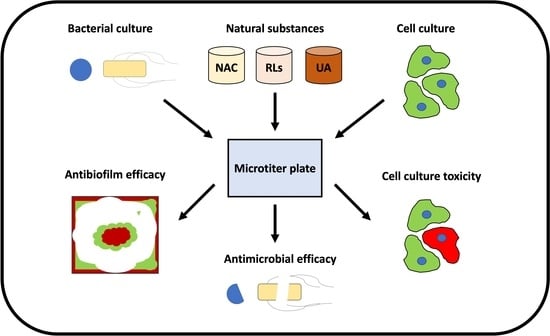
Evaluation of the Antimicrobial Efficacy of N-Acetyl-l-Cysteine, Rhamnolipids, and Usnic Acid—Novel Approaches to Fight Food-Borne Pathogens
Abstract
:1. Introduction
2. Results
2.1. The Effect of N-Acetyl-l-Cysteine
2.2. The Effect of Rhamnolipids
2.3. The Effect of Usnic Acid
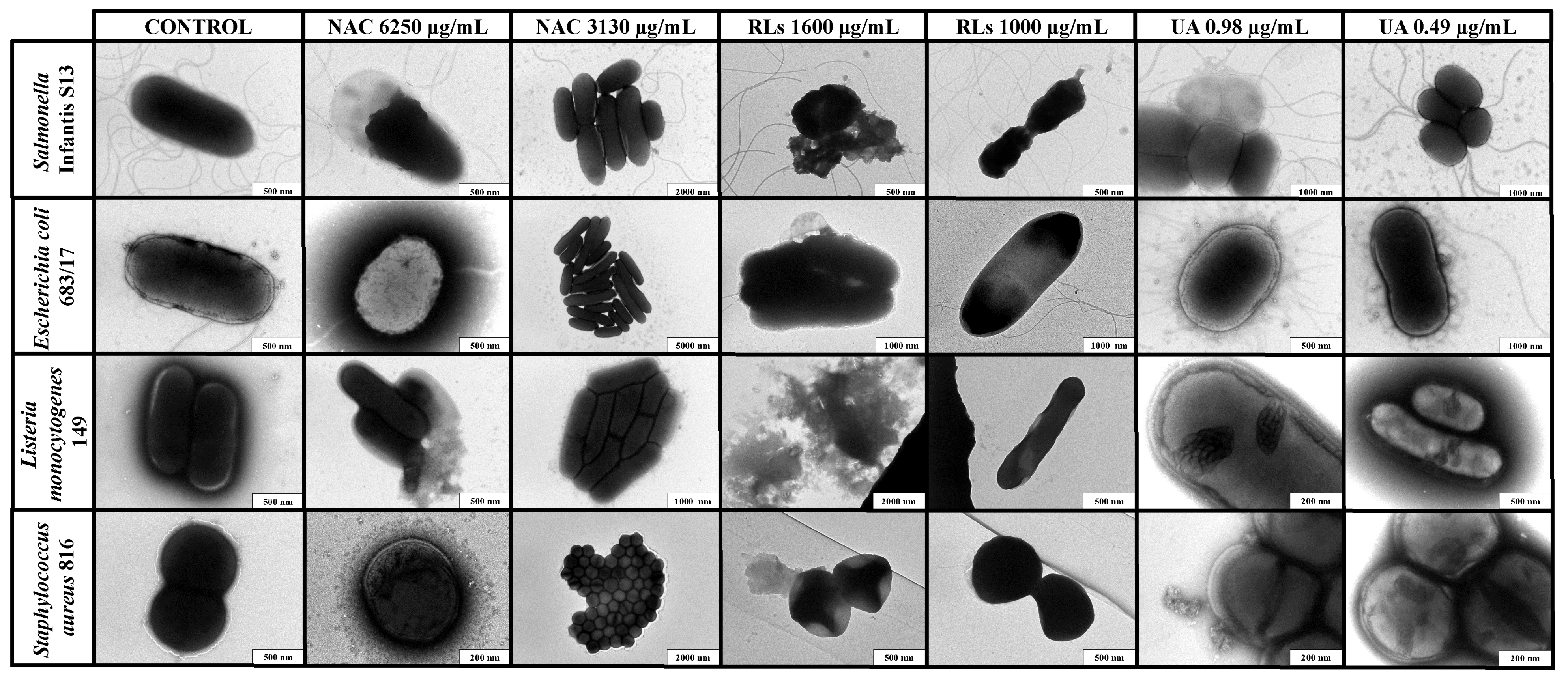
2.4. Transmission Electron Microscopy Imaging
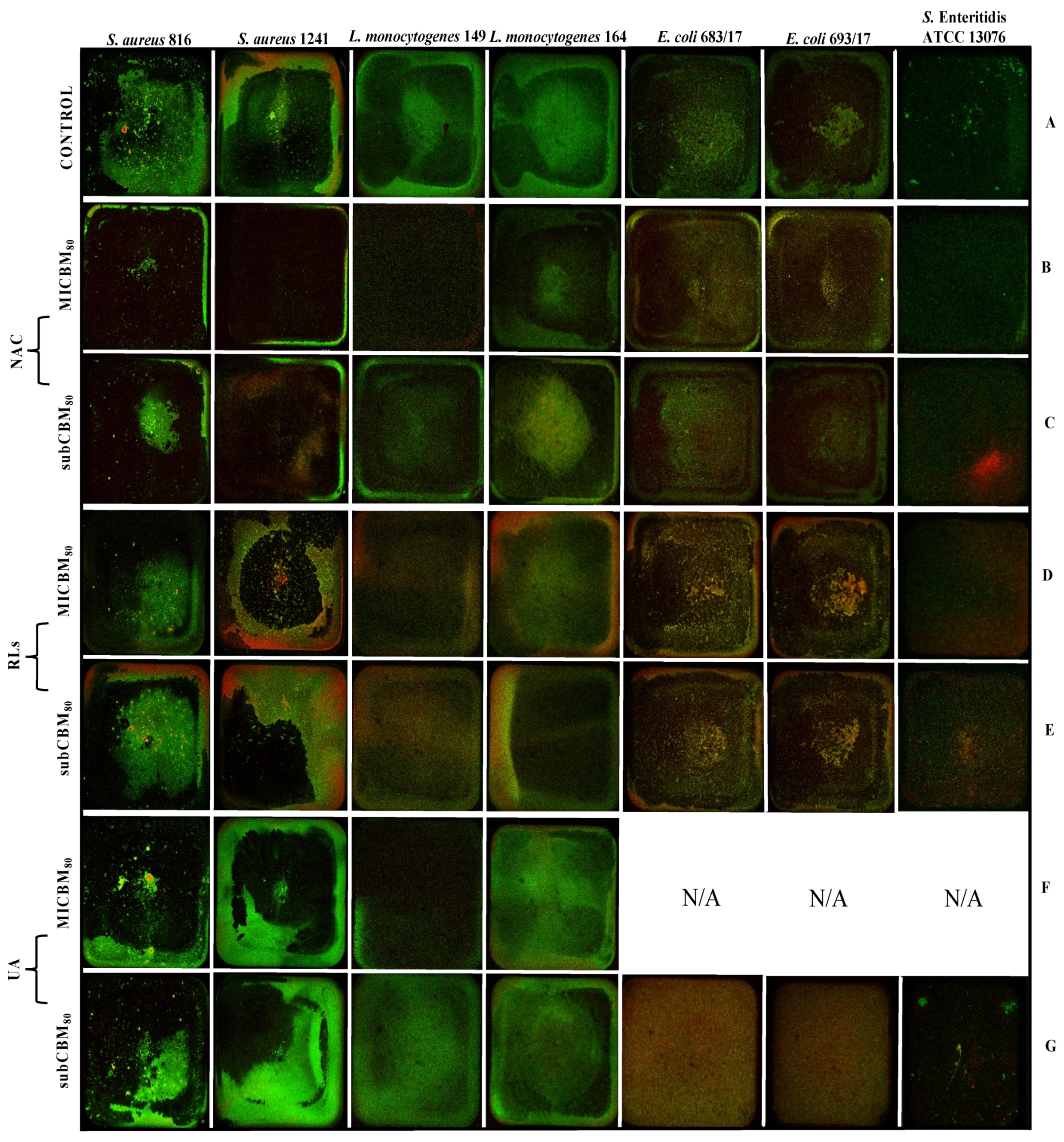
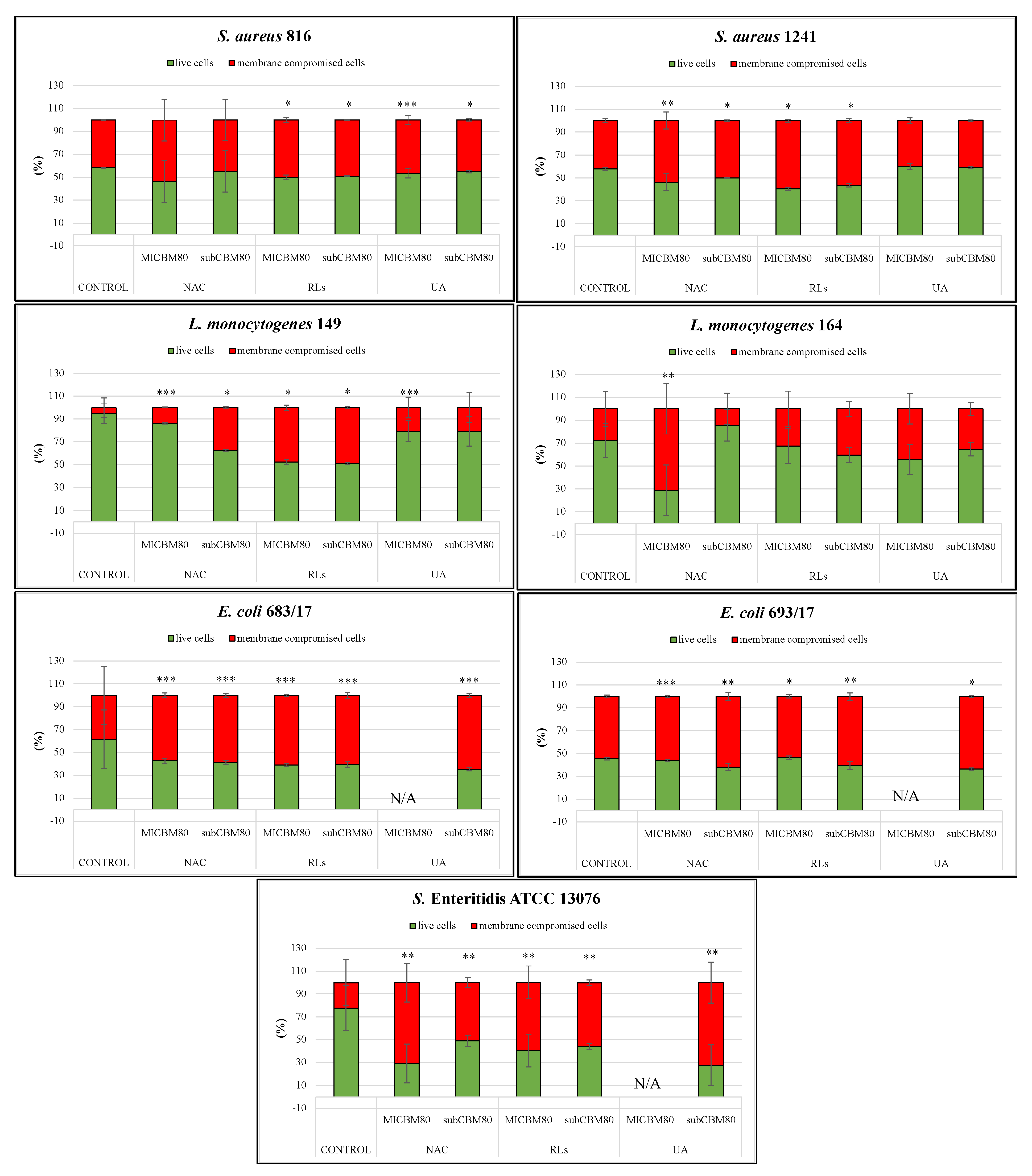
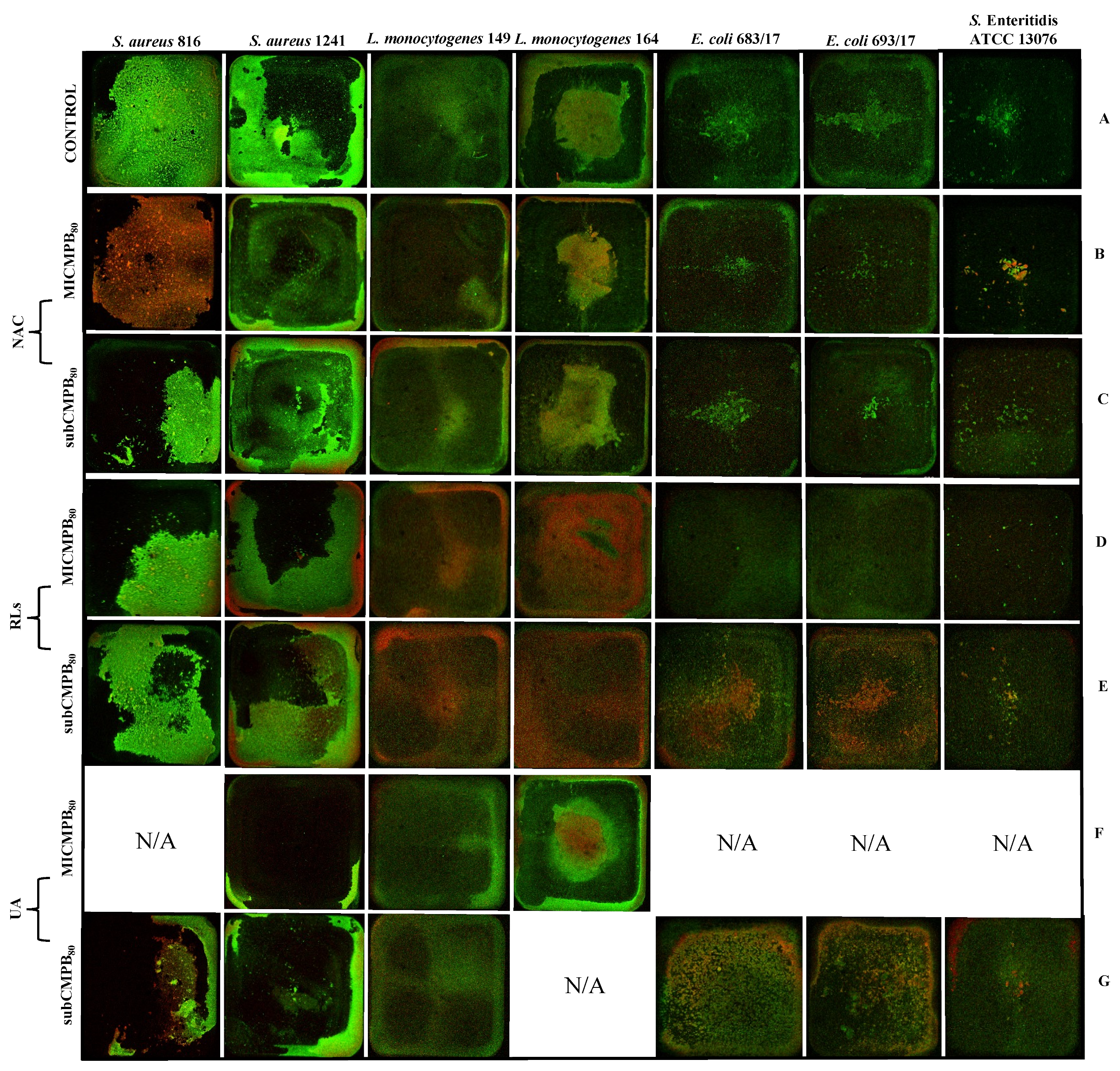
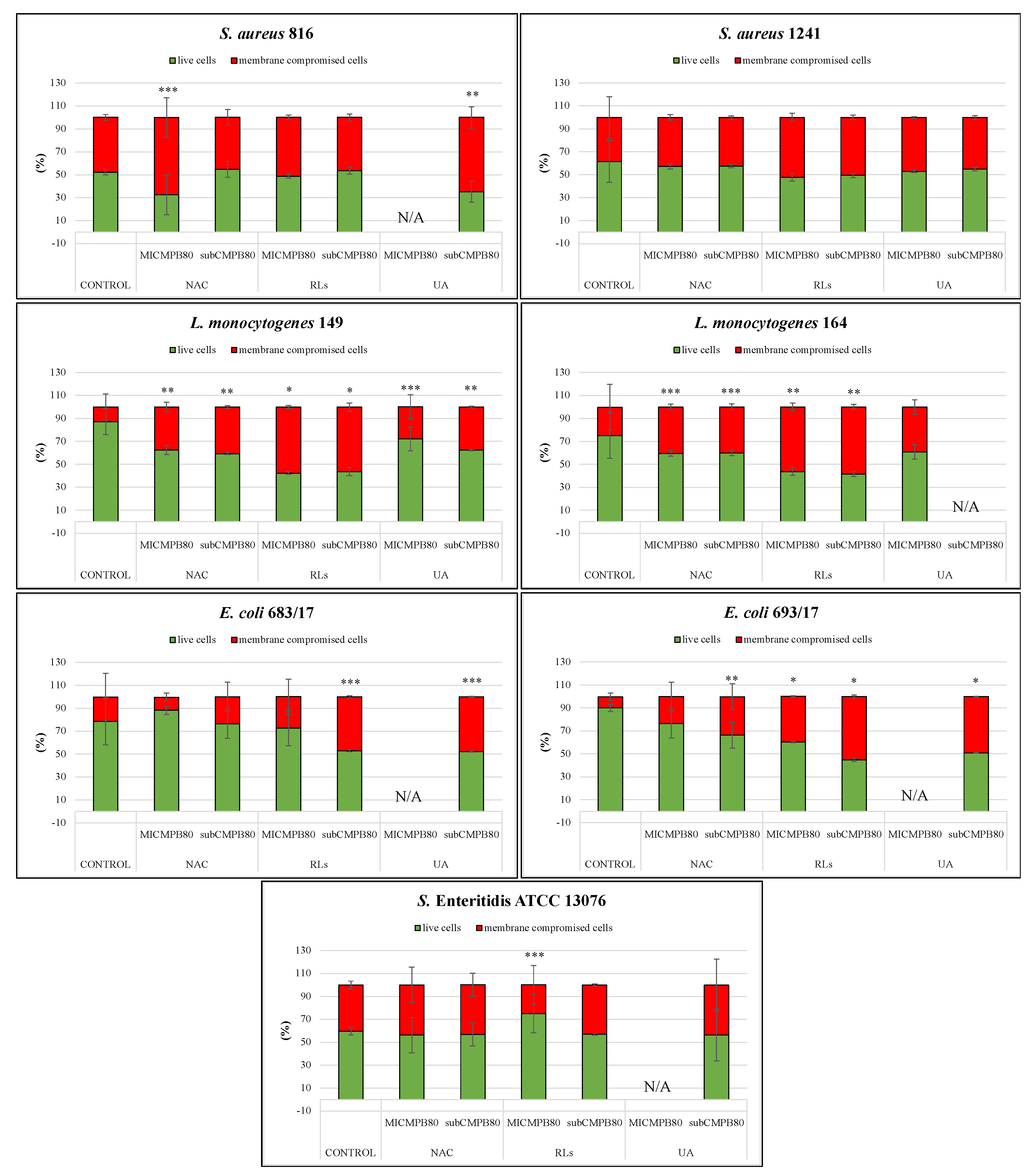
2.5. Confocal Scanning Laser Microscopy Analysis
2.5.1. Biofilm Prevention
2.5.2. Biofilm Removal
2.6. Acute Cytotoxicity of Individual Natural Substances
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Bacterial Strains
4.3. Inoculum Preparation
4.4. Preparation of Natural Substances Stock Solutions and Dilution Series
4.5. Determination of Minimum Inhibitory Concentrations
4.6. Transmission Electron Microscopy Imaging
4.7. Confocal Laser Scanning Microscopy Analysis
4.7.1. Strains and Growth Conditions
4.7.2. CLSM Imaging and Image Processing Analysis
4.8. Cytotoxicity Assay
4.9. Statistical Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- AL-Mamun, M.; Chowdhury, T.; Biswas, B.; Absar, N. Food poisoning and intoxication: A global leading concern for human health. Food Saf. Preserv. 2018, 307–352. [Google Scholar] [CrossRef]
- Bintsis, T. Foodborne pathogens. AIMS Microbiol. 2017, 3, 529–563. [Google Scholar] [CrossRef]
- Schirone, M.; Visciano, P.; Tofalo, R.; Suzzi, G. Editorial: Foodborne pathogens: Hygiene and safety. Front. Microbiol. 2019, 10, 1974. [Google Scholar] [CrossRef] [Green Version]
- Bogdanovicova, K.; Kamenik, J.; Dorotikova, K.; Strejcek, J.; Krepelova, S.; Duskova, M.; Harustiakova, D. Occurrence of foodborne agents at food service facilities in the Czech Republic. J. Food Protect. 2019, 82, 1096–1103. [Google Scholar] [CrossRef]
- Galié, S.; García-Gutiérrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food Industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 9. [Google Scholar] [CrossRef]
- Marriott, N.G.; Schilling, M.W.; Gravani, R.B. Principles of Food Sanitation; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef] [Green Version]
- Donaghy, J.A.; Jagadeesan, B.; Goodburn, K.; Grunwald, L.; Jensen, O.N.; Jespers, A.; Kanagachandran, K.; Lafforgue, H.; Seefelder, W.; Quentin, M.-C. Relationship of sanitizers, disinfectants, and cleaning agents with antimicrobial resistance. J. Food Protect. 2019, 82, 889–902. [Google Scholar] [CrossRef]
- Nahar, S.; Mizan, M.F.R.; Ha, A.J.; Ha, S.-D. Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1484–1502. [Google Scholar] [CrossRef] [Green Version]
- Flemming, H.-C.; Wingender, J.; Szewzyk, U.; Steinberg, P.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial biofilms in the food industry—A comprehensive review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef]
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96. [Google Scholar] [CrossRef]
- Founou, L.L.; Founou, R.C.; Essack, S.Y. Antibiotic resistance in the food chain: A developing country-perspective. Front. Microbiol. 2016, 7, 1881. [Google Scholar] [CrossRef]
- Sharma, D.; Misba, L.; Khan, A.U. Antibiotics versus biofilm: An emerging battleground in microbial communities. Antimicrob. Resist. Infect. Control 2019, 8, 1–10. [Google Scholar] [CrossRef]
- Costa, F.; Sousa, D.M.; Parreira, P.; Lamghari, M.; Gomes, P.; Martins, M.C.L. N-acetylcysteine-functionalized coating avoids bacterial adhesion and biofilm formation. Sci. Rep. 2017, 7, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Kregiel, D.; Rygala, A.; Kolesinska, B.; Nowacka, M.; Herc, A.S.; Kowalewska, A. Antimicrobial and antibiofilm N-Acetyl-l-cysteine grafted siloxane polymers with potential for use in water systems. Int. J. Mol. Sci. 2019, 20, 2011. [Google Scholar] [CrossRef] [Green Version]
- Domenech, M.; García, E. N-Acetyl-l-cysteine and cysteamine: New strategies against mixed biofilms of non-encapsulated Streptococcus pneumoniae and non-typeable Haemophilus influenzae. Antimicrob. Agents Chemother. 2017, 61, e01992-16. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Kim, J.; Wu, J.; Ahamed, A.I.; Wang, Y.; Martins-Green, M. N-acetyl-cysteine and mechanisms involved in resolution of chronic wound biofilm. J. Diabetes Res. 2020, 2020, 9589507. [Google Scholar] [CrossRef] [Green Version]
- Blasi, F.; Page, C.; Rossolini, G.M.; Pallecchi, L.; Matera, M.G.; Rogliani, P.; Cazzola, M. The effect of N -acetylcysteine on biofilms: Implications for the treatment of respiratory tract infections. Respir. Med. 2016, 117, 190–197. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Liu, K.; Wang, N.; Zhang, H. N-acetylcysteine induces apoptosis via the mitochondria-dependent pathway but not via endoplasmic reticulum stress in H9c2 cells. Mol. Med. Rep. 2017, 16, 6626–6633. [Google Scholar] [CrossRef]
- Nakagawa, Y.; Suzuki, T.; Nakajima, K.; Inomata, A.; Ogata, A.; Nakae, D. Effects of N-Acetyl-l-cysteine on target sites of hydroxylated fullerene-induced cytotoxicity in isolated rat hepatocytes. Arch. Toxicol. 2014, 88, 115–126. [Google Scholar] [CrossRef]
- Liu, G.; Zhong, H.; Yang, X.; Liu, Y.; Shao, B.; Liu, Z. Advances in applications of rhamnolipids biosurfactant in environmental remediation: A review. Biotechnol. Bioeng. 2018, 115, 796–814. [Google Scholar] [CrossRef]
- Soberón-Chávez, G.; González-Valdez, A.; Soto-Aceves, M.P.; Cocotl-Yañez, M. Rhamnolipids produced by Pseudomonas: From molecular genetics to the market. Microb. Biotechnol. 2020, 14, 136–146. [Google Scholar] [CrossRef]
- Aleksic, I.; Petkovic, M.; Jovanovic, M.; Milivojevic, D.; Vasiljevic, B.; Nikodinovic-Runic, J.; Senerovic, L. Anti-biofilm properties of bacterial di-rhamnolipids and their semi-synthetic amide derivatives. Front. Microbiol. 2017, 8, 2454. [Google Scholar] [CrossRef]
- Sun, L.; Forauer, E.C.; Brown, S.R.B.; D’Amico, D.J. Application of bioactive glycolipids to control Listeria monocytogenes biofilms and as post-lethality contaminants in milk and cheese. Food Microbiol. 2020, 95, 103683. [Google Scholar] [CrossRef]
- Chong, H.; Li, Q. Microbial production of rhamnolipids: Opportunities, challenges and strategies. Microb. Cell Fact. 2017, 16, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Xu, N.; Liu, S.; Xu, L.; Zhou, J.; Xin, F.; Zhang, W.; Qian, X.; Li, M.; Dong, W.; Jiang, M. Enhanced rhamnolipids production using a novel bioreactor system based on integrated foam-control and repeated fed-batch fermentation strategy. Biotechnol. Biofuels 2020, 13, 1–10. [Google Scholar] [CrossRef]
- Bakkar, M.R.; Faraag, A.H.I.; Soliman, E.R.S.; Fouda, M.S.; Sarguos, A.M.M.; McLean, G.R.; Hebishy, A.M.S.; Elkhouly, G.E.; Raya, N.R.; Abo-zeid, Y. Rhamnolipids nano-micelles as a potential hand sanitizer. Antibiotics 2021, 10, 751. [Google Scholar] [CrossRef]
- Francolini, I.; Piozzi, A.; Donelli, G. Usnic Acid: Potential role in management of wound infections. Adv. Exp. Med. Biol. 2019, 1214, 31–41. [Google Scholar] [CrossRef]
- Ahmad, R.; Borowiec, P.; Falck-Ytter, A.B.; Strætkvern, K.O. Extraction, solubility and antimicrobial activity of (-) usnic acid in ethanol, a pharmaceutically relevant solvent. Nat. Prod. Commun. 2017, 12, 1934578X1701200. [Google Scholar] [CrossRef] [Green Version]
- Maciąg-Dorszyńska, M.; Węgrzyn, G.; Guzow-Krzemińska, B. Antibacterial activity of lichen secondary metabolite usnic acid is primarily caused by inhibition of RNA and DNA synthesis. FEMS Microbiol. Lett. 2014, 353, 57–62. [Google Scholar] [CrossRef]
- Kartsev, V.; Geronikaki, A.; Petrou, A.; Lichitsky, B.; Smiljkovic, M.; Kostic, M.; Radanovic, O.; Soković, M. Design, synthesis and antimicrobial activity of usnic acid derivatives. Med. Chem. Comm. 2018, 9, 870–882. [Google Scholar] [CrossRef] [Green Version]
- Kaskatepe, B.; Yildiz, S. Rhamnolipid biosurfactants produced by Pseudomonas species. Braz. Arch. Biol. Technol. 2016, 59. [Google Scholar] [CrossRef] [Green Version]
- Culikova, H. Effect of Usnic Acid on Biofilm Formation. Bachelor’s Thesis, University of Chemistry and Technology, Prague, Czech Republic, 2015. [Google Scholar]
- Thi Nguyen, H.G.; Vinh Nguyen, N. Synthesis, structure elucidation and cytotoxicity of (+)-usnic acid derivatives on U87MG glioblastoma cells. Nat. Prod. Chem. Res. 2016, 4, 2. [Google Scholar] [CrossRef]
- Galvao, J.; Davis, B.; Tilley, M.; Normando, E.; Duchen, M.R.; Cordeiro, M.F. Unexpected low-dose toxicity of the universal solvent DMSO. FASEB J. 2014, 28, 1317–1330. [Google Scholar] [CrossRef]
- Ferrer, M.D.; Rodriguez, J.C.; Álvarez, L.; Artacho, A.; Royo, G.; Mira, A. Effect of antibiotics on biofilm inhibition and induction measured by real-time cell analysis. J. Appl. Microbiol. 2017, 122, 640–650. [Google Scholar] [CrossRef]
- Chlumsky, O.; Purkrtova, S.; Michova, H.; Sykorova, H.; Slepicka, P.; Fajstavr, D.; Ulbrich, P.; Viktorova, J.; Demnerova, K. Antimicrobial Properties of Palladium and Platinum Nanoparticles: A New Tool for Combating Food-Borne Pathogens. Int. J. Mol. Sci. 2021, 22, 7892. [Google Scholar] [CrossRef]
- Chlumsky, O.; Purkrtova, S.; Michova, T.H.; Svarcova, F.V.; Slepicka, P.; Fajstavr, D.; Ulbrich, P.; Demnerova, K. The effect of gold and silver nanoparticles, chitosan and their combinations on bacterial biofilms of food-borne pathogens. Biofouling 2020, 36, 222–233. [Google Scholar] [CrossRef] [PubMed]
- Drago, L.; Agrappi, S.; Bortolin, M.; Toscano, M.; Romanò, C.; De Vecchi, E. How to study biofilms after microbial colonization of materials used in orthopaedic implants. Int. J. Mol. Sci. 2016, 17, 293. [Google Scholar] [CrossRef] [PubMed]
- Tran, V.N.; Viktorova, J.; Augustynkova, K.; Jelenova, N.; Dobiasova, S.; Rehorova, K.; Stranska-Zachariasova, M.; Vitek, L.; Hajslova, J.; Ruml, T. In silico and In vitro studies of mycotoxins and their cocktails; their toxicity and its mitigation by silibinin pre-treatment. Toxicol. Eff. Mycotoxins Target Cells 2020, 12, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parker, A.E.; Christen, J.A.; Lorenz, L.; Smith, H. Optimal surface estimation and thresholding of confocal microscope images of biofilms using Beer’s Law. J. Microbiol. Meth. 2020, 174, 105943. [Google Scholar] [CrossRef]
- Kragh, K.N.; Alhede, M.; Kvich, L.; Bjarnsholt, T. Into the well—A close look at the complex structures of a microtiter biofilm and the crystal violet assay. Biofilm 2019, 1, 100006. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: https://www.R-project.org/2020 (accessed on 10 October 2020).
| Bacterial Strain | NAC (µg/mL) | RLs (µg/mL) | UA (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MICPC80 | MICBF80 | MICBM80 | MICPC80 | MICBF80 | MICBM80 | MICPC80 | MICBF80 | MICBM80 | |
| S. aureus 816 | 3130 | 3130 | 3130 | >1000/49% * | 62.5 | 62.5 | 0.24 | 0.24 | 0.24 |
| S. aureus 1241 | 3130 | 12,500 | 6250 | >1000/57% * | 125 | 125 | 0.24 | 0.24 | 0.49 |
| L. monocytogenes 149 | 3130 | 6250 | 12,500 | 250 | 62.5 | 62.5 | 0.98 | 0.98 | 1.96 |
| L. monocytogenes 164 | 6250 | 6250 | 6250 | >1000/75% * | 125 | 62.5 | 1.96 | 1.96 | 1.96 |
| E. coli 683/17 | 3130 | 6250 | 6250 | >1000/29% * | 250 | 500 | >31.3/20% * | >31.3/10% * | >31.3/27% * |
| E. coli 693/17 | 6250 | 6250 | 6250 | >1000/23% * | 125 | 250 | >31.3/40% * | >31.3/47% * | >31.3/16% * |
| S. Infantis S13 | 6250 | 12,500 | 12,500 | >1000/50% * | 125 | 1000 | >31.3/25% * | >31.3/45% * | >31.3/41% * |
| S. Infantis S59 | 6250 | 6250 | 6250 | >1000/39% * | 15.65 | 500 | >31.3/31% * | >31.3/34% * | >31.3/47% * |
| S. Enteritidis ATCC 13076 | 12,500 | 12,500 | 12,500 | >1000/33% * | >1000/16% * | 1000 | >31.3/42% * | >31.3/25% * | >31.3/45% * |
| Bacterial Strain | NAC (µg/mL) | RLs (µg/mL) | UA (µg/mL) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| MICBC80 | MICBR80 | MICMPB80 | MICBC80 | MICBR80 | MICMPB80 | MICBC80 | MICBR80 | MICMPB80 | |
| S. aureus 816 | 3130 | 12,500 | 12,500 | 2000 | 62.5 | 62.5 | 1.96 | >31.3/35% * | >31.3/67% * |
| S. aureus 1241 | 6250 | >100,000/31% * | 6250 | 2000 | 1000 | 250 | 3.91 | >31.3/72% * | 3.91 |
| L. monocytogenes 149 | 3130 | 100,000 | 6250 | 62.5 | 2000 | 62.5 | 1.96 | >31.3/69% * | 3.91 |
| L. monocytogenes 164 | 3130 | >100,000/46% * | 6250 | 250 | 250 | 125 | 1.96 | 1.96 | 1.96 |
| E. coli 683/17 | 6250 | >100,000/68% * | 12,500 | 2000 | 2000 | 2000 | >31.3/44% * | >31.3/0% * | >31.3/45% * |
| E. coli 693/17 | 3130 | 100,000 | 12,500 | 2000 | 2000 | 2000 | >31.3/61% * | >31.3/0% * | >31.3/20% * |
| S. Infantis S13 | 6250 | 12,500 | 12,500 | 2000 | 1000 | 2000 | >31.3/33% * | >31.3/30% * | >31.3/17% * |
| S. Infantis S59 | 12,500 | 100,000 | 12,500 | 2000 | 500 | 2000 | >31.3/31% * | >31.3/21% * | >31.3/26% * |
| S. Enteritidis ATCC 13076 | 6250 | 12,500 | 12,500 | 2000 | 2000 | 1000 | >31.3/51% * | >31.3/0% * | >31.3/53% * |
| Bacterial Strain | NAC | RLs | UA | CONTROL | |||
|---|---|---|---|---|---|---|---|
| Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) | |
| S. aureus 816 | 104 ± 67.8* | 181 ± 57.7*** | 217 ± 38.5 | 244 ± 28.5 | 237 ± 41.9 | 270 ± 21 | 244 ± 22.4 |
| S. aureus 1241 | 236 ± 50.5 | 212 ± 16.1*** | 266 ± 25.5 | 285 ± 67.4 | 367 ± 40.5 *** | 392 ± 21.7 ** | 298 ± 58 |
| L. monocytogenes 149 | 86.5 ± 2.6 * | 238 ± 32 | 239 ± 49.6 | 260 ± 33.3 | 175 ± 79.4 *** | 268 ± 68.9 | 243 ± 25.6 |
| L. monocytogenes 164 | 158 ± 93.2 *** | 288 ± 40.6 | 337 ± 74.1 | 348 ± 49.6 *** | 325 ± 70.5 | 417 ± 40.4 * | 258 ± 72.9 |
| E. coli 683/17 | 271 ± 50.7 * | 229 ± 9.9 *** | 234 ± 30.4 *** | 259 ± 37.4 ** | N/A | 269 ± 28.1 ** | 162 ± 65.2 |
| E. coli 693/17 | 273 ± 40.8 | 181 ± 21.9 | 205 ± 84.5 | 230 ± 26 | N/A | 256 ± 12.6 | 222 ± 34.3 |
| S. Enteritidis ATCC 13076 | 117 ± 41.6 | 137 ± 69.6 | 144 ± 37.1 | 150 ± 10.9 | N/A | 77.6 ± 39.1 | 139 ± 64.6 |
| Bacterial Strain | NAC | RLs | UA | CONTROL | |||
|---|---|---|---|---|---|---|---|
| Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) MICBM80 | Thickness (µm) subCBM80 | Thickness (µm) | |
| S. aureus 816 | 156 ± 80.4 ** | 247 ± 64.7 | 344 ± 87.3 | 317 ± 33.1 | N/A | 152 ± 68.6 ** | 299 ± 47.5 |
| S. aureus 1241 | 253 ± 44.4 | 266 ± 37.9 | 222 ± 23.5 | 267 ± 26.8 | 210 ± 32.5 | 251 ± 15.7 | 247 ± 58.1 |
| L. monocytogenes 149 | 338 ± 85.7 *** | 332 ± 51.2 *** | 302 ± 41 | 291 ± 52.3 | 317 ± 62.3 *** | 276 ± 26.8 | 237 ± 61.9 |
| L. monocytogenes 164 | 345 ± 62 | 335 ± 23.1 | 328 ± 84.2 | 348 ± 21.5 | 440 ± 60.8 ** | N/A | 285 ± 90.5 |
| E. coli 683/17 | 153 ± 33.3 | 128 ± 31.6 | 133 ± 49.8 | 249 ± 14.9 ** | N/A | 285 ± 38.5 ** | 178 ± 64.8 |
| E. coli 693/17 | 222 ± 68.8 ** | 181 ± 33.7 | 194 ± 7.2 | 266 ± 27.1 *** | N/A | 254 ± 81.8 *** | 149 ± 9.5 |
| S. Enteritidis ATCC 13076 | 212 ± 16.6 | 229 ± 54.9 | 128 ± 34.3 ** | 181 ± 10.7 | N/A | 203 ± 19.8 | 193 ± 17.7 |
| IC50 (µg/mL) | |||
|---|---|---|---|
| Cell Lines | NAC | RLs | UA |
| HRTEC | 1570 ± 70 b | 56.95 ± 0.65 d | 3.06 ± 0.17 g |
| HaCat | 4020 ± 470 a | 38.74 ± 4.96 de | 0.59 ± 0.14 h |
| HDF | 460 ± 20 c | 47.64 ± 2.43 de | 4.35 ± 0.41 f |
| HEK 293 | 1940 ± 250 b | 31.48 ± 4.31 e | 2.75 ± 0.22 g |
| HCAEC | 1830 ± 90 b | 27.50 ± 0.82 e | 0.03 ± 0.01 i |
| Bacterial Strain | Origin | |
|---|---|---|
| Staphylococcus aureus 816 | Frozen sea fish | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Staphylococcus aureus 1241 | Cow milk | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Listeria monocytogenes 149 | Pork ham | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Listeria monocytogenes 164 | Pork ham | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Escherichia coli 683/17 | Salt bath (cheese industry) | Veterinary Research Institute Brno, Czech Republic |
| Escherichia coli 693/17 | Floor (cheese industry) | Veterinary Research Institute Brno, Czech Republic |
| Salmonella enterica Infantis S13 | Wastewater treatment plant | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Salmonella enterica Infantis S59 | Frozen chicken meat | Department of Biochemistry and Microbiology, UCT Prague, Czech Republic |
| Salmonella enterica Enteritidis ATCC 13076 | Culture collection | American Type Culture Collection, Manassas, VA, USA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chlumsky, O.; Smith, H.J.; Parker, A.E.; Brileya, K.; Wilking, J.N.; Purkrtova, S.; Michova, H.; Ulbrich, P.; Viktorova, J.; Demnerova, K. Evaluation of the Antimicrobial Efficacy of N-Acetyl-l-Cysteine, Rhamnolipids, and Usnic Acid—Novel Approaches to Fight Food-Borne Pathogens. Int. J. Mol. Sci. 2021, 22, 11307. https://doi.org/10.3390/ijms222111307
Chlumsky O, Smith HJ, Parker AE, Brileya K, Wilking JN, Purkrtova S, Michova H, Ulbrich P, Viktorova J, Demnerova K. Evaluation of the Antimicrobial Efficacy of N-Acetyl-l-Cysteine, Rhamnolipids, and Usnic Acid—Novel Approaches to Fight Food-Borne Pathogens. International Journal of Molecular Sciences. 2021; 22(21):11307. https://doi.org/10.3390/ijms222111307
Chicago/Turabian StyleChlumsky, Ondrej, Heidi J. Smith, Albert E. Parker, Kristen Brileya, James N. Wilking, Sabina Purkrtova, Hana Michova, Pavel Ulbrich, Jitka Viktorova, and Katerina Demnerova. 2021. "Evaluation of the Antimicrobial Efficacy of N-Acetyl-l-Cysteine, Rhamnolipids, and Usnic Acid—Novel Approaches to Fight Food-Borne Pathogens" International Journal of Molecular Sciences 22, no. 21: 11307. https://doi.org/10.3390/ijms222111307
APA StyleChlumsky, O., Smith, H. J., Parker, A. E., Brileya, K., Wilking, J. N., Purkrtova, S., Michova, H., Ulbrich, P., Viktorova, J., & Demnerova, K. (2021). Evaluation of the Antimicrobial Efficacy of N-Acetyl-l-Cysteine, Rhamnolipids, and Usnic Acid—Novel Approaches to Fight Food-Borne Pathogens. International Journal of Molecular Sciences, 22(21), 11307. https://doi.org/10.3390/ijms222111307