Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber
Abstract
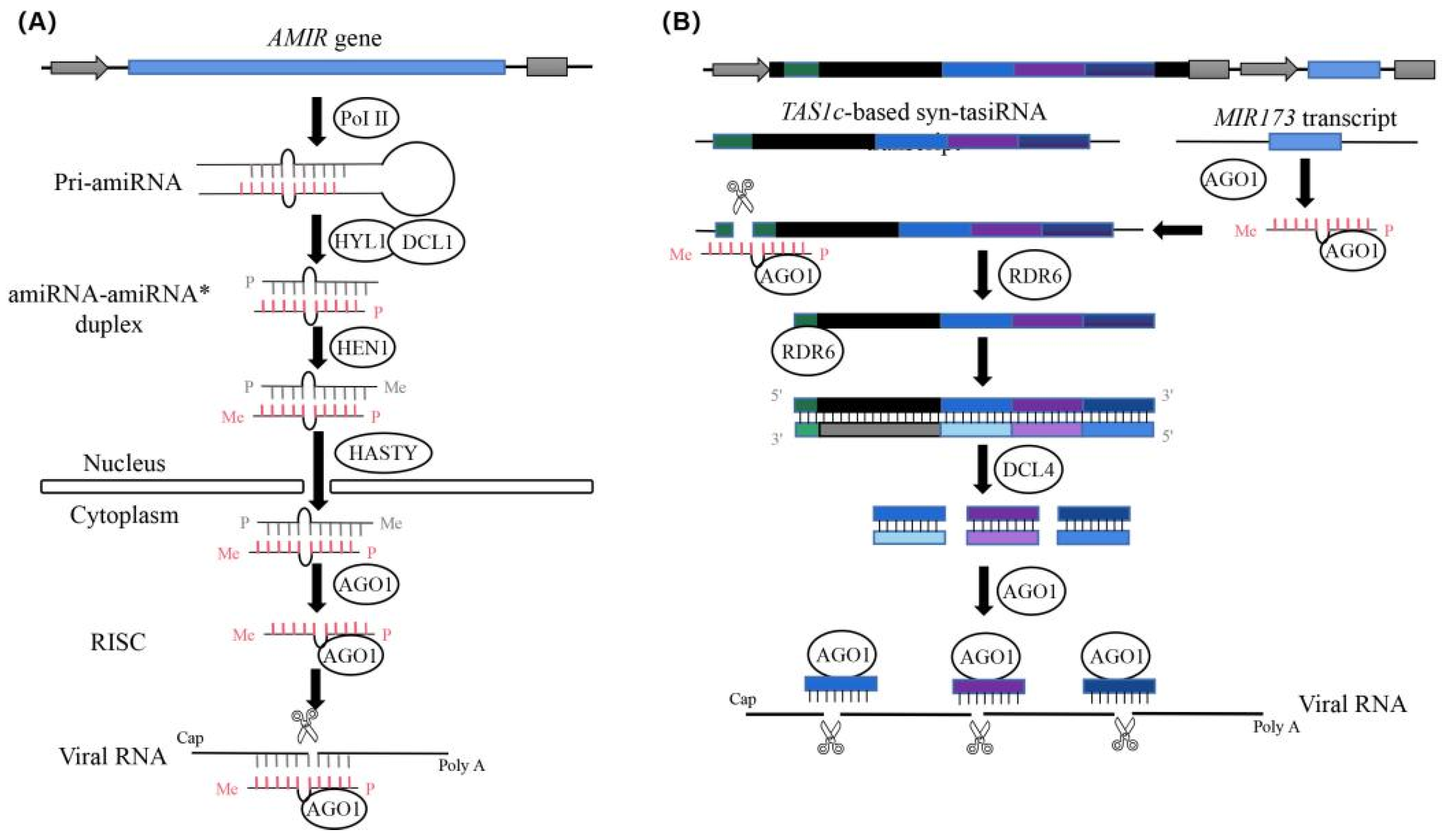
:1. Introduction
2. Results
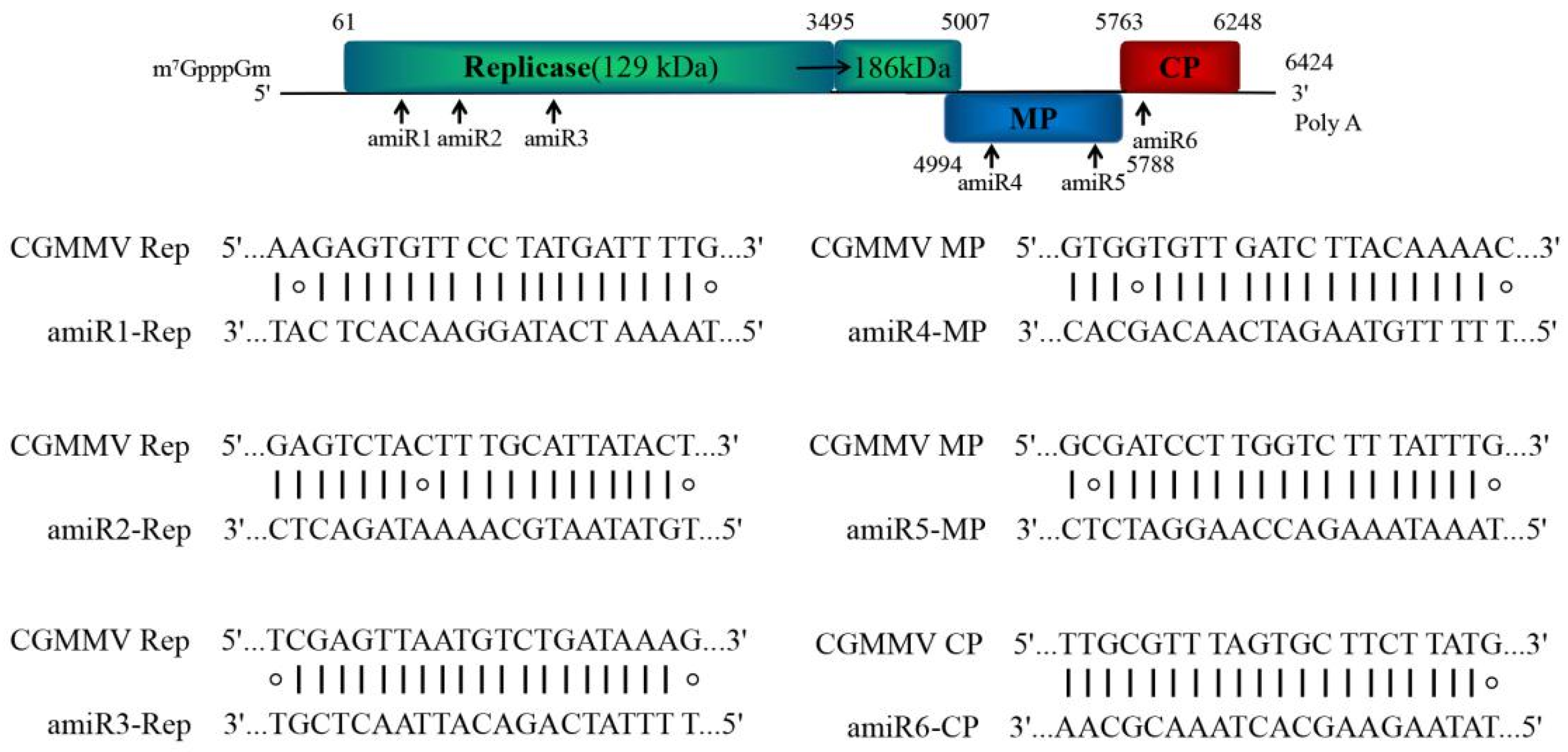
2.1. Design of amiRNAs against Multiple CGMMV Strains
2.2. Expression and Anti-CGMMV Activity of amiRNAs in Nicotiana Benthamiana
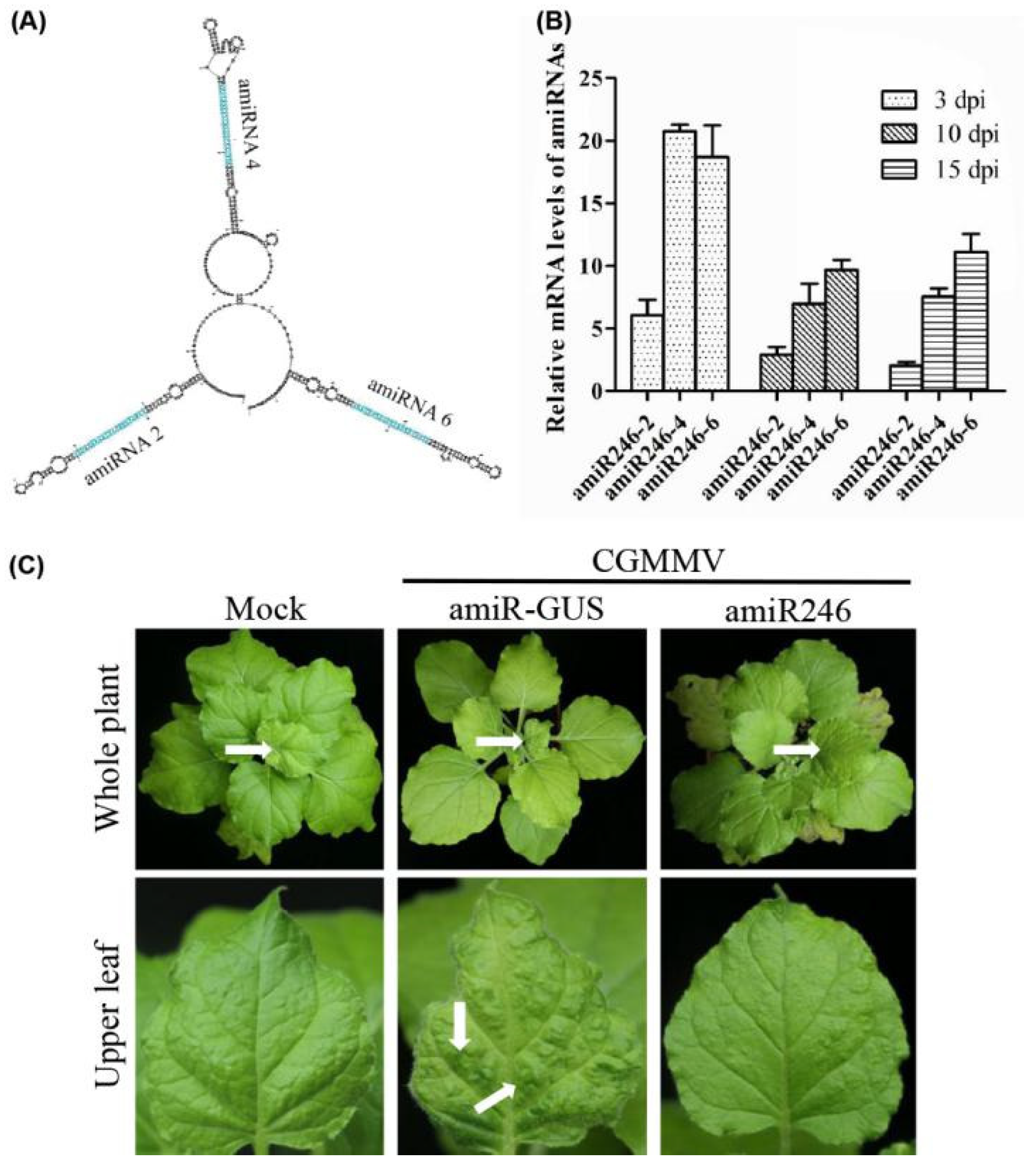
2.3. Identify the Most Effective amiRNA for Generating Polycistronic Constructs
2.4. Expression and Anti-CGMMV Activity of Polycistronic amiRNA Constructs in Protoplasts of Cucumber Infected with CGMMV
2.5. Evaluation of Transgenic Cucumber Lines’ Resistance Following CGMMV Infection
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Plasmid Construction
4.3. Agrobacterium Tumefaciens Infiltration and Viral Infection Assays
4.4. Vacuum Agroinfiltration and Co-Cultivation
4.5. Transient Expression in Cucumber Mesophyll Protoplasts
4.6. Generation of Transgenic Cucumber Plants
4.7. sRNA Gel Northern Blot Assays
4.8. qRT-PCR
4.9. Western Blot Assays
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wang, D.W.; Maule, A.J. A Model for Seed Transmission of a Plant Virus: Genetic and Structural Analyses of Pea Embryo Invasion by Pea Seed-Borne Mosaic Virus. Plant Cell 1994, 6, 777–787. [Google Scholar] [CrossRef] [PubMed]
- Aranda, M.A.; Escaler, M.; Wang, D.; Maule, A.J. Induction of HSP70 and polyubiquitin expression associated with plant virus replication. Proc. Natl. Acad. Sci. USA 1996, 93, 15289–15293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ugaki, M.; Tomiyama, M.; Kakutani, T.; Hidaka, S.; Kiguchi, T.; Nagata, R.; Sato, T.; Motoyoshi, F.; Nishiguchi, M. The complete nucleotide sequence of cucumber green mottle mosaic virus (SH strain) genomic RNA. J. Gen. Virol. 1991, 72, 1487–1495. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Y.A.; Wang, X.F.; Zhou, G.H. Molecular Characterization and Distribution of Cucumber green mottle mosaic virus in China. J. Phytopathol. 2009, 157, 393–399. [Google Scholar] [CrossRef]
- Dombrovsky, A.; Tran-Nguyen, L.T.; Jones, R.A. Cucumber green mottle mosaic virus: Rapidly Increasing Global Distribution, Etiology, Epidemiology, and Management. Annu. Rev. Phytopathol. 2017, 55, 231–256. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.W.; Luo, L.X.; Li, J.Q.; Liu, P.F.; Chen, X.Y.; Hao, J.J. Pollen and seed transmission of Cucumber green mottle mosaic virus in cucumber. Plant Pathol. 2014, 63, 72–77. [Google Scholar] [CrossRef]
- Reingold, V.; Lachman, O.; Blaosov, E.; Dombrovsky, A. Seed disinfection treatments do not sufficiently eliminate the infectivity of Cucumber green mottle mosaic virus (CGMMV) on cucurbit seeds. Plant Pathol. 2015, 64, 245–255. [Google Scholar] [CrossRef]
- Fletcher, J.T.; George, A.J.; Green, D.E. Cucumber Green Mottle Mosaic Virus, its Effect on Yield and its Control in the Lea Valley, England. Plant Pathol. 1969, 18, 16–22. [Google Scholar] [CrossRef]
- Reingold, V.; Lachman, O.; Koren, A.; Dombrovsky, A. First report of Cucumber green mottle mosaic virus (CGMMV) symptoms in watermelon used for the discrimination of non-marketable fruits in Israeli commercial fields. New Dis. Rep. 2013, 28, 11. [Google Scholar] [CrossRef] [Green Version]
- Sugiyama, M.; Ohara, T.; Sakata, Y. A New Source of Resistance to Cucumber Green Mottle Mosaic Virus in Melon. J. Jpn. Soc. Hortic. Sci. 2006, 75, 469–475. [Google Scholar] [CrossRef] [Green Version]
- Crespo, O.; Robles, C.; Ruiz, L.; Janssen, D. Antagonism of Cucumber green mottle mosaic virus against Tomato leaf curl New Delhi virus in zucchini and cucumber. Ann. Appl. Biol. 2020, 176, 147–157. [Google Scholar] [CrossRef]
- Baulcombe, D.C. RNA silencing in plants. Nat. Cell Biol. 2004, 431, 356–363. [Google Scholar] [CrossRef]
- Cillo, F.; Palukaitis, P. Transgenic resistance. Adv. Virus Res. 2014, 90, 35–146. [Google Scholar] [PubMed]
- Burch-Smith, T.; Anderson, J.C.; Martin, G.B.; Dinesh-Kumar, S.P. Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 2004, 39, 734–746. [Google Scholar] [CrossRef]
- Xu, P.; Zhang, Y.J.; Kang, L.; Roossinck, M.J.; Mysore, K.S. Computational estimation and experimental verification of off-target silencing during post transcriptional gene silencing in plants. Plant Physiol. 2006, 142, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Niu, Q.-W.; Lin, S.-S.; Reyes, J.L.; Chen, K.-C.; Wu, H.-W.; Yeh, S.-D.; Chua, N.-H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef]
- Butardo, V.; Fitzgerald, M.; Bird, A.; Gidley, M.J.; Flanagan, B.M.; Larroque, O.; Resurreccion, A.P.; Laidlaw, H.K.C.; Jobling, S.; Morell, M.K.; et al. Impact of down-regulation of starch branching enzyme IIb in rice by artificial microRNA- and hairpin RNA-mediated RNA silencing. J. Exp. Bot. 2011, 62, 4927–4941. [Google Scholar] [CrossRef] [Green Version]
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.; Su, Y.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 2014, 14, 62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jones-Rhoades, M.W.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef]
- Carbonell, A.; Lisón, P.; Daròs, J. Multi-targeting of viral RNAs with synthetic trans -acting small interfering RNAs enhances plant antiviral resistance. Plant J. 2019, 100, 720–737. [Google Scholar] [CrossRef] [Green Version]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly Specific Gene Silencing by Artificial MicroRNAs inArabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef] [Green Version]
- Khraiwesh, B.; Ossowski, S.; Weigel, D.; Reski, R.; Frank, W. Specific Gene Silencing by Artificial MicroRNAs in Physcomitrella patens: An Alternative to Targeted Gene Knockouts. Plant Physiol. 2008, 148, 684–693. [Google Scholar] [CrossRef] [Green Version]
- Kis, A.; Tholt, G.; Ivanics, M.; Varallyay, E.; Jenes, B.; Havelda, Z. Polycistronic artificial miRNA-mediated resistance to Wheat dwarf virus in barley is highly efficient at low temperature. Mol. Plant Pathol. 2016, 17, 427–437. [Google Scholar] [CrossRef] [PubMed]
- Vargason, J.M.; Szittya, G.; Burgyán, J.; Hall, T.M. Size Selective Recognition of siRNA by an RNA Silencing Suppressor. Cell 2003, 115, 799–811. [Google Scholar] [CrossRef] [Green Version]
- González, I.; Rakitina, D.; Semashko, M.; Taliansky, M.; Praveen, S.; Palukaitis, P.; Carr, J.; Kalinina, N.; Canto, T.; Semashko, M. RNA binding is more critical to the suppression of silencing function of Cucumber mosaic virus 2b protein than nuclear localization. RNA 2012, 18, 771–782. [Google Scholar] [CrossRef] [Green Version]
- Hamera, S.; Song, X.; Su, L.; Chen, X.; Fang, R. Cucumber mosaic virus suppressor 2b binds to AGO4-related small RNAs and impairs AGO4 activities. Plant J. 2012, 69, 104–115. [Google Scholar] [CrossRef] [PubMed]
- Deleris, A.; Gallego-Bartolome, J.; Bao, J.; Kasschau, K.D.; Carrington, J.C.; Voinnet, O. Hierarchical Action and Inhibition of Plant Dicer-Like Proteins in Antiviral Defense. Science 2006, 313, 68–71. [Google Scholar] [CrossRef]
- Chen, H.; Ino, M.; Shimono, M.; Wagh, S.G.; Kobayashi, K.; Yaeno, T.; Yamaoka, N.; Bai, G.; Nishiguchi, M. A Single Amino Acid Substitution in the Intervening Region of 129K Protein of Cucumber Green Mottle Mosaic Virus Resulted in Attenuated Symptoms. Phytopathology 2020, 110, 146–152. [Google Scholar] [CrossRef]
- Li, J.-F.; Chung, H.S.; Niu, Y.; Bush, J.; McCormack, M.; Sheen, J. Comprehensive Protein-Based Artificial MicroRNA Screens for Effective Gene Silencing in Plants. Plant Cell 2013, 25, 1507–1522. [Google Scholar] [CrossRef] [Green Version]
- Liang, C.Q.; Hao, J.J.; Li, J.Q.; Baker, B.; Luo, L.X. Artificial microRNA-mediated resistance to cucumber green mottle mosaic virus in Nicotiana benthamiana. Planta 2019, 250, 1591–1601. [Google Scholar] [CrossRef]
- Merchan, F.; Boualem, A.; Crespi, M.; Frugier, F. Plant polycistronic precursors containing non-homologous microRNAs target transcripts encoding functionally related proteins. Genome Biol. 2009, 10, R136. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yu, D.; Zhang, Y.; Liu, K.; Xu, K.; Zhang, F.; Wang, J.; Tan, G.; Nie, X.; Ji, Q.; et al. Vacuum and Co-cultivation Agroinfiltration of (Germinated) Seeds Results in Tobacco Rattle Virus (TRV) Mediated Whole-Plant Virus-Induced Gene Silencing (VIGS) in Wheat and Maize. Front. Plant Sci. 2017, 8, 393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Sun, J.; Li, S.; Cui, Q.; Zhang, H.; Xin, F.; Wang, H.; Lin, T.; Gao, D.; Wang, S.; et al. An ACC Oxidase Gene Essential for Cucumber Carpel Development. Mol. Plant 2016, 9, 1315–1327. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.; Sui, X.; Guo, J.; Wang, Z.; Cheng, J.; Ma, S.; Li, X.; Zhang, Z. Antisense suppression of cucumber (Cucumis sativus L.) sucrose synthase 3 (CsSUS3) reduces hypoxic stress tolerance. Plant Cell Environ. 2013, 37, 795–810. [Google Scholar] [CrossRef]
- Qu, J.; Ye, J.; Fang, R.X. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [Green Version]
- Kung, Y.-J.; Lin, S.-S.; Huang, Y.-L.; Chen, T.-C.; Harish, S.S.; Chua, N.-H.; Yeh, S.-D. Multiple artificial microRNAs targeting conserved motifs of the replicase gene confer robust transgenic resistance to negative-sense single-stranded RNA plant virus. Mol. Plant Pathol. 2012, 13, 303–317. [Google Scholar] [CrossRef]
- Duan, C.-G.; Wang, C.-H.; Fang, R.-X.; Guo, H.-S. Artificial MicroRNAs Highly Accessible to Targets Confer Efficient Virus Resistance in Plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef] [Green Version]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA Translation and Stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [Green Version]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef]
- Zhang, N.; Zhang, D.; Chen, S.L.; Gong, B.-Q.; Guo, Y.; Xu, L.; Zhang, X.-N.; Li, J.-F. Engineering Artificial MicroRNAs for Multiplex Gene Silencing and Simplified Transgenic Screen. Plant Physiol. 2018, 178, 989–1001. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yuan, Y.-R.; Pei, Y.; Lin, S.-S.; Tuschl, T.; Patel, D.J.; Chua, N.-H. Cucumber mosaic virus-encoded 2b suppressor inhibits Arabidopsis Argonaute1 cleavage activity to counter plant defense. Genes Dev. 2006, 20, 3255–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schott, G.; Marí-Ordóñez, A.; Himber, C.; Alioua, A.; Voinnet, O.; Dunoyer, P. Differential effects of viral silencing suppressors on siRNA and miRNA loading support the existence of two distinct cellular pools of ARGONAUTE1. EMBO J. 2012, 31, 2553–2565. [Google Scholar] [CrossRef] [Green Version]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef] [PubMed]
- Simón-Mateo, C.; García, J.A. MicroRNA-Guided Processing Impairs Plum Pox Virus Replication, but the Virus Readily Evolves to Escape This Silencing Mechanism. J. Virol. 2006, 80, 2429–2436. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, S.S.; Wu, H.W.; Elena, S.F.; Chen, K.C.; Niu, Q.W.; Yeh, S.D.; Chen, C.C.; Chua, N.H. Molecular evolution of a viral non-coding sequence under the selective pressure of amiRNA-mediated silencing. PLoS Pathog. 2009, 5, e1000312. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.Z.; Amin, I.; Hameed, A.; Mansoor, S. CRISPR–Cas13a: Prospects for Plant Virus Resistance. Trends Biotechnol. 2018, 36, 1207–1210. [Google Scholar] [CrossRef]
- Liu, Q.; Li, Y.; Xu, K.; Li, D.; Hu, H.; Zhou, F.; Song, P.; Yu, Y.; Wei, Q.; Liu, Q.; et al. Clay nanosheet-mediated delivery of recombinant plasmids expressing artificial miRNAs via leaf spray to prevent infection by plant DNA viruses. Hortic. Res. 2020, 7, 1–12. [Google Scholar] [CrossRef]
- Yoo, S.-D.; Cho, Y.-H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [Green Version]
- Li, F.; Pignatta, D.; Bendix, C.; Brunkard, J.; Cohn, M.M.; Tung, J.; Sun, H.; Kumar, P.; Baker, B. MicroRNA regulation of plant innate immune receptors. Proc. Natl. Acad. Sci. USA 2012, 109, 1790–1795. [Google Scholar] [CrossRef] [Green Version]




Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Miao, S.; Liang, C.; Li, J.; Baker, B.; Luo, L. Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber. Int. J. Mol. Sci. 2021, 22, 12237. https://doi.org/10.3390/ijms222212237
Miao S, Liang C, Li J, Baker B, Luo L. Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber. International Journal of Molecular Sciences. 2021; 22(22):12237. https://doi.org/10.3390/ijms222212237
Chicago/Turabian StyleMiao, Shuo, Chaoqiong Liang, Jianqiang Li, Barbara Baker, and Laixin Luo. 2021. "Polycistronic Artificial microRNA-Mediated Resistance to Cucumber Green Mottle Mosaic Virus in Cucumber" International Journal of Molecular Sciences 22, no. 22: 12237. https://doi.org/10.3390/ijms222212237






