Precise Characterization of Genetic Interactions in Cancer via Molecular Network Refining Processes
Abstract
:1. Introduction
2. Results
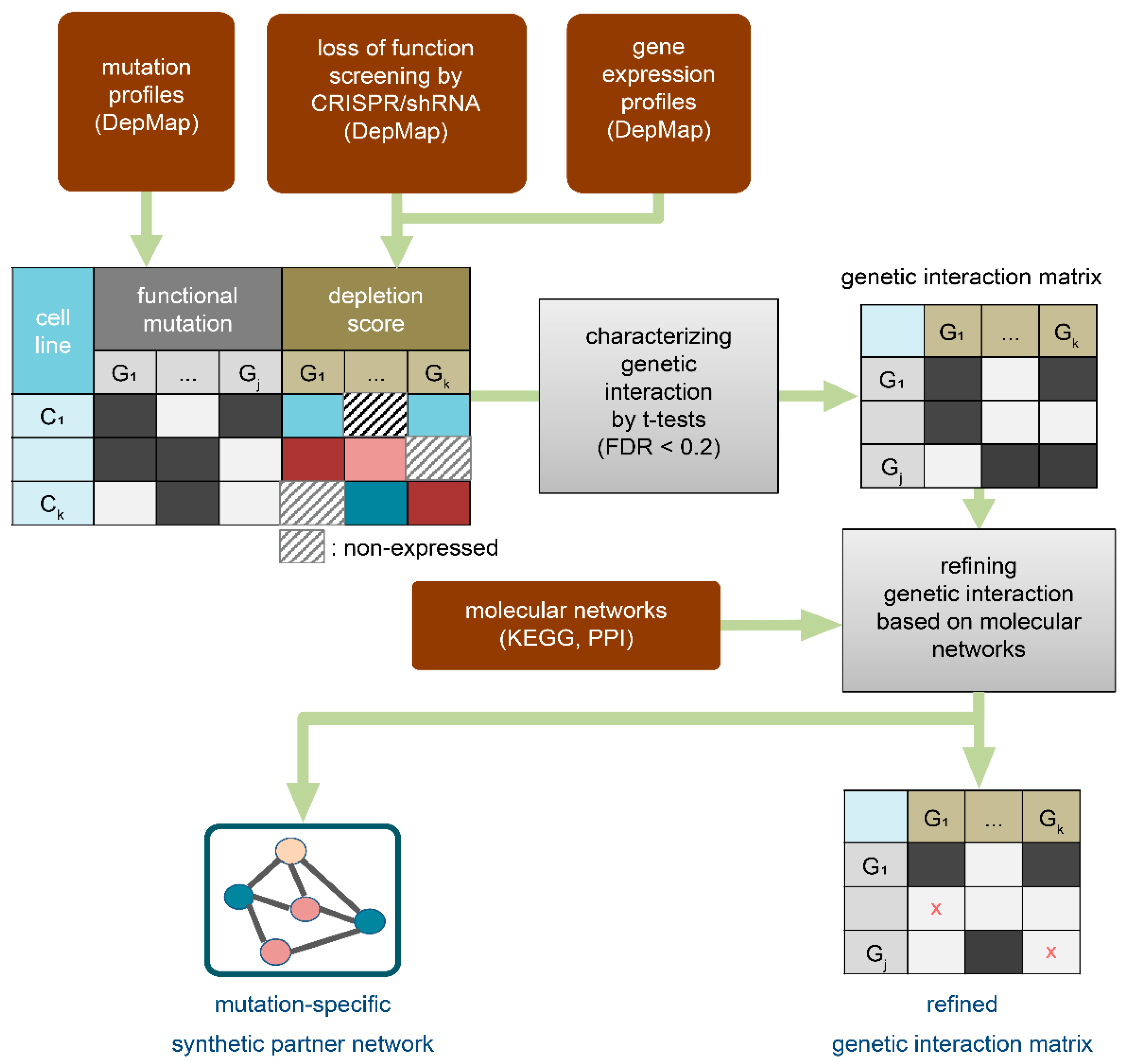
2.1. Strategy Overview
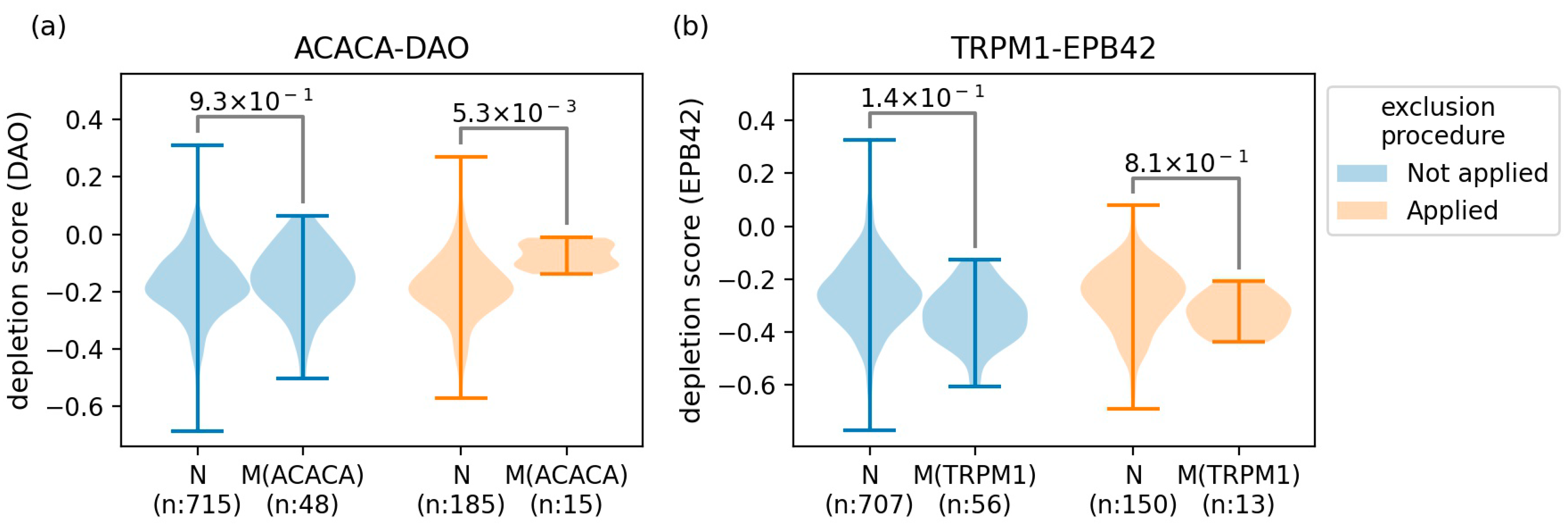
2.2. The Characterized GIs
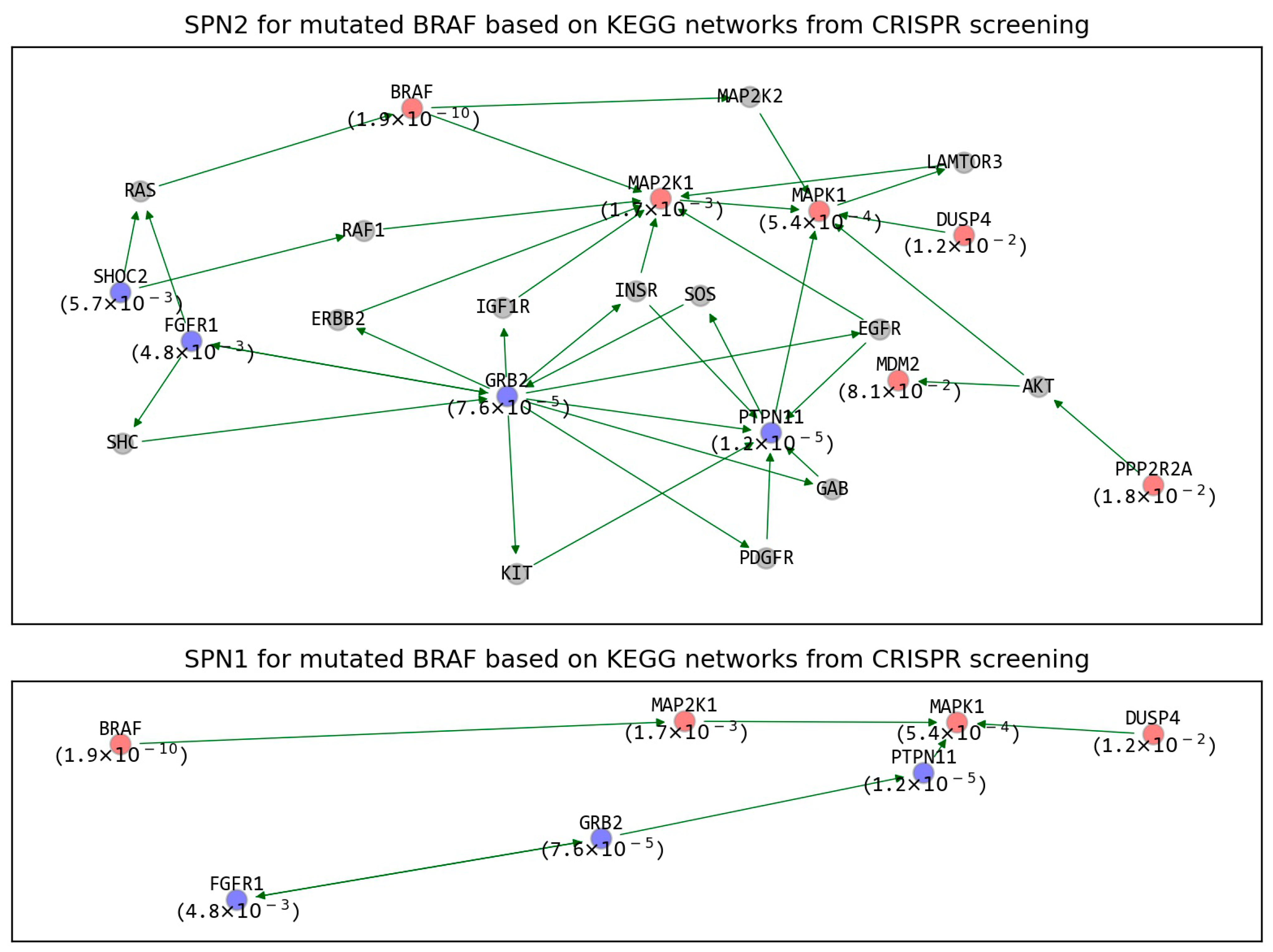
2.3. The Refined GIs Based on KEGG Network Analysis
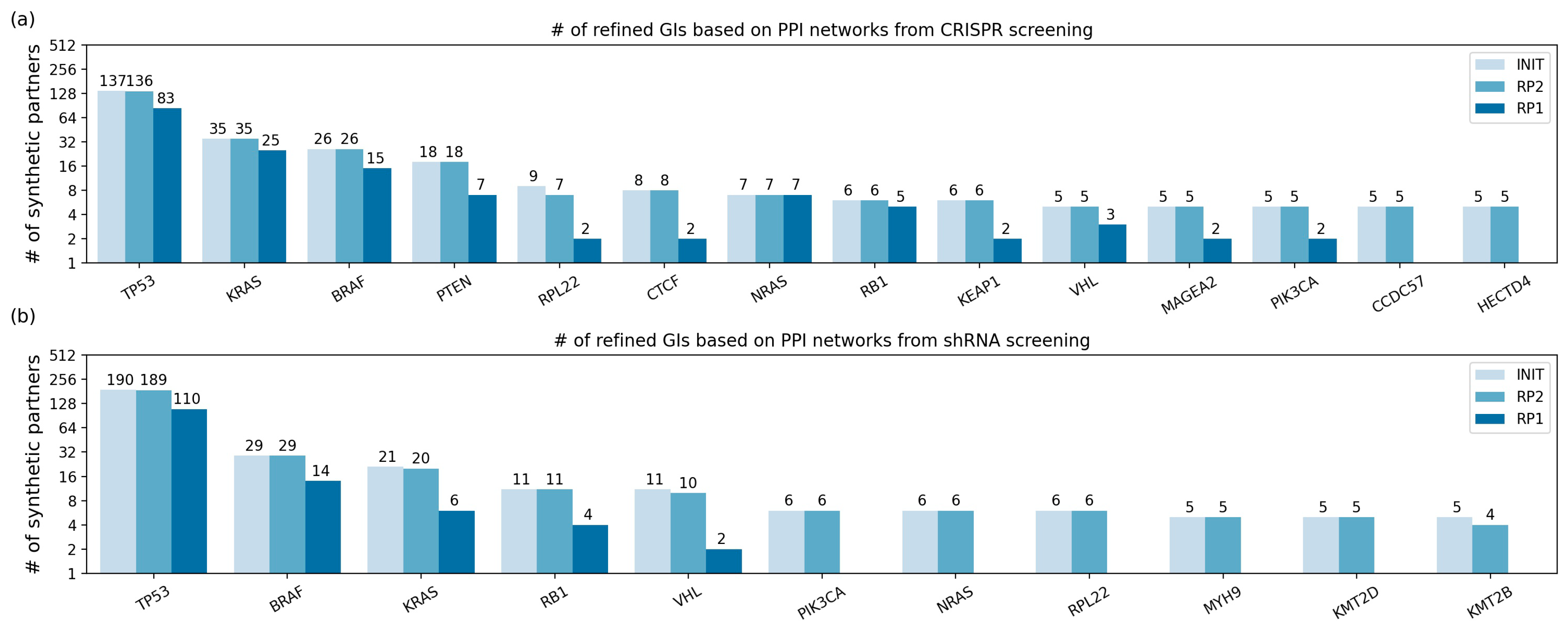
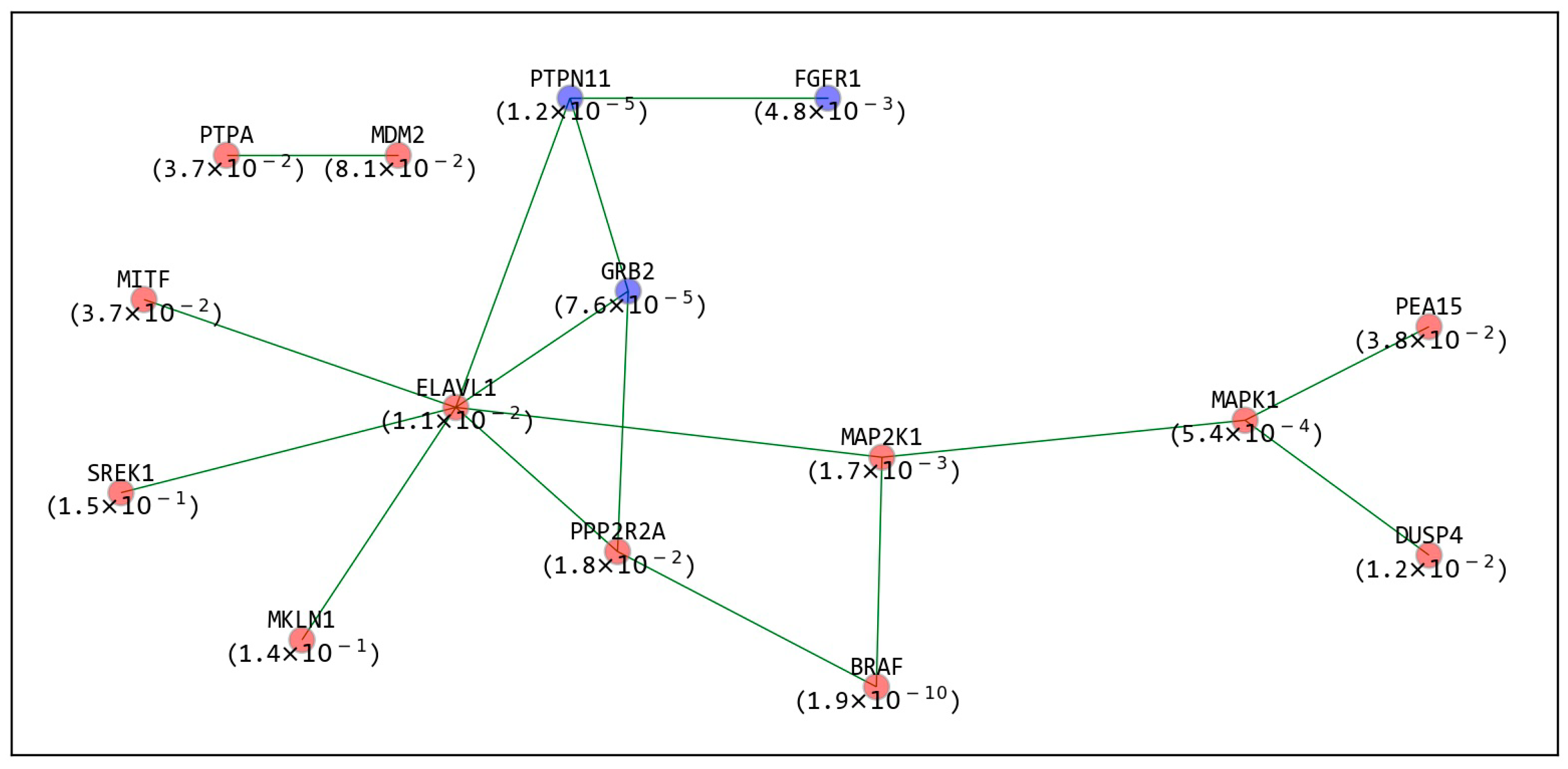
2.4. The Refined GIs Based on PPI
2.5. Evaluation with SynlethDB and MISL
3. Discussion and Conclusions
4. Materials and Methods
4.1. Data Preprocessing
4.1.1. Mutation Profiles
4.1.2. Loss-of-Function Profiles
4.1.3. Expression Profiles
4.1.4. Network Construction
4.2. Characterizing GIs
4.3. Refining GI Based on Molecular Networks
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ryan, C.J.; Bajrami, I.; Lord, C.J. Synthetic lethality and cancer–penetrance as the major barrier. Trends Cancer 2018, 4, 671–683. [Google Scholar] [CrossRef] [PubMed]
- Ashley, E.A. Towards precision medicine. Nat. Rev. Genet. 2016, 17, 507–522. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G. The concept of synthetic lethality in the context of anticancer therapy. Nat. Rev. Cancer 2005, 5, 689–698. [Google Scholar] [CrossRef] [PubMed]
- Beijersbergen, R.L.; Wessels, L.F.; Bernards, R. Synthetic lethality in cancer therapeutics. Annu. Rev. Cancer Biol. 2017, 1, 141–161. [Google Scholar] [CrossRef]
- Cristóbal, I.; González-Alonso, P.; Daoud, L.; Solano, E.; Torrejón, B.; Manso, R.; Madoz-Gúrpide, J.; Rojo, F.; García-Foncillas, J. Activation of the tumor suppressor PP2A emerges as a potential therapeutic strategy for treating prostate cancer. Mar. Drugs 2015, 13, 3276–3286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lord, C.J.; Quinn, N.; Ryan, C.J. Integrative analysis of large-scale loss-of-function screens identifies robust cancer-associated genetic interactions. Elife 2020, 9, e58925. [Google Scholar] [CrossRef]
- Lin, A.; Sheltzer, J.M. Discovering and validating cancer genetic dependencies: Approaches and pitfalls. Nat. Rev. Genet. 2020, 21, 671–682. [Google Scholar] [CrossRef]
- Behan, F.M.; Iorio, F.; Picco, G.; Gonçalves, E.; Beaver, C.M.; Migliardi, G.; Santos, R.; Rao, Y.; Sassi, F.; Pinnelli, M. Prioritization of cancer therapeutic targets using CRISPR–Cas9 screens. Nature 2019, 568, 511–516. [Google Scholar] [CrossRef]
- Marcotte, R.; Sayad, A.; Brown, K.R.; Sanchez-Garcia, F.; Reimand, J.; Haider, M.; Virtanen, C.; Bradner, J.E.; Bader, G.D.; Mills, G.B. Functional genomic landscape of human breast cancer drivers, vulnerabilities, and resistance. Cell 2016, 164, 293–309. [Google Scholar] [CrossRef] [Green Version]
- McDonald III, E.R.; De Weck, A.; Schlabach, M.R.; Billy, E.; Mavrakis, K.J.; Hoffman, G.R.; Belur, D.; Castelletti, D.; Frias, E.; Gampa, K. Project DRIVE: A compendium of cancer dependencies and synthetic lethal relationships uncovered by large-scale, deep RNAi screening. Cell 2017, 170, 577–592.e10. [Google Scholar] [CrossRef] [Green Version]
- Tsherniak, A.; Vazquez, F.; Montgomery, P.G.; Weir, B.A.; Kryukov, G.; Cowley, G.S.; Gill, S.; Harrington, W.F.; Pantel, S.; Krill-Burger, J.M. Defining a cancer dependency map. Cell 2017, 170, 564–576.e16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, H.-Q.; Tien, M.; Polychronakos, C. Statistical significance in genetic association studies. Clin. Investig. Med. Med. Clin. Exp. 2010, 33, E266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerby-Arnon, L.; Pfetzer, N.; Waldman, Y.Y.; McGarry, L.; James, D.; Shanks, E.; Seashore-Ludlow, B.; Weinstock, A.; Geiger, T.; Clemons, P.A. Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell 2014, 158, 1199–1209. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.S.; Das, A.; Jerby-Arnon, L.; Arafeh, R.; Auslander, N.; Davidson, M.; McGarry, L.; James, D.; Amzallag, A.; Park, S.G. Harnessing synthetic lethality to predict the response to cancer treatment. Nat. Commun. 2018, 9, 2546. [Google Scholar] [CrossRef]
- Michaloglou, C.; Vredeveld, L.; Mooi, W.; Peeper, D. BRAF E600 in benign and malignant human tumours. Oncogene 2008, 27, 877–895. [Google Scholar] [CrossRef] [Green Version]
- Moon, J.; Ha, J.; Park, S.H. Identification of PTPN1 as a novel negative regulator of the JNK MAPK pathway using a synthetic screening for pathway-specific phosphatases. Sci. Rep. 2017, 7, 12974. [Google Scholar] [CrossRef] [Green Version]
- Liebig, J.K.; Kuphal, S.; Bosserhoff, A.K. HuRdling senescence: HuR breaks BRAF-induced senescence in melanocytes and supports melanoma growth. Cancers 2020, 12, 1299. [Google Scholar] [CrossRef]
- Chou, S.D.; Murshid, A.; Eguchi, T.; Gong, J.; Calderwood, S.K. HSF1 regulation of β-catenin in mammary cancer cells through control of HuR/elavL1 expression. Oncogene 2015, 34, 2178–2188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, C.; Zhang, M.; Sun, W.; Dong, C. Upregulation of miR-324-5p inhibits proliferation and invasion of colorectal cancer cells by targeting ELAVL1. Oncol. Res. 2019, 27, 515. [Google Scholar] [CrossRef] [PubMed]
- Xue, F.; Li, Q.R.; Xu, Y.H.; Zhou, H.B. MicroRNA-139-3p inhibits the growth and metastasis of ovarian cancer by inhibiting ELAVL1. OncoTargets Ther. 2019, 12, 8935. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, J.; Liu, H.; Zheng, J. SynLethDB: Synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res. 2016, 44, D1011–D1017. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinha, S.; Thomas, D.; Chan, S.; Gao, Y.; Brunen, D.; Torabi, D.; Reinisch, A.; Hernandez, D.; Chan, A.; Rankin, E.B. Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun. 2017, 8, 15580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, J.; Ryan, C.J.; Brough, R.; Bajrami, I.; Pemberton, H.N.; Chong, I.Y.; Costa-Cabral, S.; Frankum, J.; Gulati, A.; Holme, H. Large-scale profiling of kinase dependencies in cancer cell lines. Cell Rep. 2016, 14, 2490–2501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kelley, R.; Ideker, T. Systematic interpretation of genetic interactions using protein networks. Nat. Biotechnol. 2005, 23, 561–566. [Google Scholar] [CrossRef]
- Liu, X.; Li, C.; Mou, C.; Dong, Y.; Tu, Y. dbNSFP v4: A comprehensive database of transcript-specific functional predictions and annotations for human nonsynonymous and splice-site SNVs. Genome Med. 2020, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Meyers, R.M.; Bryan, J.G.; McFarland, J.M.; Weir, B.A.; Sizemore, A.E.; Xu, H.; Dharia, N.V.; Montgomery, P.G.; Cowley, G.S.; Pantel, S. Computational correction of copy number effect improves specificity of CRISPR–Cas9 essentiality screens in cancer cells. Nat. Genet. 2017, 49, 1779–1784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, B.; Cheung, H.W.; Subramanian, A.; Sharifnia, T.; Okamoto, M.; Yang, X.; Hinkle, G.; Boehm, J.S.; Beroukhim, R.; Weir, B.A. Highly parallel identification of essential genes in cancer cells. Proc. Natl. Acad. Sci. USA 2008, 105, 20380–20385. [Google Scholar] [CrossRef] [Green Version]
- McFarland, J.M.; Ho, Z.V.; Kugener, G.; Dempster, J.M.; Montgomery, P.G.; Bryan, J.G.; Krill-Burger, J.M.; Green, T.M.; Vazquez, F.; Boehm, J.S. Improved estimation of cancer dependencies from large-scale RNAi screens using model-based normalization and data integration. Nat. Commun. 2018, 9, 4610. [Google Scholar] [CrossRef] [Green Version]
- Ghandi, M.; Huang, F.W.; Jané-Valbuena, J.; Kryukov, G.V.; Lo, C.C.; McDonald, E.R.; Barretina, J.; Gelfand, E.T.; Bielski, C.M.; Li, H. Next-generation characterization of the cancer cell line encyclopedia. Nature 2019, 569, 503–508. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Oughtred, R.; Stark, C.; Breitkreutz, B.-J.; Rust, J.; Boucher, L.; Chang, C.; Kolas, N.; O’Donnell, L.; Leung, G.; McAdam, R. The BioGRID interaction database: 2019 update. Nucleic Acids Res. 2019, 47, D529–D541. [Google Scholar] [CrossRef] [PubMed] [Green Version]


| Abbreviation | Meaning |
|---|---|
| GI | genetic interaction |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| PPI | protein–protein interaction |
| RP | refining process |
| RP1 | RP with distance 1 |
| RP2 | RP with distance 2 |
| SGWE | sensitive GI characterized with the exclusion procedure |
| SGOE | sensitive GI characterized without the exclusion procedure |
| SLI | synthetic lethal interaction |
| SP | synthetic partner |
| SPN | synthetic partner network |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jung, J.; Hwang, Y.; Ahn, H.; Lee, S.; Yoo, S. Precise Characterization of Genetic Interactions in Cancer via Molecular Network Refining Processes. Int. J. Mol. Sci. 2021, 22, 11114. https://doi.org/10.3390/ijms222011114
Jung J, Hwang Y, Ahn H, Lee S, Yoo S. Precise Characterization of Genetic Interactions in Cancer via Molecular Network Refining Processes. International Journal of Molecular Sciences. 2021; 22(20):11114. https://doi.org/10.3390/ijms222011114
Chicago/Turabian StyleJung, Jinmyung, Yongdeuk Hwang, Hongryul Ahn, Sunjae Lee, and Sunyong Yoo. 2021. "Precise Characterization of Genetic Interactions in Cancer via Molecular Network Refining Processes" International Journal of Molecular Sciences 22, no. 20: 11114. https://doi.org/10.3390/ijms222011114
APA StyleJung, J., Hwang, Y., Ahn, H., Lee, S., & Yoo, S. (2021). Precise Characterization of Genetic Interactions in Cancer via Molecular Network Refining Processes. International Journal of Molecular Sciences, 22(20), 11114. https://doi.org/10.3390/ijms222011114






