SQSTM1 paradigm. Essays BioChem. 2017, 61, 609–624. [Google Scholar] [CrossRef] [PubMed]
- Geng, J.; Klionsky, D.J. The Atg8 and Atg12 ubiquitin-like conjugation systems in macroautophagy. ‘Protein modifications: Beyond the usual suspects’ review series. EMBO Rep. 2008, 9, 859–864. [Google Scholar] [CrossRef] [Green Version]
- Hershko, A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005, 12, 1191–1197. [Google Scholar] [CrossRef] [Green Version]
- Cajee, U.F.; Hull, R.; Ntwasa, M. Modification by ubiquitin-like proteins: Significance in apoptosis and autophagy pathways. Int. J. Mol. Sci. 2012, 13, 11804–11831. [Google Scholar] [CrossRef] [PubMed]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef]
- Mizushima, N.; Noda, T.; Yoshimori, T.; Tanaka, Y.; Ishii, T.; George, M.D.; Klionsky, D.J.; Ohsumi, M.; Ohsumi, Y. A protein conjugation system essential for autophagy. Nature 1998, 395, 395–398. [Google Scholar] [CrossRef]
- Mizushima, N.; Sugita, H.; Yoshimori, T.; Ohsumi, Y. A new protein conjugation system in human. The counterpart of the yeast Apg12p conjugation system essential for autophagy. J. Biol. Chem. 1998, 273, 33889–33892. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohsumi, Y. Molecular mechanism of autophagy in yeast, Saccharomyces cerevisiae. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1999, 354, 1577–1581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lang, T.; Schaeffeler, E.; Bernreuther, D.; Bredschneider, M.; Wolf, D.H.; Thumm, M. Aut2p and Aut7p, two novel microtubule-associated proteins are essential for delivery of autophagic vesicles to the vacuole. EMBO J. 1998, 17, 3597–3607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirisako, T.; Baba, M.; Ishihara, N.; Miyazawa, K.; Ohsumi, M.; Yoshimori, T.; Noda, T.; Ohsumi, Y. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 1999, 147, 435–446. [Google Scholar] [CrossRef] [Green Version]
- Huang, W.P.; Scott, S.V.; Kim, J.; Klionsky, D.J. The itinerary of a vesicle component, Aut7p/Cvt5p, terminates in the yeast vacuole via the autophagy/Cvt pathways. J. Biol. Chem. 2000, 275, 5845–5851. [Google Scholar] [CrossRef] [Green Version]
- Kirisako, T.; Ichimura, Y.; Okada, H.; Kabeya, Y.; Mizushima, N.; Yoshimori, T.; Ohsumi, M.; Takao, T.; Noda, T.; Ohsumi, Y. The reversible modification regulates the membrane-binding state of Apg8/Aut7 essential for autophagy and the cytoplasm to vacuole targeting pathway. J. Cell Biol. 2000, 151, 263–276. [Google Scholar] [CrossRef]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Mann, S.S.; Hammarback, J.A. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 1994, 269, 11492–11497. [Google Scholar] [CrossRef]
- Kabeya, Y.; Mizushima, N.; Ueno, T.; Yamamoto, A.; Kirisako, T.; Noda, T.; Kominami, E.; Ohsumi, Y.; Yoshimori, T. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000, 19, 5720–5728. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdel-Aziz, A.K.; Abdelfatah, S.; Abdellatif, M.; Abdoli, A.; Abel, S.; Abeliovich, H.; Abildgaard, M.H.; Abudu, Y.P.; Acevedo-Arozena, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (4th edition). Autophagy 2021, 17, 1–382. [Google Scholar] [CrossRef] [PubMed]
- Agrotis, A.; Pengo, N.; Burden, J.J.; Ketteler, R. Redundancy of human ATG4 protease isoforms in autophagy and LC3/GABARAP processing revealed in cells. Autophagy 2019, 15, 976–997. [Google Scholar] [CrossRef] [Green Version]
- Martens, S.; Fracchiolla, D. Activation and targeting of ATG8 protein lipidation. Cell Discov. 2020, 6, 23. [Google Scholar] [CrossRef]
- Reggiori, F.; Ungermann, C. Autophagosome Maturation and Fusion. J. Mol. Biol. 2017, 429, 486–496. [Google Scholar] [CrossRef] [PubMed]
- Cebollero, E.; van der Vaart, A.; Zhao, M.; Rieter, E.; Klionsky, D.J.; Helms, J.B.; Reggiori, F. Phosphatidylinositol-3-phosphate clearance plays a key role in autophagosome completion. Curr. Biol. 2012, 22, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Furuta, N.; Amano, A. SNARE mediates autophagosome–lysosome fusion. J. Oral Biosci. 2012, 54, 83–85. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, L.; Hou, C.; Lai, Y.; Long, J.; Liu, J.; Zhong, Q.; Diao, J. SNARE-mediated membrane fusion in autophagy. Semin. Cell Dev. Biol. 2016, 60, 97–104. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, H.; Nakagawa, I.; Yamamoto, A.; Amano, A.; Noda, T.; Yoshimori, T. An initial step of GAS-containing autophagosome-like vacuoles formation requires Rab7. PLoS Pathog. 2009, 5, e1000670. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Eskelinen, E.L.; Deretic, V. Autophagosomes, phagosomes, autolysosomes, phagolysosomes, autophagolysosomes... wait, I’m confused. Autophagy 2014, 10, 549–551. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, S.; Dennemarker, J.; Reinheckel, T. Specific functions of lysosomal proteases in endocytic and autophagic pathways. Biochim. Biophys. Acta 2012, 1824, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Rudnik, S.; Damme, M. The lysosomal membrane-export of metabolites and beyond. FEBS J. 2020. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Qi, H.; Taylor, R.; Xu, W.; Liu, L.F.; Jin, S. The role of autophagy in mitochondria maintenance: Characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy 2007, 3, 337–346. [Google Scholar] [CrossRef] [Green Version]
- Devenish, R.J. Mitophagy: Growing in intricacy. Autophagy 2007, 3, 293–294. [Google Scholar] [CrossRef] [Green Version]
- Lemasters, J.J. Selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res. 2005, 8, 3–5. [Google Scholar] [CrossRef]
- Bellu, A.R.; Kiel, J.A. Selective degradation of peroxisomes in yeasts. Microsc. Res. Tech. 2003, 61, 161–170. [Google Scholar] [CrossRef]
- Farre, J.C.; Subramani, S. Peroxisome turnover by micropexophagy: An autophagy-related process. Trends Cell Biol. 2004, 14, 515–523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harding, T.M.; Morano, K.A.; Scott, S.V.; Klionsky, D.J. Isolation and characterization of yeast mutants in the cytoplasm to vacuole protein targeting pathway. J. Cell Biol. 1995, 131, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Klionsky, D.J.; Cueva, R.; Yaver, D.S. Aminopeptidase I of Saccharomyces cerevisiae is localized to the vacuole independent of the secretory pathway. J. Cell Biol. 1992, 119, 287–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hutchins, M.U.; Klionsky, D.J. Vacuolar localization of oligomeric alpha-mannosidase requires the cytoplasm to vacuole targeting and autophagy pathway components in Saccharomyces cerevisiae. J. Biol. Chem. 2001, 276, 20491–20498. [Google Scholar] [CrossRef] [Green Version]
- Yuga, M.; Gomi, K.; Klionsky, D.J.; Shintani, T. Aspartyl aminopeptidase is imported from the cytoplasm to the vacuole by selective autophagy in Saccharomyces cerevisiae. J. Biol. Chem. 2011, 286, 13704–13713. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Hochstrasser, M. Microautophagy regulates proteasome homeostasis. Curr. Genet. 2020, 66, 683–687. [Google Scholar] [CrossRef]
- Schuck, S. Microautophagy—distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 2020, 133, jcs246322. [Google Scholar] [CrossRef]
- Morshed, S.; Tasnin, M.N.; Ushimaru, T. ESCRT machinery plays a role in microautophagy in yeast. BMC Mol. Cell Biol. 2020, 21, 70. [Google Scholar] [CrossRef] [PubMed]
- Oku, M.; Maeda, Y.; Kagohashi, Y.; Kondo, T.; Yamada, M.; Fujimoto, T.; Sakai, Y. Evidence for ESCRT- and clathrin-dependent microautophagy. J. Cell Biol. 2017, 216, 3263–3274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rios, J.; Sequeida, A.; Albornoz, A.; Budini, M. Chaperone Mediated Autophagy Substrates and Components in Cancer. Front. Oncol. 2020, 10, 614677. [Google Scholar] [CrossRef] [PubMed]
- Fred Dice, J. Peptide sequences that target cytosolic proteins for lysosomal proteolysis. Trends Biochem. Sci. 1990, 15, 305–309. [Google Scholar] [CrossRef]
- Liao, Z.; Wang, B.; Liu, W.; Xu, Q.; Hou, L.; Song, J.; Guo, Q.; Li, N. Dysfunction of chaperone-mediated autophagy in human diseases. Mol. Cell BioChem. 2021, 476, 1439–1454. [Google Scholar] [CrossRef]
- Cao, D.; Shan, D.; Yan, W.; Zhang, Z.; Song, Q.; Jiang, Y.; Zhang, X.; Zhang, Z.; Wang, Z.; Wang, Y.; et al. Chaperone-mediated autophagy affects tumor cell proliferation and cisplatin resistance in esophageal squamous cell carcinoma. Thorac. Cancer 2021, 12, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Wang, Q.; Kao, Y.R.; Diaz, A.; Tasset, I.; Kaushik, S.; Thiruthuvanathan, V.; Zintiridou, A.; Nieves, E.; Dzieciatkowska, M.; et al. Chaperone-mediated autophagy sustains haematopoietic stem-cell function. Nature 2021, 591, 117–123. [Google Scholar] [CrossRef]
- Bourdenx, M.; Martin-Segura, A.; Scrivo, A.; Rodriguez-Navarro, J.A.; Kaushik, S.; Tasset, I.; Diaz, A.; Storm, N.J.; Xin, Q.; Juste, Y.R.; et al. Chaperone-mediated autophagy prevents collapse of the neuronal metastable proteome. Cell 2021, 184, 2696–2714.e2625. [Google Scholar] [CrossRef] [PubMed]
- Auzmendi-Iriarte, J.; Matheu, A. Impact of Chaperone-Mediated Autophagy in Brain Aging: Neurodegenerative Diseases and Glioblastoma. Front. Aging NeuroSci. 2020, 12, 630743. [Google Scholar] [CrossRef] [PubMed]
- Xuan, Y.; Zhao, S.; Xiao, X.; Xiang, L.; Zheng, H.C. Inhibition of chaperonemediated autophagy reduces tumor growth and metastasis and promotes drug sensitivity in colorectal cancer. Mol. Med. Rep. 2021, 23. [Google Scholar] [CrossRef]
- Andrade-Tomaz, M.; de Souza, I.; Rocha, C.R.R.; Gomes, L.R. The Role of Chaperone-Mediated Autophagy in Cell Cycle Control and Its Implications in Cancer. Cells 2020, 9, 2140. [Google Scholar] [CrossRef]
- Li, W.; Yang, Q.; Mao, Z. Chaperone-mediated autophagy: Machinery, regulation and biological consequences. Cell Mol. Life Sci. 2011, 68, 749–763. [Google Scholar] [CrossRef]
- Bandyopadhyay, U.; Sridhar, S.; Kaushik, S.; Kiffin, R.; Cuervo, A.M. Identification of regulators of chaperone-mediated autophagy. Mol. Cell 2010, 39, 535–547. [Google Scholar] [CrossRef] [Green Version]
- Arias, E.; Koga, H.; Diaz, A.; Mocholi, E.; Patel, B.; Cuervo, A.M. Lysosomal mTORC2/PHLPP1/Akt Regulate Chaperone-Mediated Autophagy. Mol. Cell 2015, 59, 270–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Endicott, S.J.; Ziemba, Z.J.; Beckmann, L.J.; Boynton, D.N.; Miller, R.A. Inhibition of class I PI3K enhances chaperone-mediated autophagy. J. Cell Biol. 2020, 219, e202001031. [Google Scholar] [CrossRef]
- Van Houten, B.; Hunter, S.E.; Meyer, J.N. Mitochondrial DNA damage induced autophagy, cell death, and disease. Front. BioSci. (Landmark Ed.) 2016, 21, 42–54. [Google Scholar] [CrossRef] [PubMed]
- Lemasters, J.J.; Nieminen, A.L.; Qian, T.; Trost, L.C.; Elmore, S.P.; Nishimura, Y.; Crowe, R.A.; Cascio, W.E.; Bradham, C.A.; Brenner, D.A.; et al. The mitochondrial permeability transition in cell death: A common mechanism in necrosis, apoptosis and autophagy. Biochim. Biophys. Acta 1998, 1366, 177–196. [Google Scholar] [CrossRef] [Green Version]
- Rambold, A.S.; Lippincott-Schwartz, J. Mechanisms of mitochondria and autophagy crosstalk. Cell Cycle 2011, 10, 4032–4038. [Google Scholar] [CrossRef] [Green Version]
- Vessoni, A.T.; Filippi-Chiela, E.C.; Menck, C.F.; Lenz, G. Autophagy and genomic integrity. Cell Death Differ. 2013, 20, 1444–1454. [Google Scholar] [CrossRef] [PubMed]
- Mathew, R.; Kongara, S.; Beaudoin, B.; Karp, C.M.; Bray, K.; Degenhardt, K.; Chen, G.; Jin, S.; White, E. Autophagy suppresses tumor progression by limiting chromosomal instability. Genes Dev. 2007, 21, 1367–1381. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, P.; Sun, Q.; Ding, W.X.; Yin, X.M.; Sobol, R.W.; Stolz, D.B.; Yu, J.; Zhang, L. Following cytochrome c release, autophagy is inhibited during chemotherapy-induced apoptosis by caspase 8-mediated cleavage of Beclin 1. Cancer Res. 2011, 71, 3625–3634. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, P.; Yu, J.; Zhang, L. Cleaving Beclin 1 to suppress autophagy in chemotherapy-induced apoptosis. Autophagy 2011, 7, 1239–1241. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; Ye, F.; Li, D.; He, C.; He, H.; Zhang, J. p62 overexpression promotes neoplastic stromal cell proliferation and is associated with the recurrence of giant cell tumor of bone. Oncol. Lett. 2020, 20, 86. [Google Scholar] [CrossRef]
- Yao, R.Q.; Ren, C.; Xia, Z.F.; Yao, Y.M. Organelle-specific autophagy in inflammatory diseases: A potential therapeutic target underlying the quality control of multiple organelles. Autophagy 2021, 17, 385–401. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Shen, J.; Ran, Z. Emerging views of mitophagy in immunity and autoimmune diseases. Autophagy 2020, 16, 3–17. [Google Scholar] [CrossRef]
- Son, S.M.; Park, S.J.; Fernandez-Estevez, M.; Rubinsztein, D.C. Autophagy regulation by acetylation-implications for neurodegenerative diseases. Exp. Mol. Med. 2021, 53, 30–41. [Google Scholar] [CrossRef]
- Acevo-Rodriguez, P.S.; Maldonado, G.; Castro-Obregon, S.; Hernandez, G. Autophagy Regulation by the Translation Machinery and Its Implications in Cancer. Front. Oncol. 2020, 10, 322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mizushima, N.; Levine, B. Autophagy in Human Diseases. N. Engl. J. Med. 2020, 383, 1564–1576. [Google Scholar] [CrossRef]
- Ryu, H.Y.; Kim, L.E.; Jeong, H.; Yeo, B.K.; Lee, J.W.; Nam, H.; Ha, S.; An, H.K.; Park, H.; Jung, S.; et al. GSK3B induces autophagy by phosphorylating ULK1. Exp. Mol. Med. 2021, 53, 369–383. [Google Scholar] [CrossRef] [PubMed]
- Yuan, F.; Jin, X.; Li, D.; Song, Y.; Zhang, N.; Yang, X.; Wang, L.; Zhu, W.G.; Tian, C.; Zhao, Y. ULK1 phosphorylates Mad1 to regulate spindle assembly checkpoint. Nucleic Acids Res. 2019, 47, 8096–8110. [Google Scholar] [CrossRef] [PubMed]
- Emanuele, S.; Lauricella, M.; D’Anneo, A.; Carlisi, D.; De Blasio, A.; Di Liberto, D.; Giuliano, M. p62: Friend or Foe? Evidences for OncoJanus and NeuroJanus Roles. Int. J. Mol. Sci. 2020, 21, 5029. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; DiFiglia, M.; Heintz, N.; Nixon, R.A.; Qin, Z.H.; Ravikumar, B.; Stefanis, L.; Tolkovsky, A. Autophagy and its possible roles in nervous system diseases, damage and repair. Autophagy 2005, 1, 11–22. [Google Scholar] [CrossRef] [Green Version]
- Sarkar, S.; Ravikumar, B.; Floto, R.A.; Rubinsztein, D.C. Rapamycin and mTOR-independent autophagy inducers ameliorate toxicity of polyglutamine-expanded huntingtin and related proteinopathies. Cell Death Differ. 2009, 16, 46–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef]
- Komatsu, M.; Waguri, S.; Chiba, T.; Murata, S.; Iwata, J.; Tanida, I.; Ueno, T.; Koike, M.; Uchiyama, Y.; Kominami, E.; et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature 2006, 441, 880–884. [Google Scholar] [CrossRef]
- Ravikumar, B.; Duden, R.; Rubinsztein, D.C. Aggregate-prone proteins with polyglutamine and polyalanine expansions are degraded by autophagy. Hum. Mol. Genet. 2002, 11, 1107–1117. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.T.; Kumsta, C.; Hellman, A.B.; Adams, L.M.; Hansen, M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 2017, 6, e18459. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.T.; Hansen, M. Age-associated and tissue-specific decline in autophagic activity in the nematode C. elegans. Autophagy 2018, 14, 1276–1277. [Google Scholar] [CrossRef] [Green Version]
- Butsch, T.J.; Ghosh, B.; Bohnert, K.A. Organelle-Specific Autophagy in Cellular Aging and Rejuvenation. Adv. Geriatr. Med. Res. 2021, 3, e210010. [Google Scholar] [CrossRef]
- de la Cruz-Ruiz, P.; Hernando-Rodriguez, B.; Perez-Jimenez, M.M.; Rodriguez-Palero, M.J.; Martinez-Bueno, M.D.; Pla, A.; Gatsi, R.; Artal-Sanz, M. Prohibitin depletion extends lifespan of a TORC2/SGK-1 mutant through autophagy and the mitochondrial UPR. Aging Cell 2021, 20, e13359. [Google Scholar] [CrossRef]
- Konstantinidis, G.; Tavernarakis, N. Molecular Basis of Neuronal Autophagy in Ageing: Insights from Caenorhabditis elegans. Cells 2021, 10, 694. [Google Scholar] [CrossRef]
- Wong, S.Q.; Kumar, A.V.; Mills, J.; Lapierre, L.R.C. elegans to model autophagy-related human disorders. Prog. Mol. Biol. Transl. Sci. 2020, 172, 325–373. [Google Scholar] [CrossRef]
- Tamargo-Gomez, I.; Fernandez, A.F.; Marino, G. Pathogenic Single Nucleotide Polymorphisms on Autophagy-Related Genes. Int. J. Mol. Sci. 2020, 21, 8196. [Google Scholar] [CrossRef]
- Du, J.X.; Chen, C.; Luo, Y.H.; Cai, J.L.; Cai, C.Z.; Xu, J.; Ni, X.J.; Zhu, W. Establishment and validation of a novel autophagy-related gene signature for patients with breast cancer. Gene 2020, 762, 144974. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Li, Q.; Zhu, W.; Zhang, X.; Li, H. Identification of autophagy-related long non-coding RNA prognostic signature for breast cancer. J. Cell Mol. Med. 2021, 25, 4088–4098. [Google Scholar] [CrossRef]
- Li, X.; Chen, J.; Yu, Q.; Huang, H.; Liu, Z.; Wang, C.; He, Y.; Zhang, X.; Li, W.; Li, C.; et al. A Signature of Autophagy-Related Long Non-coding RNA to Predict the Prognosis of Breast Cancer. Front. Genet. 2021, 12, 569318. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Zhu, Q.; Yin, D.; Yang, Z.; Guo, J.; Zhang, J.; Zhou, Y.; Yu, J.J. Identification and Validation of an Autophagy-Related lncRNA Signature for Patients With Breast Cancer. Front. Oncol. 2020, 10, 597569. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Zhao, F.; Yu, X.; Guan, S.; Suo, H.; Tao, Z.; Qiu, Y.; Wu, Y.; Cao, Y.; Jin, F. Immune-related lncRNAs as predictors of survival in breast cancer: A prognostic signature. J. Transl. Med. 2020, 18, 442. [Google Scholar] [CrossRef]
- Li, X.; Jin, F.; Li, Y. A novel autophagy-related lncRNA prognostic risk model for breast cancer. J. Cell Mol. Med. 2021, 25, 4–14. [Google Scholar] [CrossRef]
- Wu, L.; Wen, Z.; Song, Y.; Wang, L. A novel autophagy-related lncRNA survival model for lung adenocarcinoma. J. Cell Mol. Med. 2021. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, S.; Li, H.B.; Cai, Z.; Tang, S.; Chen, L.X.; Lang, J.Y.; Chen, Z.; Chen, X.L. Development of Prognostic Indicator Based on Autophagy-Related lncRNA Analysis in Colon Adenocarcinoma. Biomed. Res. Int. 2020, 2020, 9807918. [Google Scholar] [CrossRef]
- Cheng, L.; Han, T.; Zhang, Z.; Yi, P.; Zhang, C.; Zhang, S.; Peng, W. Identification and Validation of Six Autophagy-related Long Non-coding RNAs as Prognostic Signature in Colorectal Cancer. Int. J. Med. Sci. 2021, 18, 88–98. [Google Scholar] [CrossRef]
- Jiang, A.; Liu, N.; Bai, S.; Wang, J.; Gao, H.; Zheng, X.; Fu, X.; Ren, M.; Zhang, X.; Tian, T.; et al. Identification and validation of an autophagy-related long non-coding RNA signature as a prognostic biomarker for patients with lung adenocarcinoma. J. Thorac. Dis. 2021, 13, 720–734. [Google Scholar] [CrossRef]
- Jin, L.; Hong, N.; Ai, X.; Wang, J.; Li, Z.; Han, Z.; Zhang, Q.; Yu, Y.; Sun, K. LncRNAs as Therapeutic Targets for Autophagy-involved Cardiovascular Diseases: A Review of Molecular Mechanism and T herapy Strategy. Curr. Med. Chem. 2021, 28, 1796–1814. [Google Scholar] [CrossRef]
- Gao, J.; Chen, X.; Shan, C.; Wang, Y.; Li, P.; Shao, K. Autophagy in cardiovascular diseases: Role of noncoding RNAs. Mol. Ther. Nucleic Acids 2021, 23, 101–118. [Google Scholar] [CrossRef]
- Rikihisa, Y. Glycogen autophagosomes in polymorphonuclear leukocytes induced by rickettsiae. Anat. Rec. 1984, 208, 319–327. [Google Scholar] [CrossRef]
- Gutierrez, M.G.; Master, S.S.; Singh, S.B.; Taylor, G.A.; Colombo, M.I.; Deretic, V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell 2004, 119, 753–766. [Google Scholar] [CrossRef] [Green Version]
- Nakagawa, I.; Amano, A.; Mizushima, N.; Yamamoto, A.; Yamaguchi, H.; Kamimoto, T.; Nara, A.; Funao, J.; Nakata, M.; Tsuda, K.; et al. Autophagy defends cells against invading group A Streptococcus. Science 2004, 306, 1037–1040. [Google Scholar] [CrossRef]
- Ogawa, M.; Yoshimori, T.; Suzuki, T.; Sagara, H.; Mizushima, N.; Sasakawa, C. Escape of intracellular Shigella from autophagy. Science 2005, 307, 727–731. [Google Scholar] [CrossRef] [PubMed]
- Amer, A.O.; Swanson, M.S. Autophagy is an immediate macrophage response to Legionella pneumophila. Cell MicroBiol. 2005, 7, 765–778. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riebisch, A.K.; Muhlen, S.; Beer, Y.Y.; Schmitz, I. Autophagy-A Story of Bacteria Interfering with the Host Cell Degradation Machinery. Pathogens 2021, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Orvedahl, A.; Alexander, D.; Talloczy, Z.; Sun, Q.; Wei, Y.; Zhang, W.; Burns, D.; Leib, D.A.; Levine, B. HSV-1 ICP34.5 confers neurovirulence by targeting the Beclin 1 autophagy protein. Cell Host Microbe 2007, 1, 23–35. [Google Scholar] [CrossRef] [Green Version]
- Talloczy, Z.; Virgin, H.W., IV; Levine, B. PKR-dependent autophagic degradation of herpes simplex virus type 1. Autophagy 2006, 2, 24–29. [Google Scholar] [CrossRef] [Green Version]
- Munz, C. Viral evasion of autophagy. Cell Host Microbe 2007, 1, 9–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexander, D.E.; Leib, D.A. Xenophagy in herpes simplex virus replication and pathogenesis. Autophagy 2008, 4, 101–103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dengjel, J.; Schoor, O.; Fischer, R.; Reich, M.; Kraus, M.; Muller, M.; Kreymborg, K.; Altenberend, F.; Brandenburg, J.; Kalbacher, H.; et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 7922–7927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paludan, C.; Schmid, D.; Landthaler, M.; Vockerodt, M.; Kube, D.; Tuschl, T.; Munz, C. Endogenous MHC class II processing of a viral nuclear antigen after autophagy. Science 2005, 307, 593–596. [Google Scholar] [CrossRef]
- Schmid, D.; Pypaert, M.; Munz, C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity 2007, 26, 79–92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klein, L.; Munz, C.; Lunemann, J.D. Autophagy-mediated antigen processing in CD4(+) T cell tolerance and immunity. FEBS Lett. 2010, 584, 1405–1410. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munz, C. Enhancing immunity through autophagy. Annu. Rev. Immunol. 2009, 27, 423–449. [Google Scholar] [CrossRef]
- Zhang, Y.; Sun, H.; Pei, R.; Mao, B.; Zhao, Z.; Li, H.; Lin, Y.; Lu, K. The SARS-CoV-2 protein ORF3a inhibits fusion of autophagosomes with lysosomes. Cell Discov. 2021, 7, 31. [Google Scholar] [CrossRef]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Xu, M.; Pradhan, M.; Tran, B.N.; Zhu, W.; Shamim, K.; Huang, W.; et al. The SARS-CoV-2 cytopathic effect is blocked with autophagy modulators. bioRxiv 2020. [Google Scholar] [CrossRef]
- Shojaei, S.; Suresh, M.; Klionsky, D.J.; Labouta, H.I.; Ghavami, S. Autophagy and SARS-CoV-2 infection: Apossible smart targeting of the autophagy pathway. Virulence 2020, 11, 805–810. [Google Scholar] [CrossRef]
- Bello-Perez, M.; Sola, I.; Novoa, B.; Klionsky, D.J.; Falco, A. Canonical and Noncanonical Autophagy as Potential Targets for COVID-19. Cells 2020, 9, 1619. [Google Scholar] [CrossRef] [PubMed]
- Gorshkov, K.; Chen, C.Z.; Bostwick, R.; Rasmussen, L.; Tran, B.N.; Cheng, Y.S.; Xu, M.; Pradhan, M.; Henderson, M.; Zhu, W.; et al. The SARS-CoV-2 Cytopathic Effect Is Blocked by Lysosome Alkalizing Small Molecules. ACS Infect. Dis. 2020. [Google Scholar] [CrossRef]
- Blaess, M.; Kaiser, L.; Sauer, M.; Csuk, R.; Deigner, H.P. COVID-19/SARS-CoV-2 Infection: Lysosomes and Lysosomotropism Implicate New Treatment Strategies and Personal Risks. Int. J. Mol. Sci. 2020, 21, 4953. [Google Scholar] [CrossRef]
- Prestes, E.B.; Bruno, J.C.P.; Travassos, L.H.; Carneiro, L.A.M. The Unfolded Protein Response and Autophagy on the Crossroads of Coronaviruses Infections. Front. Cell Infect. MicroBiol. 2021, 11, 668034. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.; McGrath, M.E.; Hu, Z.; Ariannejad, S.; Weston, S.; Frieman, M.; Jackson, W.T. Coronavirus interactions with the cellular autophagy machinery. Autophagy 2020, 16, 2131–2139. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.; Chen, Y.C.; Hassanzadeh, S.; Han, K.; Judy, J.T.; Seifuddin, F.; Tunc, I.; Sack, M.N.; Pirooznia, M. Network Analysis and Transcriptome Profiling Identify Autophagic and Mitochondrial Dysfunctions in SARS-CoV-2 Infection. Front. Genet. 2021, 12, 599261. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Perez, B.E.; Gonzalez-Rojas, J.A.; Salazar, M.I.; Torres-Torres, C.; Castrejon-Jimenez, N.S. Taming the Autophagy as a Strategy for Treating COVID-19. Cells 2020, 9, 2679. [Google Scholar] [CrossRef]
- Okuyan, H.M.; Dogan, S.; Bal, T.; Cabalak, M. Beclin-1, an autophagy-related protein, is associated with the disease severity of COVID-19. Life Sci. 2021, 278, 119596. [Google Scholar] [CrossRef]
- Pereira, G.; Leao, A.; Erustes, A.G.; Morais, I.B.M.; Vrechi, T.A.M.; Zamarioli, L.D.S.; Pereira, C.A.S.; Marchioro, L.O.; Sperandio, L.P.; Lins, I.V.F.; et al. Pharmacological Modulators of Autophagy as a Potential Strategy for the Treatment of COVID-19. Int. J. Mol. Sci. 2021, 22, 4067. [Google Scholar] [CrossRef]
- Zhu, H.; Chen, C.Z.; Sakamuru, S.; Zhao, J.; Ngan, D.K.; Simeonov, A.; Hall, M.D.; Xia, M.; Zheng, W.; Huang, R. Mining of high throughput screening database reveals AP-1 and autophagy pathways as potential targets for COVID-19 therapeutics. Sci. Rep. 2021, 11, 6725. [Google Scholar] [CrossRef]
- Delorme-Axford, E.; Klionsky, D.J. Highlights in the fight against COVID-19: Does autophagy play a role in SARS-CoV-2 infection? Autophagy 2020, 16, 2123–2127. [Google Scholar] [CrossRef]
- Parenti, G.; Medina, D.L.; Ballabio, A. The rapidly evolving view of lysosomal storage diseases. EMBO Mol. Med. 2021, 13, e12836. [Google Scholar] [CrossRef]
- Ren, H.; Wang, G. Autophagy and Lysosome Storage Disorders. Adv. Exp. Med. Biol. 2020, 1207, 87–102. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Schoonjans, K.; Auwerx, J. Sirtuin Functions in Health and Disease. Mol. Endocrinol. 2007, 21, 1745–1755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, C.S.S.; Cerqueira, N.; Gomes, P.; Sousa, S.F. A Molecular Perspective on Sirtuin Activity. Int. J. Mol. Sci. 2020, 21, 8609. [Google Scholar] [CrossRef] [PubMed]
- Zhao, E.; Hou, J.; Ke, X.; Abbas, M.N.; Kausar, S.; Zhang, L.; Cui, H. The Roles of Sirtuin Family Proteins in Cancer Progression. Cancers 2019, 11, 1949. [Google Scholar] [CrossRef] [Green Version]
- Lee, I.H. Mechanisms and disease implications of sirtuin-mediated autophagic regulation. Exp. Mol. Med. 2019, 51, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Sacitharan, P.K.; Bou-Gharios, G.; Edwards, J.R. SIRT1 directly activates autophagy in human chondrocytes. Cell Death Discov. 2020, 6, 41. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.K.; Singh, S.; Rizvi, S.I. Promising drug discovery strategies for sirtuin modulators: What lessons have we learnt? Expert Opin. Drug Discov. 2021, 16, 915–927. [Google Scholar] [CrossRef]
- Sugawara, K.; Suzuki, N.N.; Fujioka, Y.; Mizushima, N.; Ohsumi, Y.; Inagaki, F. The crystal structure of microtubule-associated protein light chain 3, a mammalian homologue of Saccharomyces cerevisiae Atg8. Genes Cells 2004, 9, 611–618. [Google Scholar] [CrossRef]
- Rubinsztein, D.C.; Gestwicki, J.E.; Murphy, L.O.; Klionsky, D.J. Potential therapeutic applications of autophagy. Nat. Rev. Drug Discov. 2007, 6, 304–312. [Google Scholar] [CrossRef]
- Sora, V.; Kumar, M.; Maiani, E.; Lambrughi, M.; Tiberti, M.; Papaleo, E. Structure and Dynamics in the ATG8 Family From Experimental to Computational Techniques. Front. Cell Dev. Biol. 2020, 8, 420. [Google Scholar] [CrossRef]
- Wesch, N.; Kirkin, V.; Rogov, V.V. Atg8-Family Proteins-Structural Features and Molecular Interactions in Autophagy and Beyond. Cells 2020, 9, 2008. [Google Scholar] [CrossRef] [PubMed]
- Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Vitale, I.; Djavaheri-Mergny, M.; D’Amelio, M.; Criollo, A.; Morselli, E.; Zhu, C.; Harper, F.; et al. Regulation of autophagy by cytoplasmic p53. Nat. Cell Biol. 2008, 10, 676–687. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morselli, E.; Tasdemir, E.; Maiuri, M.C.; Galluzzi, L.; Kepp, O.; Criollo, A.; Vicencio, J.M.; Soussi, T.; Kroemer, G. Mutant p53 protein localized in the cytoplasm inhibits autophagy. Cell Cycle 2008, 7, 3056–3061. [Google Scholar] [CrossRef] [PubMed]
- Morselli, E.; Shen, S.; Ruckenstuhl, C.; Bauer, M.A.; Marino, G.; Galluzzi, L.; Criollo, A.; Michaud, M.; Maiuri, M.C.; Chano, T.; et al. p53 inhibits autophagy by interacting with the human ortholog of yeast Atg17, RB1CC1/FIP200. Cell Cycle 2011, 10, 2763–2769. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Munoz-Alarcon, A.; Ajayi, A.; Webling, K.E.; Steinhof, A.; Langel, U.; Strom, A.L. Inhibition of autophagy via p53-mediated disruption of ULK1 in a SCA7 polyglutamine disease model. J. Mol. NeuroSci. 2013, 50, 586–599. [Google Scholar] [CrossRef]
- Tavernarakis, N.; Pasparaki, A.; Tasdemir, E.; Maiuri, M.C.; Kroemer, G. The effects of p53 on whole organism longevity are mediated by autophagy. Autophagy 2008, 4, 870–873. [Google Scholar] [CrossRef] [Green Version]
- Crighton, D.; Wilkinson, S.; O’Prey, J.; Syed, N.; Smith, P.; Harrison, P.R.; Gasco, M.; Garrone, O.; Crook, T.; Ryan, K.M. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006, 126, 121–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Zhang, H.; Levine, A.J.; Jin, S. The coordinate regulation of the p53 and mTOR pathways in cells. Proc. Natl. Acad. Sci. USA 2005, 102, 8204–8209. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Levine, B.; Cuervo, A.M.; Klionsky, D.J. Autophagy fights disease through cellular self-digestion. Nature 2008, 451, 1069–1075. [Google Scholar] [CrossRef] [Green Version]
- Wickman, G.; Julian, L.; Olson, M.F. How apoptotic cells aid in the removal of their own cold dead bodies. Cell Death Differ. 2012, 19, 735–742. [Google Scholar] [CrossRef] [Green Version]
- Arienti, S.; Barth, N.D.; Dorward, D.A.; Rossi, A.G.; Dransfield, I. Regulation of Apoptotic Cell Clearance during Resolution of Inflammation. Front. Pharmacol. 2019, 10, 891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, C.; Heit, B. Cellular Responses to the Efferocytosis of Apoptotic Cells. Front. Immunol. 2021, 12, 631714. [Google Scholar] [CrossRef]
- Kloditz, K.; Chen, Y.Z.; Xue, D.; Fadeel, B. Programmed cell clearance: From nematodes to humans. BioChem. Biophys. Res. Commun. 2017, 482, 491–497. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.S.; He, L.; Lemasters, J.J. Mitochondrial permeability transition: A common pathway to necrosis and apoptosis. BioChem. Biophys. Res. Commun. 2003, 304, 463–470. [Google Scholar] [CrossRef]
- Vargas, J.L.; Roche, E.; Knecht, E.; Grisolía, S. Differences in the half-lives of some mitochondrial rat liver enzymes may derive partially from hepatocyte heterogeneity. FEBS Lett. 1987, 224, 182–186. [Google Scholar] [CrossRef] [Green Version]
- Pfeifer, U. Inhibition by insulin of the formation of autophagic vacuoles in rat liver. A morphometric approach to the kinetics of intracellular degradation by autophagy. J. Cell Biol. 1978, 78, 152–167. [Google Scholar] [CrossRef]
- Menzies, R.A.; Gold, P.H. The turnover of mitochondria in a variety of tissues of young adult and aged rats. J. Biol. Chem. 1971, 246, 2425–2429. [Google Scholar] [CrossRef]
- Dunn, W.A., Jr. Studies on the mechanisms of autophagy: Formation of the autophagic vacuole. J. Cell Biol. 1990, 110, 1923–1933. [Google Scholar] [CrossRef] [Green Version]
- Elmore, S. Detection of mitochondrial depolarization during autophagy by confocal fluorescence resonance energy transfer (CFRET). Cell Vis. 1997, 4, 170–171. [Google Scholar]
- Weiss, J.N.; Korge, P.; Honda, H.M.; Ping, P. Role of the mitochondrial permeability transition in myocardial disease. Circ. Res. 2003, 93, 292–301. [Google Scholar] [CrossRef] [Green Version]
- Boya, P.; Gonzalez-Polo, R.A.; Casares, N.; Perfettini, J.L.; Dessen, P.; Larochette, N.; Metivier, D.; Meley, D.; Souquere, S.; Yoshimori, T.; et al. Inhibition of macroautophagy triggers apoptosis. Mol. Cell Biol. 2005, 25, 1025–1040. [Google Scholar] [CrossRef] [Green Version]
- Yu, L.; Alva, A.; Su, H.; Dutt, P.; Freundt, E.; Welsh, S.; Baehrecke, E.H.; Lenardo, M.J. Regulation of an ATG7-beclin 1 program of autophagic cell death by caspase-8. Science 2004, 304, 1500–1502. [Google Scholar] [CrossRef]
- Booth, L.A.; Tavallai, S.; Hamed, H.A.; Cruickshanks, N.; Dent, P. The role of cell signalling in the crosstalk between autophagy and apoptosis. Cell Signal 2014, 26, 549–555. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eisenberg-Lerner, A.; Bialik, S.; Simon, H.U.; Kimchi, A. Life and death partners: Apoptosis, autophagy and the cross-talk between them. Cell Death Differ. 2009, 16, 966–975. [Google Scholar] [CrossRef] [PubMed]
- Kapuy, O.; Vinod, P.K.; Mandl, J.; Banhegyi, G. A cellular stress-directed bistable switch controls the crosstalk between autophagy and apoptosis. Mol. Biosyst. 2013, 9, 296–306. [Google Scholar] [CrossRef] [Green Version]
- Lai, M.; Amato, R.; La Rocca, V.; Bilgin, M.; Freer, G.; Spezia, P.; Quaranta, P.; Piomelli, D.; Pistello, M. Acid ceramidase controls apoptosis and increases autophagy in human melanoma cells treated with doxorubicin. Sci. Rep. 2021, 11, 11221. [Google Scholar] [CrossRef]
- Norman, J.M.; Cohen, G.M.; Bampton, E.T. The in vitro cleavage of the hAtg proteins by cell death proteases. Autophagy 2010, 6, 1042–1056. [Google Scholar] [CrossRef] [Green Version]
- Ojha, R.; Ishaq, M.; Singh, S.K. Caspase-mediated crosstalk between autophagy and apoptosis: Mutual adjustment or matter of dominance. J. Cancer Res. Ther. 2015, 11, 514–524. [Google Scholar] [CrossRef]
- Wu, H.; Che, X.; Zheng, Q.; Wu, A.; Pan, K.; Shao, A.; Wu, Q.; Zhang, J.; Hong, Y. Caspases: A molecular switch node in the crosstalk between autophagy and apoptosis. Int. J. Biol. Sci. 2014, 10, 1072–1083. [Google Scholar] [CrossRef] [PubMed]
- Betin, V.M.; Lane, J.D. Atg4D at the interface between autophagy and apoptosis. Autophagy 2009, 5, 1057–1059. [Google Scholar] [CrossRef] [Green Version]
- You, M.; Savaraj, N.; Kuo, M.T.; Wangpaichitr, M.; Varona-Santos, J.; Wu, C.; Nguyen, D.M.; Feun, L. TRAIL induces autophagic protein cleavage through caspase activation in melanoma cell lines under arginine deprivation. Mol. Cell BioChem. 2013, 374, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Djavaheri-Mergny, M.; Maiuri, M.C.; Kroemer, G. Cross talk between apoptosis and autophagy by caspase-mediated cleavage of Beclin 1. Oncogene 2010, 29, 1717–1719. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Su, J.; Xia, M.; Li, H.; Xu, Y.; Ma, C.; Ma, L.; Kang, J.; Yu, H.; Zhang, Z.; et al. Caspase-mediated cleavage of Beclin1 inhibits autophagy and promotes apoptosis induced by S1 in human ovarian cancer SKOV3 cells. Apoptosis 2016, 21, 225–238. [Google Scholar] [CrossRef]
- Siddiqui, M.A.; Mukherjee, S.; Manivannan, P.; Malathi, K. RNase L Cleavage Products Promote Switch from Autophagy to Apoptosis by Caspase-Mediated Cleavage of Beclin-1. Int. J. Mol. Sci. 2015, 16, 17611–17636. [Google Scholar] [CrossRef] [PubMed]
- Pagliarini, V.; Wirawan, E.; Romagnoli, A.; Ciccosanti, F.; Lisi, G.; Lippens, S.; Cecconi, F.; Fimia, G.M.; Vandenabeele, P.; Corazzari, M.; et al. Proteolysis of Ambra1 during apoptosis has a role in the inhibition of the autophagic pro-survival response. Cell Death Differ. 2012, 19, 1495–1504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, Y.; Zhao, L.; Liu, L.; Gao, P.; Tian, W.; Wang, X.; Jin, H.; Xu, H.; Chen, Q. Beclin 1 cleavage by caspase-3 inactivates autophagy and promotes apoptosis. Protein Cell 2010, 1, 468–477. [Google Scholar] [CrossRef] [Green Version]
- Tsapras, P.; Nezis, I.P. Caspase involvement in autophagy. Cell Death Differ. 2017, 24, 1369–1379. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wirawan, E.; Vande Walle, L.; Kersse, K.; Cornelis, S.; Claerhout, S.; Vanoverberghe, I.; Roelandt, R.; De Rycke, R.; Verspurten, J.; Declercq, W.; et al. Caspase-mediated cleavage of Beclin-1 inactivates Beclin-1-induced autophagy and enhances apoptosis by promoting the release of proapoptotic factors from mitochondria. Cell Death Dis. 2010, 1, e18. [Google Scholar] [CrossRef]
- Betin, V.M.; Lane, J.D. Caspase cleavage of Atg4D stimulates GABARAP-L1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 2009, 122, 2554–2566. [Google Scholar] [CrossRef] [Green Version]
- Han, J.; Hou, W.; Goldstein, L.A.; Stolz, D.B.; Watkins, S.C.; Rabinowich, H. A Complex between Atg7 and Caspase-9: A Novel Mechanism of Cross-Regulation Between Autophagy and Apoptosis. J. Biol. Chem. 2014, 289, 6485–6497. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Gao, W.; Loughran, P.; Shapiro, R.; Fan, J.; Billiar, T.R.; Scott, M.J. Caspase 1 activation is protective against hepatocyte cell death by up-regulating beclin 1 protein and mitochondrial autophagy in the setting of redox stress. J. Biol. Chem. 2013, 288, 15947–15958. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamy, L.; Ngo, V.N.; Emre, N.C.; Shaffer, A.L., 3rd; Yang, Y.; Tian, E.; Nair, V.; Kruhlak, M.J.; Zingone, A.; Landgren, O.; et al. Control of autophagic cell death by caspase-10 in multiple myeloma. Cancer Cell 2013, 23, 435–449. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, J.R.; Li, C.; Yang, Q.; Li, G.; Garcia, I.G.; Ju, S.; Roodman, D.G.; Windle, J.J.; Zhang, X.; Lu, B. Autophagy promotes T-cell survival through degradation of proteins of the cell death machinery. Cell Death Differ. 2012, 19, 144–152. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Shoji-Kawata, S.; Sumpter, R.M., Jr.; Wei, Y.; Ginet, V.; Zhang, L.; Posner, B.; Tran, K.A.; Green, D.R.; Xavier, R.J.; et al. Autosis is a Na+,K+-ATPase-regulated form of cell death triggered by autophagy-inducing peptides, starvation, and hypoxia-ischemia. Proc. Natl. Acad. Sci. USA 2013, 110, 20364–20371. [Google Scholar] [CrossRef] [Green Version]
- Nah, J.; Zablocki, D.; Sadoshima, J. Autosis: A New Target to Prevent Cell Death. JACC Basic Transl. Sci. 2020, 5, 857–869. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Levine, B. Autosis and autophagic cell death: The dark side of autophagy. Cell Death Differ. 2015, 22, 367–376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munoz-Pinedo, C.; Martin, S.J. Autosis: A new addition to the cell death Tower of Babel. Cell Death Dis. 2014, 5, e1319. [Google Scholar] [CrossRef] [Green Version]
- Kovacs, A.L. The application of traditional transmission electron microscopy for autophagy research in Caenorhabditis elegans. Biophys. Rep. 2015, 1, 99–105. [Google Scholar] [CrossRef] [Green Version]


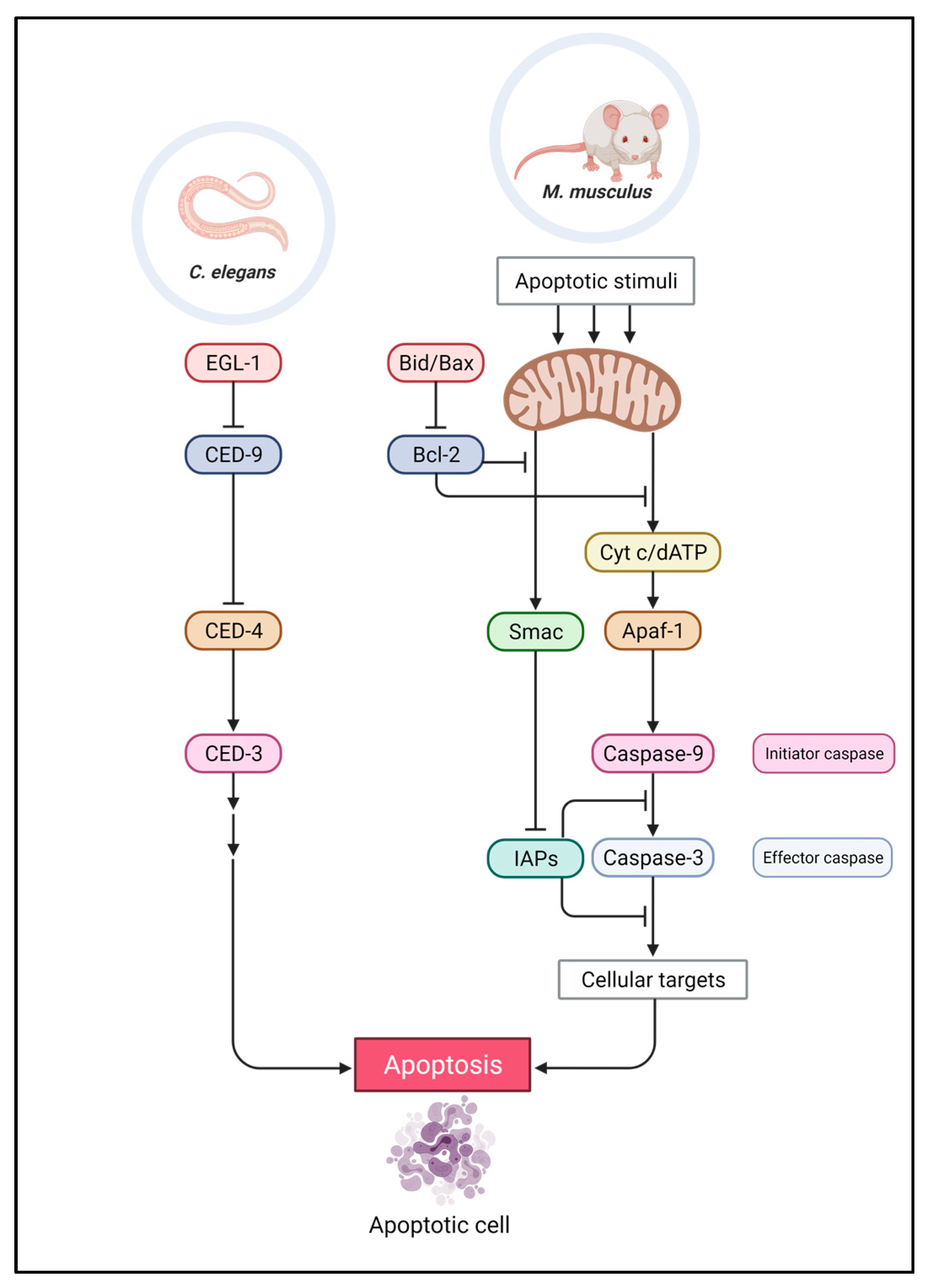
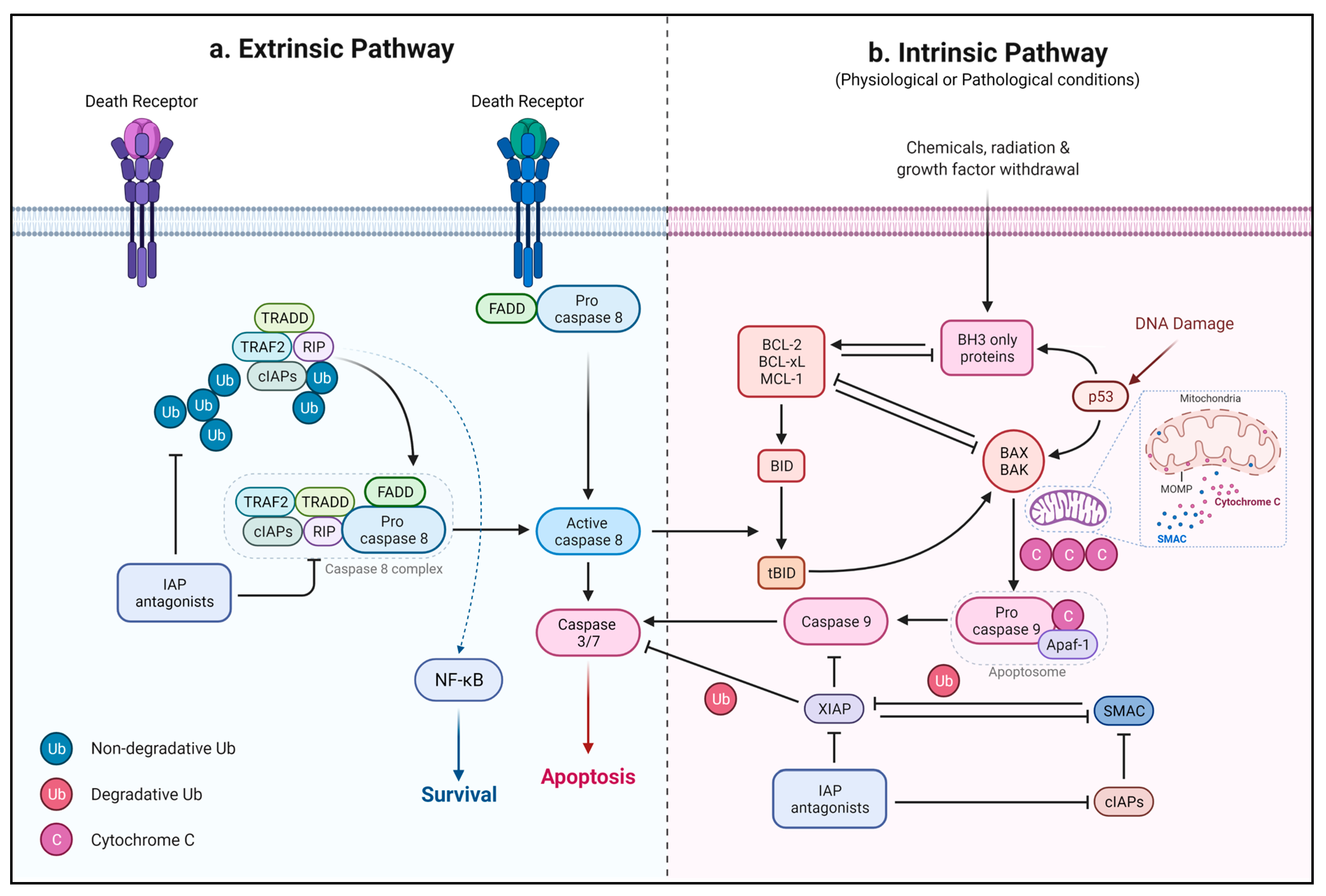

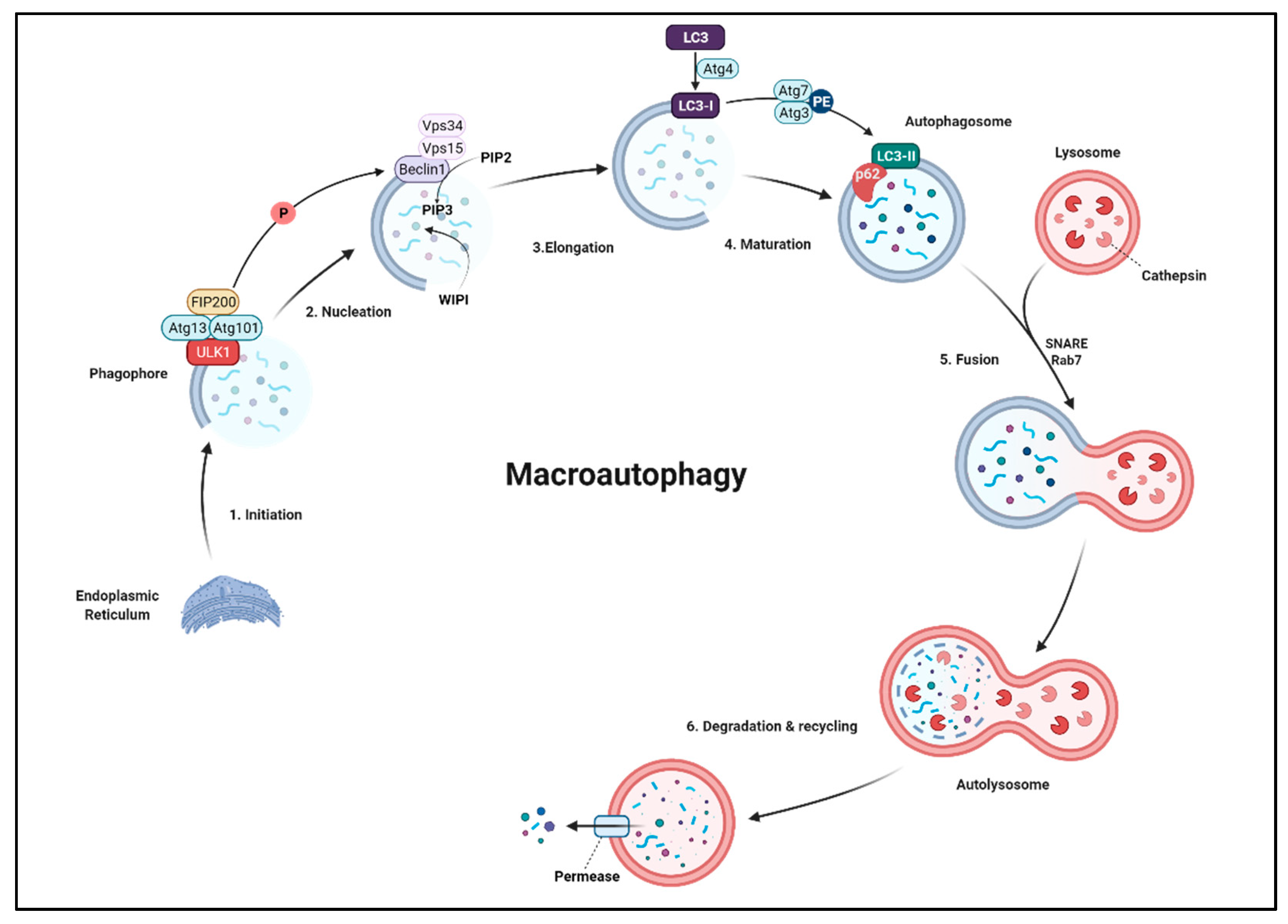
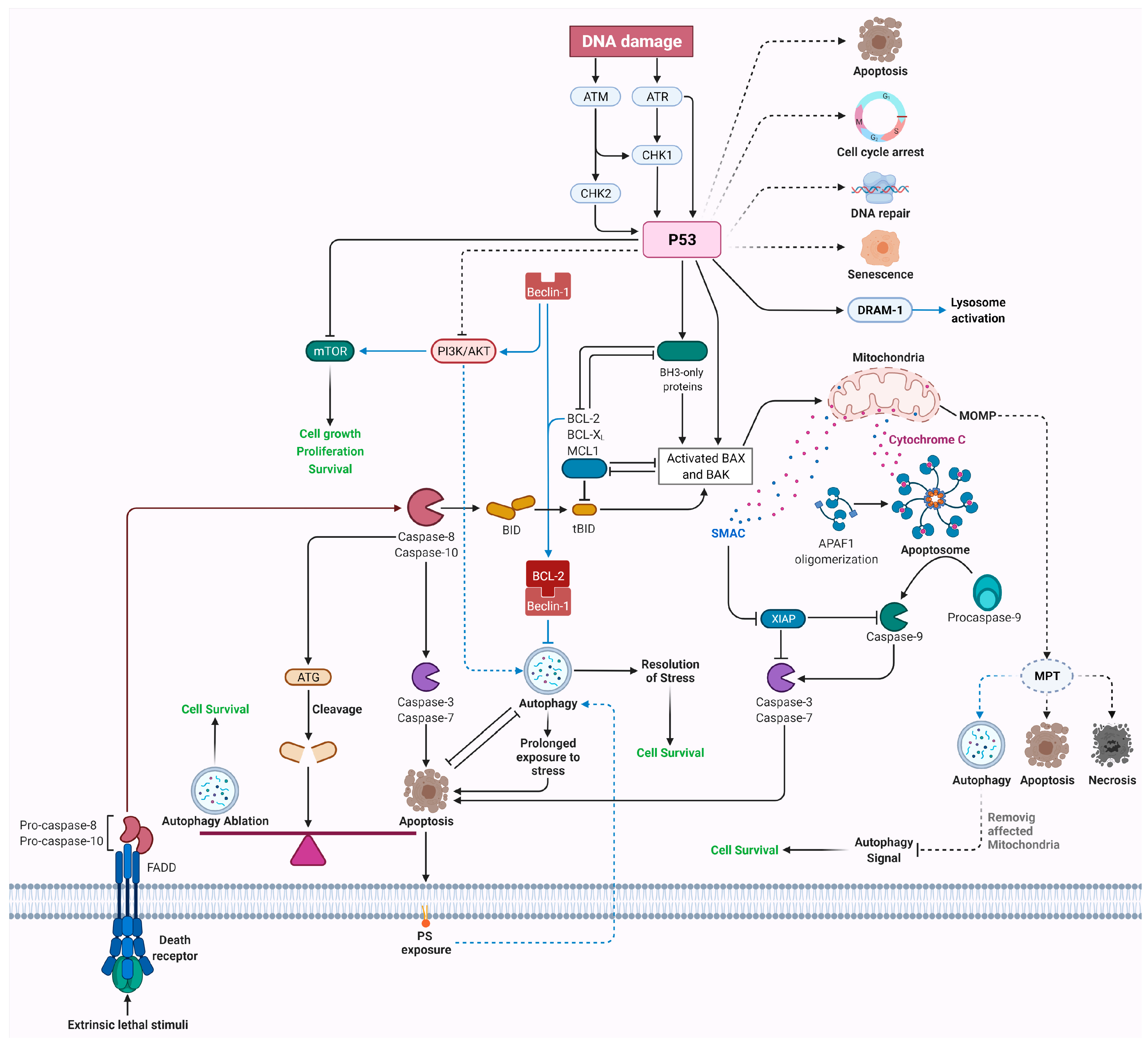
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Izadi, M.; Ali, T.A.; Pourkarimi, E. Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy. Int. J. Mol. Sci. 2021, 22, 12466. https://doi.org/10.3390/ijms222212466
Izadi M, Ali TA, Pourkarimi E. Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy. International Journal of Molecular Sciences. 2021; 22(22):12466. https://doi.org/10.3390/ijms222212466
Chicago/Turabian StyleIzadi, Mahmoud, Tayyiba Akbar Ali, and Ehsan Pourkarimi. 2021. "Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy" International Journal of Molecular Sciences 22, no. 22: 12466. https://doi.org/10.3390/ijms222212466
APA StyleIzadi, M., Ali, T. A., & Pourkarimi, E. (2021). Over Fifty Years of Life, Death, and Cannibalism: A Historical Recollection of Apoptosis and Autophagy. International Journal of Molecular Sciences, 22(22), 12466. https://doi.org/10.3390/ijms222212466






