New Approaches to Assess Mechanisms of Action of Selective Vitamin D Analogues
Abstract
:1. Background and Review Rationale
2. Transcriptional Mechanism of Action of 1,25(OH)2D3
3. Analogues of Vitamin D
4. Pharmacology of Vitamin D Analogues
5. New Insights into Transcriptional Activation of Vitamin D
6. Current Advances in Transcriptional Regulation
7. Recent Work at the Frontier for Enhancing Vitamin D Analogue SAR Potential
8. Vitamin D Analogues
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- DeLuca, H.F. Overview of general physiologic features and functions of vitamin D. Am. J. Clin. Nutr. 2004, 80, 1689S–1696S. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Peng, X.; Porta, A.; Takanaga, H.; Peng, J.-B.; Hediger, M.A.; Fleet, J.C.; Christakos, S. Calcium Transporter 1 and Epithelial Calcium Channel Messenger Ribonucleic Acid Are Differentially Regulated by 1,25 Dihydroxyvitamin D3 in the Intestine and Kidney of Mice. Endocrinology 2003, 144, 3885–3894. [Google Scholar] [CrossRef]
- Hoenderop, J.G.J.; Nilius, B.; Bindels, R.J.M. Calcium Absorption across Epithelia. Physiol. Rev. 2005, 85, 373–422. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Cromphaut, S.J.; Dewerchin, M.; Hoenderop, J.G.J.; Stockmans, I.; Van Herck, E.; Kato, S.; Bindels, R.J.M.; Collen, D.; Carmeliet, P.; Bouillon, R.; et al. Duodenal calcium absorption in vitamin D receptor-knockout mice: Functional and molecular aspects. Proc. Natl. Acad. Sci. USA 2001, 98, 13324–13329. [Google Scholar] [CrossRef] [Green Version]
- Van Abel, M.; Hoenderop, J.G.; Bindels, R.J. The epithelial calcium channels TRPV5 and TRPV6: Regulation and implications for disease. Naunyn-Schmiedeberg’s Arch. Pharmacol. 2005, 371, 295–306. [Google Scholar] [CrossRef] [Green Version]
- Nijenhuis, T.; Hoenderop, J.G.J.; Bindels, R.J.M. TRPV5 and TRPV6 in Ca2+ (re)absorption: Regulating Ca2+ entry at the gate. Pflug. Arch. 2005, 451, 181–192. [Google Scholar] [CrossRef] [PubMed]
- Shimada, T.; Hasegawa, H.; Yamazaki, Y.; Muto, T.; Hino, R.; Takeuchi, Y.; Fujita, T.; Nakahara, K.; Fukumoto, S.; Yamashita, T. FGF-23 Is a Potent Regulator of Vitamin D Metabolism and Phosphate Homeostasis. J. Bone Miner. Res. 2004, 19, 429–435. [Google Scholar] [CrossRef] [Green Version]
- Hu, M.C.; Shiizaki, K.; Kuro-O, M.; Moe, O.W. Fibroblast Growth Factor 23 and Klotho: Physiology and Pathophysiology of an Endocrine Network of Mineral Metabolism. Annu. Rev. Physiol. 2013, 75, 503–533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suda, T.; Takahashi, N.; Udagawa, N.; Jimi, E.; Gillespie, M.; Martin, T. Modulation of osteoclast differentiation and function by the new members of the tumor necrosis factor receptor and ligand families. Endocr. Rev. 1999, 20, 345–357. [Google Scholar] [CrossRef]
- Kondo, M.; Yamaoka, K.; Sakata, K.; Sonomoto, K.; Lin, L.; Nakano, K.; Tanaka, Y. Contribution of the Interleukin-6/STAT-3 Signaling Pathway to Chondrogenic Differentiation of Human Mesenchymal Stem Cells. Arthritis Rheumatol. 2015, 67, 1250–1260. [Google Scholar] [CrossRef]
- Kim, S.; Shevde, N.K.; Pike, J.W. 1,25-Dihydroxyvitamin D3 Stimulates Cyclic Vitamin D Receptor/Retinoid X Receptor DNA-Binding, Co-activator Recruitment, and Histone Acetylation in Intact Osteoblasts. J. Bone Miner. Res. 2004, 20, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Jagga, S.; Martins, J.S.; Rana, R.; Pajevic, P.D.; Liu, E.S. Impaired 1,25 dihydroxyvitamin D3 action and hypophosphatemia underlie the altered lacuno-canalicular remodeling observed in the Hyp mouse model of XLH. PLoS ONE 2021, 16, e0252348. [Google Scholar] [CrossRef]
- St John, H.C.; Bishop, K.A.; Meyer, M.B.; Benkusky, N.A.; Leng, N.; Kendziorski, C.; Bonewald, L.F.; Pike, J.W. The osteoblast to osteocyte transition: Epigenetic changes and response to the vitamin D3 hormone. Mol. Endocrinol. 2014, 28, 1150–1165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, M.B.; Goetsch, P.D.; Pike, J.W. Genome-wide analysis of the VDR/RXR cistrome in osteoblast cells provides new mechanistic insight into the actions of the vitamin D hormone. J. Steroid Biochem. Mol. Biol. 2010, 121, 136–141. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Lee, C.-H.; Pike, J. Genomic Determinants of Gene Regulation by 1,25-Dihydroxyvitamin D3 during Osteoblast-lineage Cell Differentiation. J. Biol. Chem. 2014, 289, 19539–19554. [Google Scholar] [CrossRef] [Green Version]
- Martin, A.; Quarles, L.D. Evidence for FGF23 Involvement in a Bone-Kidney Axis Regulating Bone Mineralization and Systemic Phosphate and Vitamin D Homeostasis. Adv. Exp. Med. Biol. 2012, 728, 65–83. [Google Scholar] [CrossRef]
- Quarles, L.D. Role of FGF23 in vitamin D and phosphate metabolism: Implications in chronic kidney disease. Exp. Cell Res. 2012, 318, 1040–1048. [Google Scholar] [CrossRef] [Green Version]
- Saini, R.K.; Kaneko, I.; Jurutka, P.; Forster, R.; Hsieh, A.; Hsieh, J.-C.; Haussler, M.R.; Whitfield, G.K. 1,25-Dihydroxyvitamin D3 Regulation of Fibroblast Growth Factor-23 Expression in Bone Cells: Evidence for Primary and Secondary Mechanisms Modulated by Leptin and Interleukin-6. Calcif. Tissue Int. 2012, 92, 339–353. [Google Scholar] [CrossRef] [Green Version]
- Andrukhova, O.; Zeitz, U.; Goetz, R.; Mohammadi, M.; Lanske, B.; Erben, R.G. FGF23 acts directly on renal proximal tubules to induce phosphaturia through activation of the ERK1/2-SGK1 signaling pathway. Bone 2012, 51, 621–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrukhova, O.; Slavic, S.; Smorodchenko, A.; Zeitz, U.; Shalhoub, V.; Lanske, B.; Pohl, E.E.; Erben, R.G. FGF 23 regulates renal sodium handling and blood pressure. EMBO Mol. Med. 2014, 6, 744–759. [Google Scholar] [CrossRef]
- Jones, G.; Prosser, D.E.; Kaufmann, M. Cytochrome P450-mediated metabolism of vitamin D. J. Lipid Res. 2014, 55, 13–31. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Pike, J.W. Mechanistic homeostasis of vitamin D metabolism in the kidney through reciprocal modulation of Cyp27b1 and Cyp24a1 expression. J. Steroid Biochem. Mol. Biol. 2019, 196, 105500. [Google Scholar] [CrossRef] [PubMed]
- Adams, J.S.; Hewison, M. Extrarenal expression of the 25-hydroxyvitamin D-1-hydroxylase. Arch. Biochem. Biophys. 2012, 523, 95–102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hewison, M.; Burke, F.; Evans, K.N.; Lammas, D.A.; Sansom, D.; Liu, P.; Modlin, R.; Adams, J. Extra-renal 25-hydroxyvitamin D3-1α-hydroxylase in human health and disease. J. Steroid Biochem. Mol. Biol. 2007, 103, 316–321. [Google Scholar] [CrossRef]
- Chun, R.; Liu, P.T.; Modlin, R.L.; Adams, J.S.; Hewison, M. Impact of vitamin D on immune function: Lessons learned from genome-wide analysis. Front. Physiol. 2014, 5, 151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikle, D.D. Vitamin D and the skin. J. Bone Min. Metab. 2010, 28, 117–130. [Google Scholar] [CrossRef]
- Hewison, M.; Adams, J.S. Extrareanl 1Alpha-Hydroxylase, 3rd ed.; Academic Press: San Diego, CA, USA, 2018; Volume 1. [Google Scholar]
- Adams, J.S.; Rafison, B.; Witzel, S.; Reyes, R.E.; Shieh, A.; Chun, R.; Zavala, K.; Hewison, M.; Liu, P.T. Regulation of the extrarenal CYP27B1-hydroxylase. J. Steroid Biochem. Mol. Biol. 2014, 144 Pt A, 22–27. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Kaufmann, M.; Lee, S.M.; Onal, M.; Jones, G.; Pike, J.W. A kidney-specific genetic control module in mice governs endocrine regulation of the cytochrome P450 gene Cyp27b1 essential for vitamin D3 activation. J. Biol. Chem. 2017, 292, 17541–17558. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Lee, S.M.; Carlson, A.H.; Benkusky, N.A.; Kaufmann, M.; Jones, G.; Pike, J.W. A chromatin-based mechanism controls differential regulation of the cytochrome P450 gene Cyp24a1 in renal and non-renal tissues. J. Biol. Chem. 2019, 294, 14467–14481. [Google Scholar] [CrossRef] [Green Version]
- Welsh, J. Vitamin D and breast cancer: Insights from animal models. Am. J. Clin. Nutr. 2004, 80, 1721S–1724S. [Google Scholar] [CrossRef] [Green Version]
- Pike, J.; Haussler, M. Characteristics and purification of the intstinal receptor for 1,25-dihydroxyvitamin D. Methods Enzymol. 1980, 67, 508–522. [Google Scholar]
- McDonnell, D.P.; Mangelsdorf, D.J.; Pike, J.W.; Haussler, M.R.; O’Malley, B.W. Molecular Cloning of Complementary DNA Encoding the Avian Receptor for Vitamin, D. Science 1987, 235, 1214–1217. [Google Scholar] [CrossRef]
- Baker, A.R.; McDonnell, D.P.; Hughes, M.; Crisp, T.M.; Mangelsdorf, D.; Haussler, M.R.; Pike, J.W.; Shine, J.; O’Malley, B.W. Cloning and expression of full-length cDNA encoding human vitamin D receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 3294–3298. [Google Scholar] [CrossRef] [Green Version]
- Mangelsdorf, D.J.; Thummel, C.; Beato, M.; Herrlich, P.; Schütz, G.; Umesono, K.; Blumberg, B.; Kastner, P.; Mark, M.; Chambon, P.; et al. The nuclear receptor superfamily: The second decade. Cell 1995, 83, 835–839. [Google Scholar] [CrossRef] [Green Version]
- Rochel, N.; Wurtz, J.-M.; Mitschler, A.; Klaholz, B.; Moras, D. The Crystal Structure of the Nuclear Receptor for Vitamin D Bound to Its Natural Ligand. Mol. Cell 2000, 5, 173–179. [Google Scholar] [CrossRef]
- Vanhooke, J.L.; Prahl, J.M.; Kimmel-Jehan, C.; Mendelsohn, M.; Danielson, E.W.; Healy, K.D.; DeLuca, H.F. CYP27B1 null mice with LacZreporter gene display no 25-hydroxyvitamin D3-1 -hydroxylase promoter activity in the skin. Proc. Natl. Acad. Sci. USA 2005, 103, 75–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rochel, N.; Ciesielski, F.; Godet, J.; Moman, E.; Rössle, M.; Peluso-Iltis, C.; Moulin, M.; Haertlein, M.; Callow, P.; Mély, Y.; et al. Common architecture of nuclear receptor heterodimers on DNA direct repeat elements with different spacings. Nat. Struct. Mol. Biol. 2011, 18, 564–570. [Google Scholar] [CrossRef]
- Liao, J.; Ozono, K.; Sone, T.; McDonnell, D.P.; Pike, J.W. Vitamin D receptor interaction with specific DNA requires a nuclear protein and 1,25-dihydroxyvitamin D3. Proc. Natl. Acad. Sci. USA 1990, 87, 9751–9755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kliewer, S.A.; Umesono, K.; Mangelsdorf, D.; Evans, R. Retinoid X receptor interacts with nuclear receptors in retinoic acid, thyroid hormone and vitamin D3 signalling. Nature 1992, 355, 446–449. [Google Scholar] [CrossRef] [PubMed]
- Kerner, S.A.; Scott, R.A.; Pike, J.W. Sequence elements in the human osteocalcin gene confer basal activation and inducible response to hormonal vitamin D3. Proc. Natl. Acad. Sci. USA 1989, 86, 4455–4459. [Google Scholar] [CrossRef] [Green Version]
- Sone, T.; Kerner, S.; Pike, J. Vitamin D receptor interaction with specific DNA. Association as a 1,25-dihydroxyvitamin D3-modulated heterodimer. J. Biol. Chem. 1991, 266, 23296–23305. [Google Scholar] [CrossRef]
- Ozono, K.; Liao, J.; Kerner, S.A.; Scott, R.A.; Pike, J.W. The vitamin D-responsive element in the human osteocalcin gene. Association with a nuclear proto-oncogene enhancer. J. Biol. Chem. 1990, 265, 21881–21888. [Google Scholar] [CrossRef]
- Meyer, M.B.; Pike, J.W. Corepressors (NCoR and SMRT) as well as coactivators are recruited to positively regulated 1α,25-dihydroxyvitamin D3-responsive genes. J. Steroid Biochem. Mol. Biol. 2013, 136, 120–124. [Google Scholar] [CrossRef] [Green Version]
- Pike, J.W.; Lee, S.M.; Meyer, M.B. Regulation of gene expression by 1,25-dihydroxyvitamin D3 in bone cells: Exploiting new approaches and defining new mechanisms. BoneKEy Rep. 2014, 3, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- St-Arnaud, R.; Messerlian, S.; Moir, J.M.; Omdahl, J.L.; Glorieux, F.H. The 25-Hydroxyvitamin D 1-Alpha-Hydroxylase Gene Maps to the Pseudovitamin D-Deficiency Rickets (PDDR) Disease Locus. J. Bone Miner. Res. 1997, 12, 1552–1559. [Google Scholar] [CrossRef]
- Haussler, M.R.; McCain, T.A. Basic and clinical concepts related to vitamin D metabolism and action (first of two parts). N. Engl. J. Med. 1977, 297, 974–983. [Google Scholar] [CrossRef] [PubMed]
- Verstuyf, A.; Segaert, S.; Verlinden, L.; Bouillon, R.; Mathieu, C. Recent developments in the use of vitamin D analogues. Expert Opin. Investig. Drugs 2000, 9, 443–455. [Google Scholar] [CrossRef]
- Bouillon, R.; Eelen, G.; Verlinden, L.; Mathieu, C.; Carmeliet, G.; Verstuyf, A. Vitamin D and cancer. J. Steroid Biochem. Mol. Biol. 2006, 102, 156–162. [Google Scholar] [CrossRef]
- Eelen, G.; Verlinden, L.; Meyer, M.; Gijsbers, R.; Pike, J.W.; Bouillon, R.; Verstuyf, A. 1,25-Dihydroxyvitamin D3 and the aging-related Forkhead Box O and Sestrin proteins in osteoblasts. J. Steroid Biochem. Mol. Biol. 2013, 136, 112–119. [Google Scholar] [CrossRef] [PubMed]
- Nigwekar, S.U.; Thadhani, R.I. Shining light on vitamin D trials in chronic kidney disease. Kidney Int. 2013, 83, 198–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nigwekar, S.U.; Tamez, H.; Thadhani, R.I. Vitamin D and chronic kidney disease-mineral bone disease (CKD-MBD). Bonekey Rep. 2014, 3, 498. [Google Scholar] [CrossRef] [Green Version]
- Nigwekar, S.U.; Thadhani, R. Vitamin D receptor activation: Cardiovascular and renal implications. Kidney Int. Suppl. 2013, 3, 427–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pittas, A.G.; Jorde, R.; Kawahara, T.; Dawson-Hughes, B. Vitamin D Supplementation for Prevention of Type 2 Diabetes Mellitus: To D or Not to D? J. Clin. Endocrinol. Metab. 2020, 105, 3721–3733. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Brodsky, I.G.; Chatterjee, R.; Kashyap, S.R.; Knowler, W.C.; Liao, E.; Nelson, J.; Pratley, R.; Rasouli, N.; Vickery, E.M.; et al. Effect of Vitamin D Supplementation on Kidney Function in Adults with Prediabetes: A Secondary Analysis of a Randomized Trial. Clin. J. Am. Soc. Nephrol. 2021, 16, 1201–1209. [Google Scholar] [CrossRef] [PubMed]
- Rasouli, N.; Brodsky, I.G.; Chatterjee, R.; Kim, S.H.; Pratley, R.E.; Staten, M.A.; Pittas, A.G.; Ceglia, L.; Chadha, C.; Dawson-Hughes, B.; et al. Effects of Vitamin D Supplementation on Insulin Sensitivity and Secretion in Prediabetes. J. Clin. Endocrinol. Metab. 2021. [Google Scholar] [CrossRef] [PubMed]
- Pittas, A.G.; Dawson-Hughes, B.; Sheehan, P.R.; Rosen, C.J.; Ware, J.H.; Knowler, W.C.; Staten, M.A.; The D2d Research Group. Rationale and Design of the Vitamin D and Type 2 Diabetes (D2d) Study: A Diabetes Prevention Trial. Diabetes Care 2014, 37, 3227–3234. [Google Scholar] [CrossRef] [Green Version]
- Pittas, A.G.; Dawson-Hughes, B.; Sheehan, P.; Ware, J.H.; Knowler, W.C.; Aroda, V.R.; Brodsky, I.; Ceglia, L.; Chadha, C.; Chatterjee, R. Vitamin D Supplementation and Prevention of Type 2 Diabetes. N. Engl. J. Med. 2019, 381, 520–530. [Google Scholar] [CrossRef] [Green Version]
- Arroyo-Díaz, J.A.; Julve, J.; Vlacho, B.; Corcoy, R.; Ponte, P.; Román, E.; Navas-Méndez, E.; Llauradó, G.; Franch-Nadal, J.; Domingo, P. Previous Vitamin D Supplementation and Morbidity and Mortality Outcomes in People Hospitalised for COVID19: A Cross-Sectional Study. Front. Public Health 2021, 9, 758347. [Google Scholar] [CrossRef]
- Sato, Y.; Ramalanjaona, N.; Huet, T.; Potier, N.; Osz, J.; Antony, P.; Peluso-Iltis, C.; Poussin-Courmontagne, P.; Ennifar, E.; Mély, Y.; et al. The “Phantom Effect” of the Rexinoid LG100754: Structural and Functional Insights. PLoS ONE 2010, 5, e15119. [Google Scholar] [CrossRef] [Green Version]
- Tocchini-Valentini, G.; Rochel, N.; Wurtz, J.-M.; Moras, D. Crystal Structures of the Vitamin D Nuclear Receptor Liganded with the Vitamin D Side Chain Analogues Calcipotriol and Seocalcitol, Receptor Agonists of Clinical Importance. Insights into a Structural Basis for the Switching of Calcipotriol to a Receptor Antagonist by Further Side Chain Modification. J. Med. Chem. 2004, 47, 1956–1961. [Google Scholar] [CrossRef]
- Tocchini-Valentini, G.; Rochel, N.; Wurtz, J.-M.; Mitschler, A.; Moras, D. Crystal structures of the vitamin D receptor complexed to superagonist 20-epi ligands. Proc. Natl. Acad. Sci. USA 2001, 98, 5491–5496. [Google Scholar] [CrossRef] [Green Version]
- Orlov, I.; Rochel, N.; Moras, D.; Klaholz, B.P. Structure of the full human RXR/VDR nuclear receptor heterodimer complex with its DR3 target DNA. EMBO J. 2011, 31, 291–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hourai, S.; Rodrigues, L.C.; Antony, P.; Reina-San-Martin, B.; Ciesielski, F.; Magnier, B.C.; Schoonjans, K.; Mouriño, A.; Rochel, N.; Moras, D. Structure-Based Design of a Superagonist Ligand for the Vitamin D Nuclear Receptor. Chem. Biol. 2008, 15, 383–392. [Google Scholar] [CrossRef]
- Cherepanova, O.A.; Gomez, D.; Shankman, L.S.; Swiatlowska, P.; Williams, J.; Sarmento, O.F.; Alencar, G.F.; Hess, D.L.; Bevard, M.H.; Greene, E.S.; et al. Activation of the pluripotency factor OCT4 in smooth muscle cells is atheroprotective. Nat. Med. 2016, 22, 657–665. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nettles, K.W.; Greene, G.L. Ligand control of coregulator recruitment to nuclear receptors. Annu. Rev. Physiol. 2005, 67, 309–333. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Pike, A.C.W.; Dauter, Z.; Hubbard, R.E.; Bonn, T.; Engström, O.; Öhman, L.; Greene, G.L.; Gustafsson, J.A.; Carlquist, M. Molecular basis of agonism and antagonism in the oestrogen receptor. Nature 1997, 389, 753–758. [Google Scholar] [CrossRef]
- Wu, Y.-L.; Yang, X.; Ren, Z.; McDonnell, D.P.; Norris, J.; Willson, T.M.; Greene, G.L. Structural Basis for an Unexpected Mode of SERM-Mediated ER Antagonism. Mol. Cell 2005, 18, 413–424. [Google Scholar] [CrossRef] [PubMed]
- Peleg, S.; Sastry, M.; Collins, E.D.; Bishop, J.E.; Norman, A.W. Distinct Conformational Changes Induced by 20-epi Analogues of 1α,25-Dihydroxyvitamin D3 Are Associated with Enhanced Activation of the Vitamin D Receptor. J. Biol. Chem. 1995, 270, 10551–10558. [Google Scholar] [CrossRef] [Green Version]
- Posner, G.H.; Crawford, K.R.; Peleg, S.; Welsh, J.E.; Romu, S.; Gewirtz, D.A.; Gupta, M.S.; Dolan, P.; Kensler, T.W. A non-calcemic sulfone version of the vitamin D(3) analogue seocalcitol (EB 1089): Chemical synthesis, biological evaluation and potency enhancement of the anticancer drug adriamycin. Bioorgan. Med. Chem. 2001, 9, 2365–2371. [Google Scholar] [CrossRef]
- Igarashi, M.; Yoshimoto, N.; Yamamoto, K.; Shimizu, M.; Ishizawa, M.; Makishima, M.; DeLuca, H.F.; Yamada, S. Identification of a highly potent vitamin D receptor antagonist: (25S)-26-Adamantyl-25-hydroxy-2-methylene-22,23-didehydro-19,27-dinor-20-epi-vitamin D3 (ADMI3). Arch. Biochem. Biophys. 2007, 460, 240–253. [Google Scholar] [CrossRef]
- Choi, M.; Yamamoto, K.; Itoh, T.; Makishima, M.; Mangelsdorf, D.; Moras, D.; DeLuca, H.F.; Yamada, S. Interaction between Vitamin D Receptor and Vitamin D Ligands: Two-Dimensional Alanine Scanning Mutational Analysis. Chem. Biol. 2003, 10, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Pike, J.W. 1,25-Dihydroxyvitamin D3 induced histone profiles guide discovery of VDR action sites. J. Steroid Biochem. Mol. Biol. 2013, 144, 19–21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacLeod, R.S.; Meyer, M.B.; Xiong, J.; Cawley, K.M.; Liu, Y.; Onal, M.; Benkusky, N.A.; Thostenson, J.D.; Pike, J.W.; O’Brien, C.A. Deletion of a putative promoter-proximal Tnfsf11 regulatory region in mice does not alter bone mass or Tnfsf11 expression in vivo. PLoS ONE 2021, 16, e0250974. [Google Scholar] [CrossRef]
- Onal, M.; John, H.C.S.; Danielson, A.L.; Pike, J.W. Deletion of the Distal Tnfsf11 RL-D2 Enhancer That Contributes to PTH-Mediated RANKL Expression in Osteoblast Lineage Cells Results in a High Bone Mass Phenotype in Mice. J. Bone Miner. Res. 2015, 31, 416–429. [Google Scholar] [CrossRef] [Green Version]
- Onal, M.; Bishop, K.A.; John, H.C.S.; Danielson, A.L.; Riley, E.M.; Piemontese, M.; Xiong, J.; Goellner, J.J.; O’Brien, C.A.; Pike, J.W. A DNA Segment Spanning the Mouse Tnfsf11 Transcription Unit and Its Upstream Regulatory Domain Rescues the Pleiotropic Biologic Phenotype of the RANKL Null Mouse. J. Bone Miner. Res. 2014, 30, 855–868. [Google Scholar] [CrossRef] [Green Version]
- Pike, J.W.; Meyer, M.B.; Benkusky, N.A.; Lee, S.M.; John, H.S.; Carlson, A.; Onal, M.; Shamsuzzaman, S. Genomic Determinants of Vitamin D-Regulated Gene Expression. Vitam Horm. 2015, 100, 21–44. [Google Scholar] [CrossRef] [Green Version]
- Mangelsdorf, D.; Umesono, K.; Kliewer, S.A.; Borgmeyer, U.; Ong, E.S.; Evans, R. A direct repeat in the cellular retinol-binding protein type II gene confers differential regulation by RXR and RAR. Cell 1991, 66, 555–561. [Google Scholar] [CrossRef]
- Kaneko, I.; Sabir, M.S.; Dussik, C.M.; Whitfield, G.K.; Karrys, A.; Hsieh, J.-C.; Haussler, M.R.; Meyer, M.B.; Pike, J.W.; Jurutka, P.W. 1,25-Dihydroxyvitamin D regulates expression of the tryptophan hydroxylase 2 and leptin genes: Implication for behavioral influences of vitamin D. FASEB J. 2015, 29, 4023–4035. [Google Scholar] [CrossRef]
- Gerstein, M.B.; Kundaje, A.; Hariharan, M.; Landt, S.G.; Yan, K.-K.; Cheng, C.; Mu, X.J.; Khurana, E.; Rozowsky, J.; Alexander, R.P.; et al. Architecture of the human regulatory network derived from ENCODE data. Nature 2012, 489, 91–100. [Google Scholar] [CrossRef]
- Lan, X.; Witt, H.; Katsumura, K.; Ye, Z.; Wang, Q.; Bresnick, E.H.; Farnham, P.J.; Jin, V.X. Integration of Hi-C and ChIP-seq data reveals distinct types of chromatin linkages. Nucleic Acids Res. 2012, 40, 7690–7704. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mouse ENCODE Consortium; Stamatoyannopoulos, J.A.; Snyder, M.; Hardison, R.; Ren, B.; Gingeras, T.; Gilbert, D.M.; Groudine, M.; Bender, M.; Kaul, R.; et al. An encyclopedia of mouse DNA elements (Mouse ENCODE). Genome Biol. 2012, 13, 1–5. [Google Scholar] [CrossRef] [Green Version]
- Thurman, R.E.; Day, N.; Noble, W.S.; Stamatoyannopoulos, J.A. Identification of higher-order functional domains in the human ENCODE regions. Genome Res. 2007, 17, 917–927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bikle, D.D. Vitamin D: Newer Concepts of Its Metabolism and Function at the Basic and Clinical Level. J. Endocr. Soc. 2020, 4, bvz038. [Google Scholar] [CrossRef]
- Yoon, Y.S.; Tsai, W.W.; Van de Velde, S.; Chen, Z.; Lee, K.F.; Morgan, D.A.; Rahmouni, K.; Matsumura, S.; Wiater, E.; Song, Y.; et al. cAMP-inducible coactivator CRTC3 attenuates brown adipose tissue thermogenesis. Proc. Natl. Acad. Sci. USA 2018, 115, E5289–E5297. [Google Scholar] [CrossRef] [Green Version]
- Montminy, M.R.; Gonzalez, G.A.; Yamamoto, K.K. Characteristics of the cAMP response unit. Metabolism 1990, 39, 6–12. [Google Scholar] [CrossRef]
- Meyer, M.B.; Benkusky, N.A.; Sen, B.; Rubin, J.; Pike, J.W. Epigenetic Plasticity Drives Adipogenic and Osteogenic Differentiation of Marrow-derived Mesenchymal Stem Cells. J. Biol. Chem. 2016, 291, 17829–17847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pike, J.W.; Lee, S.M.; Benkusky, N.A.; Meyer, M.B. Genomic Mechanisms Governing Mineral Homeostasis and the Regulation and Maintenance of Vitamin D Metabolism. JBMR Plus 2020, 5, e10433. [Google Scholar] [CrossRef]
- Van de Velde, S.; Wiater, E.; Tran, M.; Hwang, Y.; Cole, P.A.; Montminy, M. CREB Promotes Beta Cell Gene Expression by Targeting Its Coactivators to Tissue-Specific Enhancers. Mol. Cell. Biol. 2019, 39, e00200-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Kaufmann, M.; Lee, S.M.; Redfield, R.R.; Jones, G.; Pike, J.W. Targeted genomic deletions identify diverse enhancer functions and generate a kidney-specific, endocrine-deficient Cyp27b1 pseudo-null mouse. J. Biol. Chem. 2019, 294, 9518–9535. [Google Scholar] [CrossRef]
- Meyer, M.B.; Benkusky, N.A.; Pike, J.W. Selective Distal Enhancer Control of the Mmp13 Gene Identified through Clustered Regularly Interspaced Short Palindromic Repeat (CRISPR) Genomic Deletions. J. Biol. Chem. 2015, 290, 11093–11107. [Google Scholar] [CrossRef] [Green Version]
- Domcke, S.; Hill, A.J.; Daza, R.M.; Cao, J.; O’Day, D.R.; Pliner, H.A.; Aldinger, K.A.; Pokholok, D.; Zhang, F.; Milbank, J.H.; et al. A human cell atlas of fetal chromatin accessibility. Science 2020, 370. [Google Scholar] [CrossRef] [PubMed]
- Cao, J.; Cusanovich, D.A.; Ramani, V.; Aghamirzaie, D.; Pliner, H.A.; Hill, A.J.; Daza, R.M.; McFaline-Figueroa, J.L.; Packer, J.S.; Christiansen, L.; et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 2018, 361, 1380–1385. [Google Scholar] [CrossRef] [Green Version]
- Cusanovich, D.; Hill, A.J.; Aghamirzaie, D.; Daza, R.M.; Pliner, H.; Berletch, J.B.; Filippova, G.N.; Huang, X.; Christiansen, L.; DeWitt, W.S.; et al. A Single-Cell Atlas of In Vivo Mammalian Chromatin Accessibility. Cell 2018, 174, 1309–1324.e18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swarthout, J.T.; D’Alonzo, R.C.; Selvamurugan, N.; Partridge, N.C. Parathyroid hormone-dependent signaling pathways regulating genes in bone cells. Gene 2001, 282, 1–17. [Google Scholar] [CrossRef]
- St John, H.C.; Meyer, M.B.; Benkusky, N.A.; Carlson, A.H.; Prideaux, M.; Bonewald, L.F.; Wesley Pike, J. The parathyroid hormone-regulated transcriptome in osteocytes: Parallel actions with 1,25-dihydroxyvitamin D3 to oppose gene expression changes during differentiation and to promote mature cell function. Bone 2014, 72, 81–91. [Google Scholar] [CrossRef] [Green Version]
- Meyer, M.B.; Benkusky, N.A.; Pike, J.W. The RUNX2 cistrome in osteoblasts: Characterization, down-regulation following differentiation, and relationship to gene expression. J. Biol. Chem. 2014, 289, 16016–16031. [Google Scholar] [CrossRef] [Green Version]
- Galli, C.; Zella, L.A.; Fretz, J.A.; Fu, Q.; Pike, J.W.; Weinstein, R.S.; Manolagas, S.C.; O’Brien, C.A. Targeted deletion of a distant transcriptional enhancer of the receptor activator of nuclear factor-kappaB ligand gene reduces bone remodeling and increases bone mass. Endocrinology 2008, 149, 146–153. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Carlson, A.H.; Onal, M.; Benkusky, N.A.; Meyer, M.B.; Pike, J.W. A Control Region Near the Fibroblast Growth Factor 23 Gene Mediates Response to Phosphate, 1,25(OH)2D3, and LPS In Vivo. Endocrinology 2019, 160, 2877–2891. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.M.; Riley, E.M.; Meyer, M.B.; Benkusky, N.A.; Plum, L.A.; DeLuca, H.F.; Pike, J.W. 1,25-Dihydroxyvitamin D3 Controls a Cohort of Vitamin D Receptor Target Genes in the Proximal Intestine That Is Enriched for Calcium-regulating Components. J. Biol. Chem. 2015, 290, 18199–18215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ii, Z.K.C.; Bhasin, N.; Di Rienzi, S.C.; Rajan, A.; Deans-Fielder, K.; Swaminathan, G.; Kamyabi, N.; Zeng, X.-L.; Doddapaneni, H.; Menon, V.K.; et al. Drivers of Transcriptional Variance in Human Intestinal Epithelial Organoids. Physiol. Genom. 2021. [Google Scholar] [CrossRef]
- Christakos, S.; Li, S.; De La Cruz, J.; Shroyer, N.F.; Criss, Z.K.; Verzi, M.P.; Fleet, J.C. Vitamin D and the intestine: Review and update. J. Steroid Biochem. Mol. Biol. 2020, 196, 105501. [Google Scholar] [CrossRef] [PubMed]
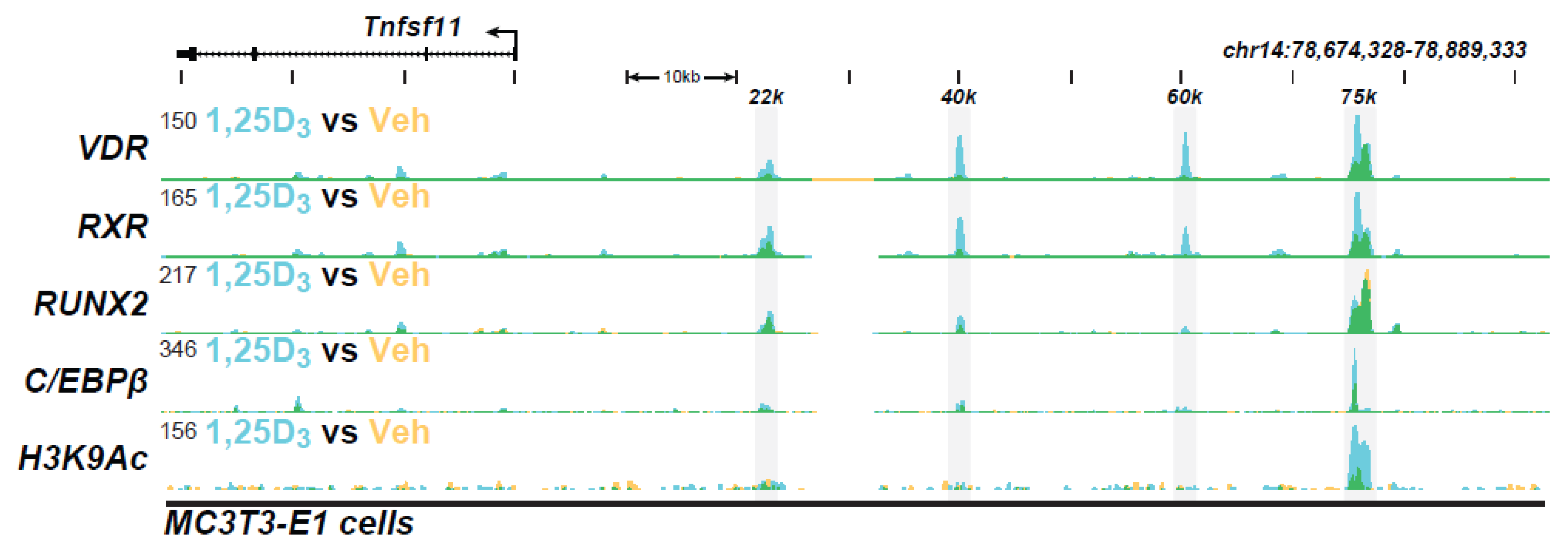
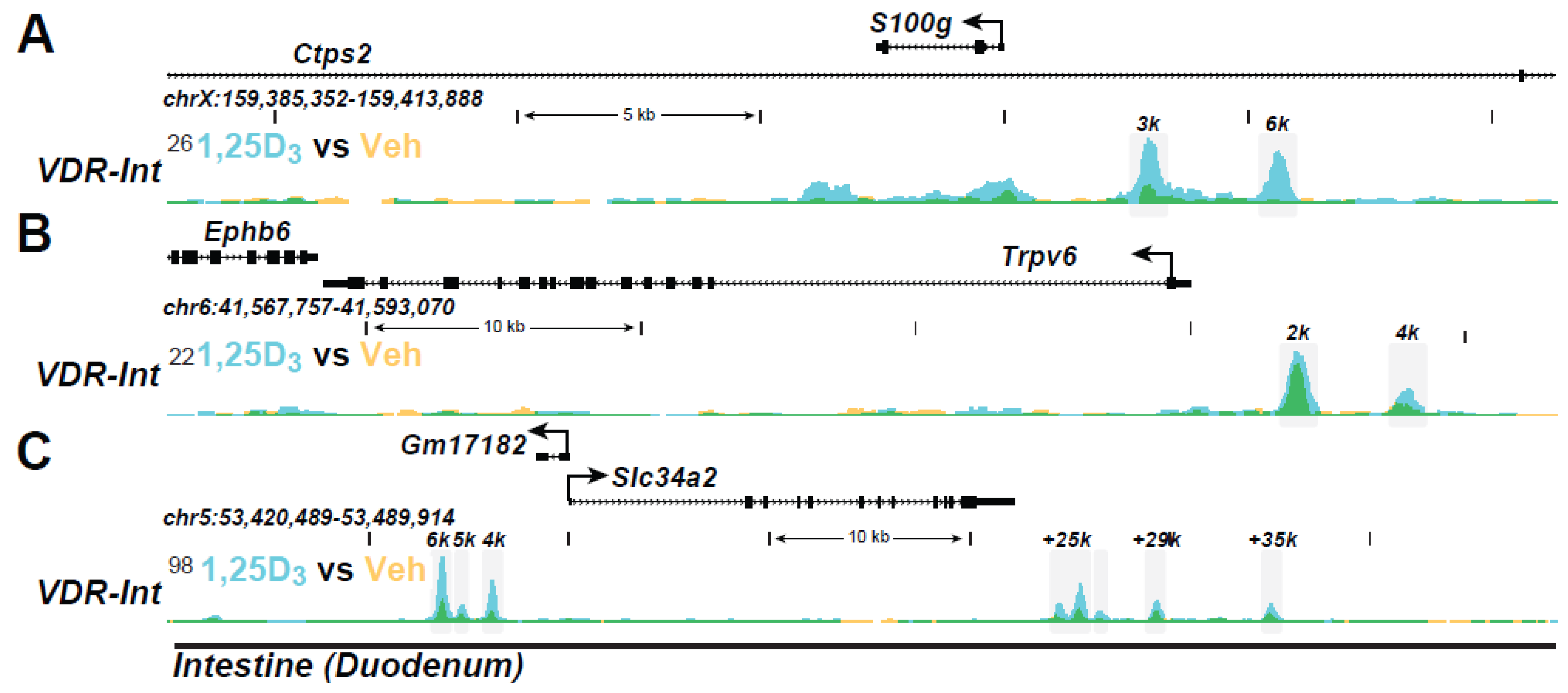
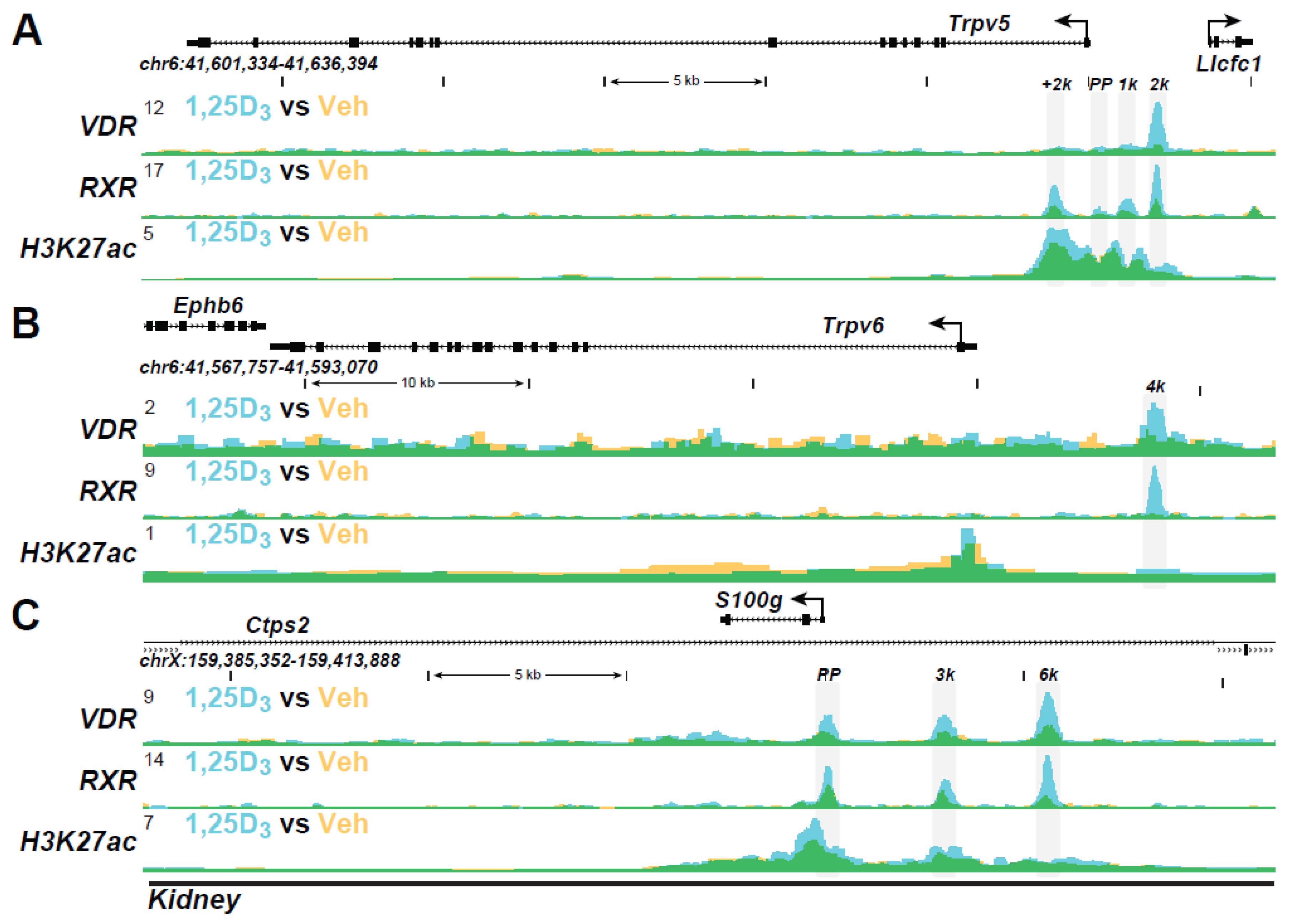
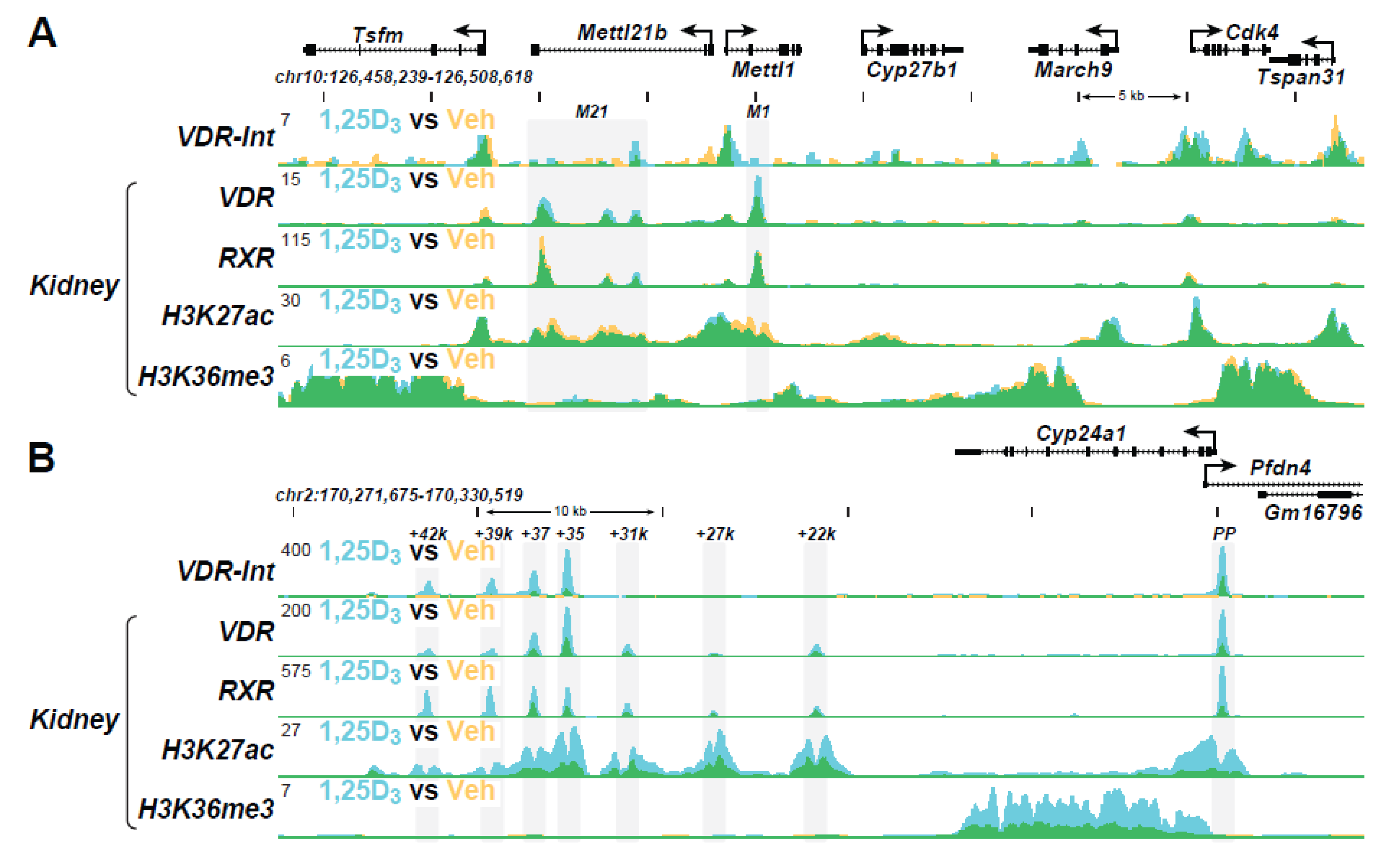
| Active Transcription Unit for Induction: The VDR/RXR heterodimer |
| VDR Binding Sites (The VDR Cistrome): 2000–8000 1,25(OH)2D3-sensitive binding sites/genome whose number and location are chromatin dependent and a function of cell-type |
| Mode of DNA Binding: Predominantly, but not exclusively, 1,25(OH)2D3-dependent |
| VDR/RXR Binding Site Sequence (VDRE): Induction mediated by classic hexameric half-sites (AGGTCA) separated by 3 base pairs; Repression mediated by divergent sites |
| Distal Binding Site Locations: Dispersed in cis-regulatory modules (CRMs or enhancers) across the genome; located in a cell-type specific manner near promoters, but predominantly within introns and distal intergenic regions; frequently located in clusters of elements |
| Epigenetic CRM Signatures: Defined by the dynamically regulated post-translational histone H3 and H4 modifications |
| Modular Features of CRMs: Contain binding sites for multiple transcription factors that facilitate either independent or synergistic interaction |
| VDR Cistromes: Dynamic alterations in the cellular epigenome during differentiation, maturation, and disease provoke changes to the VDR cistrome that qualitatively and quantitatively affect the vitamin D regulated transcriptome |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pike, J.W.; Meyer, M.B. New Approaches to Assess Mechanisms of Action of Selective Vitamin D Analogues. Int. J. Mol. Sci. 2021, 22, 12352. https://doi.org/10.3390/ijms222212352
Pike JW, Meyer MB. New Approaches to Assess Mechanisms of Action of Selective Vitamin D Analogues. International Journal of Molecular Sciences. 2021; 22(22):12352. https://doi.org/10.3390/ijms222212352
Chicago/Turabian StylePike, John Wesley, and Mark B. Meyer. 2021. "New Approaches to Assess Mechanisms of Action of Selective Vitamin D Analogues" International Journal of Molecular Sciences 22, no. 22: 12352. https://doi.org/10.3390/ijms222212352
APA StylePike, J. W., & Meyer, M. B. (2021). New Approaches to Assess Mechanisms of Action of Selective Vitamin D Analogues. International Journal of Molecular Sciences, 22(22), 12352. https://doi.org/10.3390/ijms222212352






